Abstract
Hepatocellular carcinoma (HCC) is the major primary malignant tumor in the human liver, but the molecular changes leading to liver cell transformation remain largely unknown. The Wnt-β-catenin pathway is activated in colon cancers and some melanoma cell lines, but has not yet been investigated in HCC. We have examined the status of the β-catenin gene in different transgenic mouse lines of HCC obtained with the oncogenes c-myc or H-ras. Fifty percent of the hepatic tumors in these transgenic mice had activating somatic mutations within the β-catenin gene similar to those found in colon cancers and melanomas. These alterations in the β-catenin gene (point mutations or deletions) lead to a disregulation of the signaling function of β-catenin and thus to carcinogenesis. We then analyzed human HCCs and found similar mutations in eight of 31 (26%) human liver tumors tested and in HepG2 and HuH6 hepatoma cells. The mutations led to the accumulation of β-catenin in the nucleus. Thus alterations in the β-catenin gene frequently are selected for during liver tumorigenesis and suggest that disregulation of the Wnt-β-catenin pathway is a major event in the development of HCC in humans and mice.
Hepatocellular carcinoma (HCC) is the major primary malignant tumor of the liver. Epidemiological studies indicate that hepatitis B virus is a major causal agent of liver cancer, but other etiological factors also have been found (see ref. 1 for review). However, the molecular mechanisms that contribute to tumor progression in hepatocarcinogenesis remain unknown. To identify genetic alterations that could be involved in tumor progression during hepatocarcinogenesis, we analyzed the status of the β-catenin gene. The protein β-catenin is involved in two major functions: cell adhesion and the transmission of the proliferating signal of the Wingless/Wnt pathway. Disregulation of this pathway has been implicated in carcinogenesis (see refs. 2 and 3 for reviews). β-catenin usually is found in the lateral cell membrane, as part of the E-cadherin/catenin adherens complex, and in the cytoplasm and nucleus where it acts as a mediator of Wingless/Wnt-dependent signal transduction (2, 4). Signal transduction via β-catenin involves its posttranslational stabilization and passage into the nucleus where it interacts with transcription factors of the T cell factor/lymphoid enhancer factor family to activate target genes involved in cell growth control and apoptosis (3, 5, 6). The amount of free cytoplasmic β-catenin seems to be regulated by the opposing actions of the adenomatous polyposis coli (APC) suppressor gene and the Wnt-1 protooncogene (7, 8). Activation of the Wnt-1 pathway results in inhibition of glycogen synthase kinase-3β (GSK-3β), and a lack of phosphorylation of β-catenin, which then will not be degraded by the ubiquitin/proteasome system, and thus accumulate in the cell (2, 9). The APC protein appears to be a negative regulator of cytoplasmic β-catenin, acting by an as yet unknown mechanism. The APC gene is the gene most commonly mutated in colorectal cancer, and it is inactivated early in the initiation of colon cancer. These mutations are associated with a rise in the free intracellular β-catenin, which results in loss of control of normal β-catenin signaling. This event has been demonstrated in 85% of colorectal cancers (10). The major initiating event in the remaining 15% of colorectal cancers with an intact APC gene involves mutations in the β-catenin gene, which then lead to accumulation of the protein in the cytosolic-nuclear compartment (11, 12). Similar activating mutations of the β-catenin gene recently were found in melanoma cell lines (13), implying that β-catenin acts as an oncogene in cancers without any clear association with APC mutations. We therefore have looked for such mutations in the β-catenin gene in different types of HCCs, first in transgenic mice and then in humans.
MATERIALS AND METHODS
Tumor Samples.
Transgenic mice were maintained in accordance with the Ministère de l’Agriculture et de la forêt guidelines for the care and use of laboratory animals.
The human hepatocellular samples were obtained from patients of various geographical origin who underwent surgery.
Western Blot.
Tissues were homogenized with a Polytron in 1× Laemli sample buffer (1:10 wt/vol) (Sigma). Samples of extract containing approximately 50 μg of total protein were resolved on 7.5% (wt/vol) SDS/PAGE and transferred to nitrocellulose filters. The membranes were blocked with 5% (wt/vol) milk powder in Tris-buffered saline containing 0.05% (vol/vol) Tween 20 (Sigma). β-catenin protein was detected by using a rabbit polyclonal anti-β-catenin (M. Mareels, University Hospital, Gent, Belgium; dilution 1:10,000). Blots were standardized by staining with ponceau red staining. Immunostaining was performed with peroxidase-coupled anti-rabbit IgG (Amersham) (dilution 1:10,000) and ECL (Amersham).
Reverse Transcription–PCR (RT-PCR) Amplification of β-Catenin mRNA.
Total RNAs were isolated from frozen samples of mouse and human HCCs. The corresponding normal RNAs were obtained from histologically normal tissue surrounding the tumor. Total RNAs were reverse-transcribed to obtain the cDNA that was amplified by RT-PCR. The sequences of the primers were chosen to amplify both the human and the mouse β-catenin cDNAs (21, 22). Five pairs of primers (F1-R1, F2-R2, F3-R3, F4-R4, and F5-R5) were designed to amplify overlapping PCR products, which covered the whole β-catenin ORF. The sequences of primers used to amplify β-catenin were: F1, 5′-GCGTGGACAATGGCTACTCAAG-3′; R1, 5′-TATTAACTACCACCTGGTCCTC-3′; F2, 5′-ACGCGGAACTTGCCACACGTGC-3′; R2, 5′-TTCAGCACTCTGCTTGTGGTC-3′; F3, 5′-ATCAAGAGAGCAAGCTCATCAT-3′; R3, 5′-TGAAGGCGAACGGCATTCTGGG-3′; F4, 5′-GCTCTTCGTCATCTGACCAGCC-3′; R4, 5′-GAGCAAGTTCACAGAGGACCCC-3′; F5, 5′-GGACTCAATACCATTCCATTGT-3′; and R5, 5′-TTACAGGTCAGTATCAAACCAG-3′.
The PCR fragments were separated by electrophoresis on 1.5% (wt/vol) nusieve agarose gels, transferred to a nylon membrane, and hybridized with internal oligonucleotide probes. All deleted PCR products then were sequenced.
Mutation Analysis.
Denaturing gradient gel electrophoresis (DGGE) analysis was carried out on PCR products corresponding to exon 2 of the the mouse β-catenin gene and exon 3 of the human gene (40). These exons contain the GSK-3β phosphorylation site. DGGE analysis was done by using Lerman’s algorithm (41) and psoralen clamps (22). The structure and sequence of the mouse β-catenin gene was kindly provided by R. Kemler (Max-Planck Institut, Freiburg, Germany). The sequences of the mouse β-catenin gene primers flanking the intron-exon junctions of the second exon used for DGGE analysis were: PM1, 5′Pso-TACAGGTAGCATTTTCAGTTCAC-3′ and PM2, 5′TAGCTTCCAAACACAAATGC-3′. The sequences of the human β-catenin exonic primers located in exon 3 were: PH1, 5′Pso-TACAGCTACTTGTTCTTGAGTG-3′ and PH2, 5′CTGATTTGATGGAGTTGGAC-3′. The PCR products were checked on standard agarose gels before analysis by DGGE. When the pattern of migration was abnormal, the corresponding PCR product was purified and directly sequenced on both strands by using the Dye Terminator Cycle Sequencing kit protocol. When the mutated products were underrepresented, bands were purified and amplified again by PCR before sequencing. Mutations were checked by restriction enzyme analysis whenever possible.
Immunofluorescence Staining of Hepatoma Cell Lines.
HepG2, Hep3B, Huh6, and PLC/Prf hepatoma cell lines were grown in DMEM (GIBCO/BRL) supplemented with 10% (vol/vol) fetal calf serum (GIBCO/BRL), 100 units/ml of penicillin, and 100 μg/ml of streptomycin. The cells were washed in PBS-0.1 M glycine, then fixed and permeabilized in methanol at −20°C. Cells were incubated for 1 h in blocking solution [PBS, 3% BSA (wt/vol)], and then with a rabbit polyclonal anti-β-catenin antibody (dilution 1:200) at room temperature in a moist chamber. The cells were washed several times in PBS-0.1% (vol/vol) Tween 20, and incubated for 45 min with anti-rabbit fluorescein isothiocyanate (Dako) (dilution 1:200). The cells were given a final wash in PBS-0.1% (vol/vol) Tween 20, examined under a Zeiss microscope equipped with fluorescence optics, and photographed.
RESULTS
Synthesis of Truncated β-Catenin in HCCs.
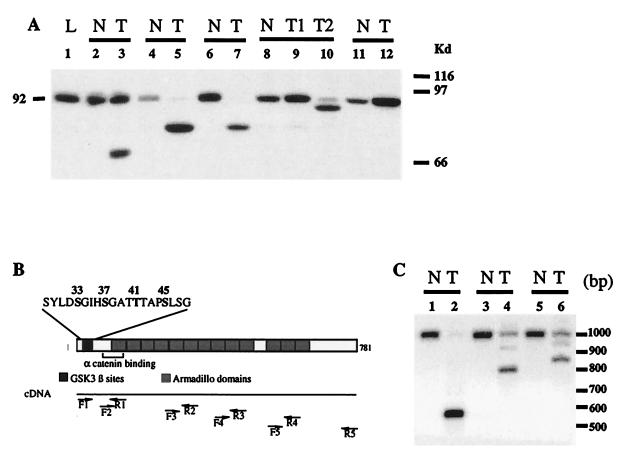
The regulation of β-catenin turnover requires the NH2-terminal region of the protein containing the potential GSK-3β phosphorylation site, as phosphorylated β-catenin is targeted for degradation (9, 14). Deletion of the potential GSK-3β phosphorylation site or a missense mutation of the serine or threonine residues therein results in the accumulation of truncated or normal-sized β-catenin and then in activation of β-catenin-mediated signal (11, 13, 15–17). We used immunoblotting to evaluate the status of β-catenin in hepatic tumors developed in several transgenic mouse models. The L-type pyruvate kinase (L-PK)/c-myc and the L-PK/H-ras transgenic mice express the oncogene c-myc and H-ras in the liver under the control of the rat L-PK regulatory regions, resulting in the development of multifocal HCCs (18, 19). Similarly, WHV/c-myc mice express a transgene carrying the c-myc oncogene under the control of the woodchuck hepatitis virus (WHV) regulatory sequences and also develop liver tumors (20). Isolated hepatic tumor nodules and samples of adjacent nontumoral tissues were collected from transgenic mice and analyzed by Western blotting using an anti-β-catenin antibody (21). In addition to the 92-kDa full-length β-catenin, several truncated β-catenins were present in 12 of 20 tumor samples taken from L-PK/c-myc transgenic mice (Fig. 1A). β-catenin was detected at increased levels in three other tumor samples (Fig. 1A, lanes 9–12). Different tumor nodules taken from a single mouse synthesize different forms of β-catenin (Fig. 1A, lanes 9–10), indicating that these multiple nodules had a distinct clonal origin.
Figure 1.
Analysis of β-catenin in hepatocarcinomas developed in transgenic mice. (A) Immunoblot analysis for β-catenin in L-PK/c-myc transgenic mice. The expected 92-kDa band for β-catenin was detected in normal tissue (N, lanes 2, 4, 6, and 8–11) surrounding the tumor, whereas various truncated proteins were found in tumor nodules (T, lanes 3, 5, 7, 9, and 10). T1 and T2 (lanes 9 and 10) are two independent nodules from the same animal. L, sample from a nontransgenic mouse (lane 1). (B) RT-PCR strategy used to study the β-catenin mRNA. The diagram shows the β-catenin functional domains. The N-terminal domain is involved in the posttranslational stabilization via the GSK-3β phosphorylation site and in binding to α-catenin. The GSK-3β site includes the S33, S37, T41, and S45 phosphorylable residues. The 13 armadillo repeats are shown. Pairs of primers were designed to amplify overlapping PCR products that covered the whole ORF of β-catenin (F1-R1, F2-R2, F3-R3, F4-R4, and F5-R5, see Materials and Methods for the sequence). (C) Southern analysis of RT-PCR showing deleted β-catenin mRNA in L-PK/c-myc transgenic mice. mRNAs obtained from tumor tissue (lanes 2, 4, and 6) together with their nontumor counterparts (lanes 1, 3, and 5) were analyzed by RT-PCR with primers F1-R2, followed by Southern analysis.
We identified the molecular abnormalities involved in the production of truncated β-catenin by analyzing the entire coding region of the β-catenin mRNA by using a RT-PCR strategy (Fig. 1B). The mouse tumor samples showed additional smaller bands when analyzed with the primers F1/R1 or F1/R2, which amplify the 5′ part of the β-catenin mRNA (Fig. 1 B and C). All of the PCR-amplified products obtained with the other primer sets were of normal size (not shown). The deletions all were mapped in the 5′terminal part of the β-catenin mRNA. The deleted PCR products then were sequenced. We analyzed tumor samples from each of the transgenic mouse models (Table 1). The most frequent deletion was that of exon 2 containing the potential GSK-3β phosphorylation site. Whether deletion of exon 2 was caused by a point mutation in the splice site or a genomic deletion is presently unclear. The other deletions were in-frame deletions that removed at least the residues of the protein containing the potential GSK-3β phosphorylation site, except for one deletion (amino acids 12–37) that removed only the first phosphorylatable serine residue (S33) (Table 1).
Table 1.
Summary of the β-catenin gene mutations in mouse and human hepatocarcinomas
| Deletions | Mouse
|
Human
|
|||
|---|---|---|---|---|---|
| PK/c-myc | WHV/c-myc | PK/H-ras | Samples | Cell lines§ | |
| m exon 2 (aa 12-37)* | 1/30 | ||||
| m exon 2 (aa 19-74)* | 1/30 | ||||
| m exon 2 (aa 5-80)*† | 9/30 | 2/20 | 3/11 | ||
| m exons 2-4 (aa 22-172)* | 1/30 | ||||
| m exons 2-5 (aa 32-249)* | 1/11 | ||||
| h exons 3-4 (aa 25-140)* | HepG2 | ||||
| h exons 3-4 (aa 20-151)* | 1/26 | ||||
| Point mutations | |||||
| D32N | D32V, D32Y | ||||
| S33C, S33Y | S33F | ||||
| G34V | G34V (2)‡, G34E | G34E | G34V(HuH6) | ||
| S37P | S37F | S37Y | |||
| T41A | T41I (2)‡ | ||||
| S45P | |||||
| Total mutations | 17/30 | 10/20 | 4/11 | 6/26 | 2/5 |
The exon encoding the GSK-3β phosphorylation site is exon 2 for the mouse (m) and exon 3 for human (h).
Amino acid deleted according to mRNA analysis.
m exon 2 encodes aa 5-80 in the mouse β-catenin gene.
Mutations detected on two independent tumors.
The different cell lines tested were the HepG2, HuH6, PLC/PRF/5, Hep3B, and Huh7.
Because the deletions were very frequent in the various mouse tumors, we looked for similar events in human HCCs. We found deletions in the β-catenin mRNA in one of the 26 HCC tumor samples examined and in the HepG2 hepatoma cell line (Table 1).
Point Mutations in the Potential GSK-3β Phosphorylation Site of β-Catenin in HCCs.
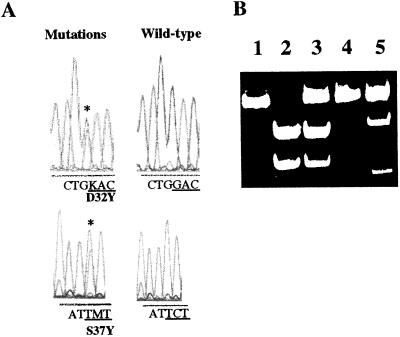
We then looked for activating point mutations in the potential GSK-3β phosphorylation site in all of the human and mouse tumor samples that had no deletions in the β-catenin gene. This search first was done by DGGE of PCR products obtained by using primers amplifying the potential GSK-3β phosphorylation site (22). Abnormal shifts in mobility were detected only in tumor samples and not in nontumoral tissues (data not shown). We then sequenced the PCR products that had an abnormal migration pattern on DGGE. The mutations were confirmed by appropriate restriction enzyme digestion whenever possible (Fig. 2 and data not shown). The point mutations (Table 1) all were somatic and appeared to affect only one allele of the β-catenin gene. The mutations were missense mutations that fell within the potential GSK-3β phosphorylation site (14). All serine or threonine residues implicated in the down-regulation of β-catenin by the GSK-3β kinase (14) were mutated: S33 (to C or F), S37 (to P or F), T41 (to A or I), and S45 (to P). These mutations also have been found in colon cancers and melanomas (11, 13). There also were mutations that modified a residue contiguous to the phosphorylatable amino acid S33: D32 (to N or V, or Y) and G34 [to V in four tumors or to E in two tumors. The same types of mutations have been described very recently in rat colon tumors (23 and 24).]. It has been suggested that these two residues (D32 and G34) are important for β-catenin ubiquitination and therefore proteasome-dependent degradation (9).
Figure 2.
Point mutations in the β-catenin gene in tumors developed in L-PK/c-myc mice. (A) Sequence analysis showing the D32Y and S37Y mutations. (B) Digestion of the RT-PCR products with appropriate restriction enzyme. Undigested wild-type amplified cDNA fragment (lane 1). Wild-type amplified cDNA fragments digested with XmnI (lane 2) and RsaI (lane 4). Digested amplified cDNA fragments from tumor sample showing a β-catenin gene mutation, the S37Y mutation led to the absence of digestion by the XmnI enzyme (lane 3), and the D32Y mutation led to the appearance of a RsaI restriction site (lane 5) on the mutated allele.
In summary, we have detected activating mutations in the human β-catenin gene in 26% (8/31) of the human liver tumor samples examined and in all of the mouse tumor models. Fifty-five percent (17/30) of the liver tumors developed in the L-PK/c-myc transgenic mice had mutational activation of the β-catenin gene, and similar mutations were found in 50% (10/20) of the liver tumors developed in the WHW/c-myc and 41% (4/11) of those in L-PK/H-ras transgenic mice (Table 1). Thus, the activation of β-catenin is a frequent feature of liver tumorigenesis.
Activation of the Wnt-β-Catenin Pathways in Cultured Human Hepatoma Cells.
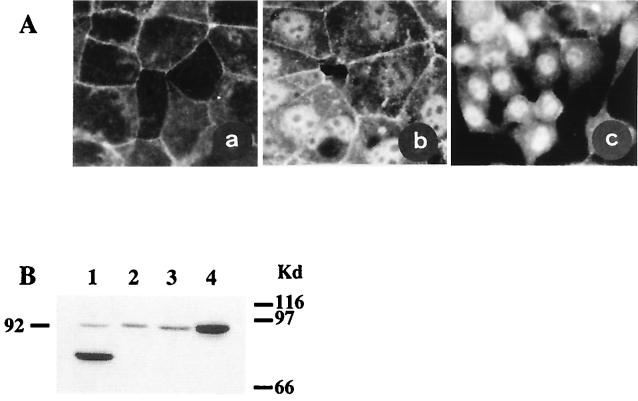
One of the current models implicating β-catenin in oncogenic signaling suggests that activating mutations lead to redistribution of the subcellular protein so that free β-catenin accumulates in the cytoplasm and interacts with a transcription factor of the T cell factor (TCF)/lymphoid enhancer factor family, and the complex translocates in the nucleus where it activates transcription of the Wnt-responsive genes (2, 3). We have used immunofluorescence staining to study the effect of the mutations identified in the HepG2 and HuH6 hepatoma cell lines. The PLC/PRF/5 cells containing a wild-type β-catenin were uniformly stained along the membrane. In contrast, HepG2 cells were intensely stained in the nucleus, with very weak membrane staining, whereas both membrane and nucleus were stained in the HuH6 cells (Fig. 3A). This finding confirms that the mutations in the HepG2 and HuH6 cell lines were activating mutations. Accordingly, a TCF-dependent reporter gene has been found to be constitutively activated in the HepG2 cell line (ref. 25 and H. Clevers, personal communication). These results strongly suggest that, as in colon cancers and melanoma cells, activating mutations of β-catenin in hepatoma cell lines lead to transcriptional activation of TCF-regulated genes. The mutation G34V, observed in the Huh6 hepatoma cell line, led to accumulation of β-catenin both in the cytoplasm and nucleus (Fig. 3 A and B). This finding strongly suggests that the mutations of amino acids surrounding Ser-33, which are very frequent in both mouse and human HCCs (see Table 1), all are activating mutations. The large deletion (amino acids 25–140) in HepG2 cells (Table 1 and Fig. 3B, lane 1) removes the potential GSK-3β regulatory site and probably the binding site for α-catenin, the β-catenin partner in the E-cadherin-mediated cell adhesion (4). This could explain the poor membrane staining (Fig. 3A). The point mutation in the HuH6, which probably does not alter the α-catenin binding site, may not impair binding to the membrane cytoskeleton (Fig. 3A).
Figure 3.
β-catenin in human hepatoma cell lines. (A) β-catenin immunolocalization. Cultured human hepatoma cells were immunolabeled with rabbit polyclonal anti-β-catenin antibody. (a) PLC/PRF/5 hepatoma cells containing wild-type β-catenin and showing only membrane staining. Hepatoma cells with activated β-catenin showed nuclear staining. (b) Huh6 cells. (c) HepG2 cells. There was no labeling when the first anti-β-catenin antibody was omitted (not shown). (B) Western blot analysis. Truncated β-catenin in HepG2 cells (lane 1). Full-length β-catenin in PLC/PRF/5 and Hep3B cells (lanes 2 and 3). The G34V β-catenin mutation identified in the Huh6 cells led to accumulation of the protein (lane 4).
DISCUSSION
We have provided direct evidence that somatic mutations of the β-catenin gene are selected for during development of liver cancer. Defining genetic alterations associated with the progression of liver tumor is a potentially useful step toward identifying the mechanisms of hepatocarcinogenesis. The activation of cellular oncogenes [i.e., c-myc (26, 27)], or growth factors [i.e., insulin-like growth factor II (28, 29) and transforming growth factor type α (30)] and mutations of tumor suppressor gene [i.e., P53 (31–34), RB (35), and IGFIIR (36)] all have been described in HCC developed in humans and mice. However, it generally is not clear whether these alterations are early, putatively causal factors, or late changes that can be viewed as markers of tumor progression. Only P53-specific mutations in hepatomas that develop in people in countries with frequent dietary aflatoxin intoxication most likely play a causal role (31, 32). We have analyzed several models of HCC: human HCC associated with hepatitis B virus or hepatitis C virus and mouse HCC developed in transgenic mice expressing the oncogenes c-myc or H-ras in the liver (18). All types of HCC contained similar activating mutations of the β-catenin gene, which strongly suggests that mutations of the β-catenin gene are essential for the development of HCC. These results suggest that β-catenin activation could constitute a privileged genetic modification in the cooperation with various oncogenes (e.g., myc and ras) for hepatic tumor progression and indicate that disregulation of the Wnt-β-catenin pathway may well be one of the major molecular mechanisms of liver cell transformation.
The mutations found in the liver tumors are all activating mutations located in the potential GSK-3β phosphorylation site (14). The serine or threonine residues mutated are thought to regulate β-catenin turnover: phosphorylated β-catenin is degraded by the ubiquitin-proteasome pathway (9). Similar mutations also have been found in colon cancers and melanoma cell lines (11, 13). In addition, we found mutations in the HCC samples studied that affect residues other than the serine-threonine residues that are potential targets of the GSK-3β kinase. These frequent mutations involved amino acid residues that may play a role in the phosphorylation-dependent ubiquitination and degradation (9).
Disregulation of the Wnt-APC-β-catenin pathway occurs in almost all colorectal tumors (10, 3). Our data show that 26% of human HCC and 50% of mouse HCC have β-catenin-activating mutations. We now need to know whether other partners of this signaling pathway can be activated in these tumors with normal β-catenin. Indeed, we have found some cases in which the intracellular concentration of wild-type β-catenin is elevated (not shown). Although such an accumulation of wild-type β-catenin could be explained by APC gene mutation, as in colon cancers (7), this should not be frequent in HCCs because no predisposition to HCC has been found in patients with familial adenomatous polyposis carrying a germ-line mutation of the APC gene (37), and there appears to be no genetic alteration in the APC locus in HCCs (38, 39). We therefore should look for other types of genetic modifications that result in β-catenin stabilization.
Acknowledgments
We are grateful to Dr. M. Mareels (Gent, Belgium) for his generous gift of the rabbit polyclonal antibody against β-catenin and to Dr. R. Kemler (Freiburg, Germany) for providing us with the structure and partial sequence of the mouse β-catenin gene. The English text was edited by Dr. Owen Parkes. This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), la Ligue Nationale contre le Cancer, l’Association pour la Recherche contre le Cancer (ARC), and le Ministère de la Recherche et de l’Enseignement Supérieur.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HCC, hepatocellular carcinoma; APC, adenomatous polyposis coli; GSK-3β, glycogen synthase kinase-3β; RT-PCR, reverse transcription–PCR; DGGE, denaturing gradient gel electrophoresis; L-PK, L-type pyruvate kinase.
References
- 1.Buendia M A. Adv Cancer Res. 1992;59:167–226. doi: 10.1016/s0065-230x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 2.Miller J R, Moon R T. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 3.Peifer M. Science. 1997;275:1752–1753. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- 4.Barth A, Näthke I S, Nelson W J. Curr Opin Cell Biol. 1997;9:683–690. doi: 10.1016/s0955-0674(97)80122-6. [DOI] [PubMed] [Google Scholar]
- 5.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 6.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 7.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 11.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 12.Ilyas M, Tomlinson I P, Rowan A, Pignatelli M, Bodmer W F. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 14.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 15.Oyama T, Kanai Y, Ochiai A, Akimoto S, Oda T, Yanagihara K, Nagafuchi A, Tsukita S, Shibamoto S, Ito F, et al. Cancer Res. 1994;54:6282–6287. [PubMed] [Google Scholar]
- 16.Kawanishi J, Kato J, Sasaki K, Fujii S, Watanabe N, Niitsu Y. Mol Cell Biol. 1995;15:1175–1181. doi: 10.1128/mcb.15.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins P F, El-Gamil M, Li Y F, Kawakami Y, Loftus D, Appella E, Rosenberg S A. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cartier N, Miquerol L, Tulliez M, Lepetit N, Levrat F, Grimber G, Briand P, Kahn A. Oncogene. 1992;7:1413–1422. [PubMed] [Google Scholar]
- 19.Gilbert E, Morel A, Tulliez M, Maunoury R, Terzi F, Miquerol L, Kahn A. Int J Cancer. 1997;73:749–756. doi: 10.1002/(sici)1097-0215(19971127)73:5<749::aid-ijc23>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 20.Etiemble J, Degott C, Renard C A, Fourel G, Shamoon B, Vitvitski-Trepo L, Hsu T Y, Tiollais P, Babinet C, Buendia M A. Oncogene. 1994;9:727–737. [PubMed] [Google Scholar]
- 21.Vermeulen S J, Bruyneel E A, van Roy F M, Mareel M M, Bracke M E. Br J Cancer. 1995;72:1447–1453. doi: 10.1038/bjc.1995.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez E, Bienvenu T, Desclaux Arramond F, Beldjord K, Kaplan J C, Beldjord C. PCR Methods Appl. 1993;3:122–124. doi: 10.1101/gr.3.2.122. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi M, Fukuda K, Sugimura T, Wakabayashi K. Cancer Res. 1998;58:42–46. [PubMed] [Google Scholar]
- 24.Dashwood R H, Suzui M, Nakagama H, Sugimura T, Nagao M. Cancer Res. 1998;58:1127–1129. [PubMed] [Google Scholar]
- 25.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X K, Huang D P, Qiu D K, Chiu J F. Oncogene. 1990;5:909–914. [PubMed] [Google Scholar]
- 27.Wei Y, Fourel G, Ponzetto A, Silvestro M, Tiollais P, Buendia M A. J Virol. 1992;66:5265–5276. doi: 10.1128/jvi.66.9.5265-5276.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu X X, Su C Y, Lee Y, Hintz R, Biempica L, Snyder R, Rogler C E. J Virol. 1988;62:3422–3430. doi: 10.1128/jvi.62.9.3422-3430.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cariani E, Lasserre C, Seurin D, Hamelin B, Kemeny F, Franco D, Czech M P, Ullrich A, Brechot C. Cancer Res. 1988;48:6844–6849. [PubMed] [Google Scholar]
- 30.Yeh Y C, Tsai J F, Chuang L Y, Yeh H W, Tsai J H, Florine D L, Tam J P. Cancer Res. 1987;47:896–901. [PubMed] [Google Scholar]
- 31.Hsu I C, Metcalf R A, Sun T, Welsh J A, Wang N J, Harris C C. Nature (London) 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 32.Bressac B, Kew M, Wands J, Ozturk M. Nature (London) 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 33.Buetow K H, Sheffield V C, Zhu M, Zhou T, Shen F M, Hino O, Smith M, McMahon B J, Lanier A P, London W T, et al. Proc Natl Acad Sci USA. 1992;89:9622–9626. doi: 10.1073/pnas.89.20.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosono S, Chou M J, Lee C S, Shih C. Oncogene. 1993;8:491–496. [PubMed] [Google Scholar]
- 35.Zhang X, Xu H J, Murakami Y, Sachse R, Yashima K, Hirohashi S, Hu S X, Benedict W F, Sekiya T. Cancer Res. 1994;54:4177–4182. [PubMed] [Google Scholar]
- 36.De Souza A T, Hankins G R, Washington M K, Orton T C, Jirtle R L. Nat Genet. 1995;11:447–449. doi: 10.1038/ng1295-447. [DOI] [PubMed] [Google Scholar]
- 37.Horii A, Nakatsuru S, Miyoshi Y, Ichii S, Nagase H, Ando H, Yanagisawa A, Tsuchiya E, Kato Y, Nakamura Y. Cancer Res. 1992;52:6696–6698. [PubMed] [Google Scholar]
- 38.Ding S F, Habib N A, Dooley J, Wood C, Bowles L, Delhanty J D. Br J Cancer. 1991;64:1083–1087. doi: 10.1038/bjc.1991.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimori M, Tokino T, Hino O, Kitagawa T, Imamura T, Okamoto E, Mitsunobu M, Ishikawa T, Nakagama H, Harada H, et al. Cancer Res. 1991;51:89–93. [PubMed] [Google Scholar]
- 40.Nollet F, Berx G, Molemans F, van Roy F. Genomics. 1996;32:413–424. doi: 10.1006/geno.1996.0136. [DOI] [PubMed] [Google Scholar]
- 41.Lerman L S, Silverstein K. Methods Enzymol. 1987;155:482–501. doi: 10.1016/0076-6879(87)55032-7. [DOI] [PubMed] [Google Scholar]