Abstract
The Epstein–Barr virus (EBV) nuclear protein BS-MLF1 (SM) is expressed early after entry of EBV into the lytic cycle. SM transactivates reporter gene constructs driven by a wide variety of promoters, but the mechanism of SM action is poorly understood. In this study, we demonstrate that the SM protein inhibits expression of intron-containing genes and activates expression of intron-less genes. We demonstrate that SM has the predicted inhibitory effect on expression of a spliced EBV gene but activates an unspliced early EBV gene. SM inhibited gene expression at the post-transcriptional level by preventing the accumulation of nuclear and cytoplasmic RNA transcripts. Conversely, SM led to increased accumulation of nuclear mRNA from intron-less genes without affecting the rate of transcription, indicating that SM enhances nuclear RNA stability. The ratio of cytoplasmic to nuclear polyadenylated mRNA was increased in the presence of SM, suggesting that SM also enhances nucleo-cytoplasmic mRNA transport. The degree of transactivation by SM was dependent on the sequence of the 3′-untranslated region of the target mRNA. Finally, we demonstrate that the amino-terminal portion of SM fused to glutathione-S-transferase binds radioactively labeled RNA in vitro, indicating that SM is a single-stranded RNA binding protein. Importantly, the latent and immediate-early genes of EBV contain introns whereas many early and late genes do not. Thus, SM may down-regulate synthesis of host cell proteins and latent EBV proteins while simultaneously enhancing expression of specific lytic EBV genes by binding to mRNA and modulating its stability and transport.
The Epstein–Barr virus (EBV) nuclear protein BS-MLF1 (SM) gene is expressed early after entry of EBV into the lytic cycle of replication (1, 2). Cotransfection of SM leads to increased expression of chloramphenicol acetyltransferase (CAT) from reporter constructs (3–8). The mechanism of SM gene action, however, is poorly understood. Activation by SM is promoter-independent (5, 6), and a specific SM-binding DNA sequence, or SM response element, has not been identified. Although some studies have indicated that SM does not affect target gene transcript levels, others have observed increases in CAT mRNA (3, 7, 9, 10). Some investigators have concluded that the degree of transactivation does not correlate with mRNA levels, suggesting that the mechanism of SM action is post-transcriptional (6). In contrast, a recent study reported that SM expression leads to increased levels of cytoplasmic CAT mRNAs, which correlate well with the degree of CAT transactivation (11). It also appears that SM activates some genes but not others, which has led to the hypothesis that activation is reporter gene-dependent (6, 10). The basis for this difference in the sensitivity to enhancement by SM also remains unexplained. SM is distantly homologous to herpes simplex virus (HSV) ICP27, which is known to be a post-transcriptional activator and inhibitor of gene expression and binds RNA in vitro (12, 13).
Although several transcripts containing the SM ORF have been described, the major mRNA expressed at early times after EBV entry into the lytic cycle is a spliced mRNA in which a small upstream exon (BSLF2) is joined to the BMLF1 exon (11). Most previous studies have used vectors expressing the second exon alone. In addition, the promoters, 3′-processing signals, and plasmid vectors have varied considerably between these studies, making it difficult to compare their various findings. We have examined the effect of the spliced BSLF2–BMLF1 gene product (SM) on gene expression by using reporter constructs that vary only in promoter sequences and 3′ mRNA-processing signals. We also have examined the effect of SM on the expression of genes with natural and artificial introns. The effect of SM on RNA transcription, accumulation of cytoplasmic and nuclear RNA, and protein expression was measured in transfection experiments and in EBV-infected cells. These experiments reveal a dual repressor and activator function for SM and provide further insights into its mechanism of action.
MATERIALS AND METHODS
Plasmids and Cell Lines.
SM and aSM were constructed by cloning the SM cDNA sequence (11) in pCDNA3 (Invitrogen), in opposite orientations. SM cDNA was made by PCR amplification of the sequence from EBV genome position 84,288 (14, 15), to the BamHI site at position 84233, and ligation to the remainder of the coding sequence. Constructs were sequenced to verify fidelity of PCR amplification. Wp-CAT-i and Wp-i-CAT were constructed by insertion of nucleotides 13467–14368 containing the W promoter in pCATbasic or pCAT3basic (Promega). Cp-CAT-i was made by insertion of EBV nucleotides 10736–11361 in pCATbasic. Wp-CAT and SV40-CAT were constructed by deleting the 5′ intron from Wp-i-CAT and pCAT3-promoter (Promega). LGU, IEGU, and synPA plasmids (12) were kind gifts of R. Sandri-Goldin, University of California, Irvine. Gp350, viral capsid antigen (VCA), EBNA-2, and BALF-2 plasmids were made as follows. Oligonucleotides corresponding to both strands of 35 nucleotides immediately following the hexanucleotide polyadenylation signal AAUAAA from each of the EBV genes (14, 15), were annealed and cloned into the EcoRI site immediately following the polyadenylation signal in synPA. Orientation was confirmed by PCR amplification and DNA sequencing of each construct. B95–8PA and P3HR-1PA plasmids were constructed by insertion of PCR-amplified fragments consisting of the 65 nucleotides flanking the polyadenylation site of the DNA polymerase gene from B95–8 and P3HR-1 (16) into synPA, replacing the synPA polyadenylation signal and 3′-untranslated region (UTR). Cytomegalovirus (CMV)-CAT and CMV-growth hormone (GH) were made by insertion of CAT (17) and genomic GH-coding sequences (18) in pCDNA3. pXGH5 contains the genomic GH sequence driven by the human metallothionine promoter (18). pSVNaeZ has been described (19). SM-glutathione-S-transferase (GST) fusion plasmids were constructed by cloning the first 845 nucleotides of SM cDNA in pGEX3X (Pharmacia).
Transfections and Reporter Gene Assays.
BJAB cells (20) were electroporated with 10 μg of reporter plasmid and 10 μg of either SM or aSM DNA, as described (19). Cells were lysed in a total volume of 1 ml, 48 hr after electroporation. Lysates were adjusted to keep the percent acetylation in the linear range for CAT assay (21). Each data point was calculated from the mean of three independent electroporations. Human growth hormone (hGH) concentrations were measured by radioimmunoassay (Nichols Institute, San Juan Capistrano, CA) of transfected cell supernatants.
Run-on Transcription Assays and Northern Blotting.
Run-on transcription was performed with 25 × 106 nuclei per reaction. Washed cells were lysed 48 hr after transfection in 10 mM Tris (pH 8), 0.5% Nonidet P-40, and 3 mM MgCl2. Nuclei were separated by centrifugation and stored at −80°C. One microgram of hGH or CAT cDNA was slot-blotted onto Zeta-Probe membrane (Bio-Rad). Bacteriophage λ DNA and 18S ribosomal RNA cDNA were used as negative and positive controls, respectively. In vitro “run-on” nuclear transcripts were generated with 32P-labeled UTP, treated with DNase, and purified (21). Then, 1 × 107 cpm of each transcript were hybridized to immobilized cDNA for 40 hr and washed at 65°C. Radioactivity bound to each DNA was quantitated by using an instant imager (Packard Instruments).
For RNA isolation, cells were harvested 48 hr after transfection and lysed in Nonidet P-40 buffer, and nuclei were separated by centrifugation. RNA was prepared from both fractions as described (21). Ten micrograms of each RNA were used per lane for Northern blotting.
Antibodies, Immunoblotting, and RNA-Binding Assays.
Immunoblotting was performed as described (23). Protein concentration of lysates was measured by Bradford assay and equalized before use. Anti-Z and anti-BMRF1 antibodies were purchased (Dako, Virostat). RNA-probed blots of bacterial lysates were performed as previously described (24). RNA-protein crosslinking was performed by incubating 40 μg of whole bacterial lysate with 10 fmol of radiolabeled RNA for 10 min on ice, UV irradiating (25 J/cm2, 254 nM), and treating with RNase A before electrophoresis.
RESULTS
SM Reduces Expression of Genes Containing Introns.
It has been reported previously that SM transactivation is reporter gene-dependent. However these previous studies used a variety of reporter constructs, making direct comparisons difficult (6, 10). The HSV gene ICP27, which is distantly homologous to SM, has inhibitory effects on spliced genes in addition to trans-activating functions (12, 25, 26). To determine if SM has a similar inhibitory effect on multi-exon genes, we examined the effect of SM on the expression of intron-containing reporter plasmids and compared them with otherwise identical expression vectors without introns.
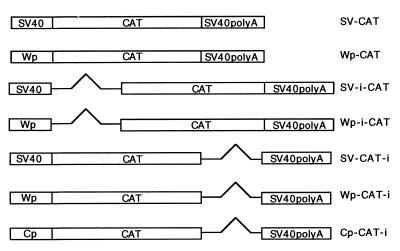
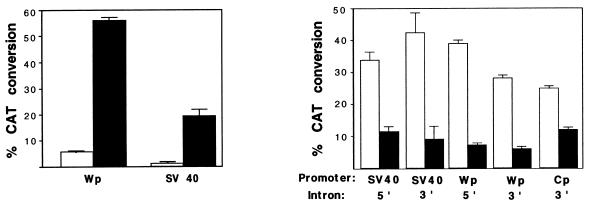
A matched series of constructs, which differed only in the promoter used to drive expression of CAT and in the presence or absence of an artificial intron, was constructed (Fig. 1). Each of these plasmids was cotransfected with the CMV-SM expression plasmid SM or the anti-sense control aSM into BJAB cells (an EBV-negative B lymphoma cell line). Regardless of the promoter used to drive CAT, SM cotransfection led to transactivation of CAT plasmids without introns (Fig. 2), as expected from previously reported studies (3–8). The degree of transactivation was ≈10-fold greater than in control transfections. However, cotransfection of SM with intron-containing reporter plasmids consistently led to a decrease in CAT activity as compared with transfection with aSM (Fig. 2). The CAT activity in the presence of SM ranged from 20 to 50% of the control. The SM inhibitory effect was independent of intron position because intron sequences located either downstream or upstream of the CAT ORF led to similar inhibition of CAT expression. Neither aSM nor empty vector had any effect on CAT expression compared with control CAT-transfected cells. Further, transfection of an irrelevant gene driven by the CMV promoter had no inhibitory effect on CAT expression (data not shown). Thus the inhibition was SM-specific and not caused by a general inhibition of transcription or “squelching.”
Figure 1.
Plasmids used in CAT assays. i, intron.
Figure 2.
Effect of SM on CAT expression from plasmids with and without introns. SM (filled bars) or aSM (open bars) was transfected into BJAB cells with intron-less plasmids Wp-CAT or SV40-CAT (Left). SM (flled bars) or aSM (open bars) was transfected with the intron-containing plasmid SV-i-CAT, SVCAT-i, Wp-i-CAT, WpCAT-i, or CpCAT-i into BJAB cells (Right).
SM Decreases Nuclear Pre-mRNA Stability.
Because SM had an inhibitory effect on intron-containing genes, it was likely that the effect was mediated at the level of the RNA transcript, either by decreasing pre-mRNA stability, nucleo-cytoplasmic transport, or protein translation. We wished therefore to determine whether SM altered the steady-state levels of cytoplasmic and nuclear RNA transcribed from intron-containing genes. We chose to use hGH as the reporter gene in this experiment for several reasons. The hGH mRNA is relatively stable; the spliced form can easily be differentiated from higher molecular weight pre-mRNA, and simultaneous measurement of secreted hGH and hGH mRNA can be performed, facilitating analysis (18).
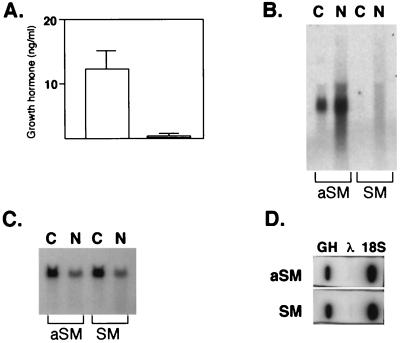
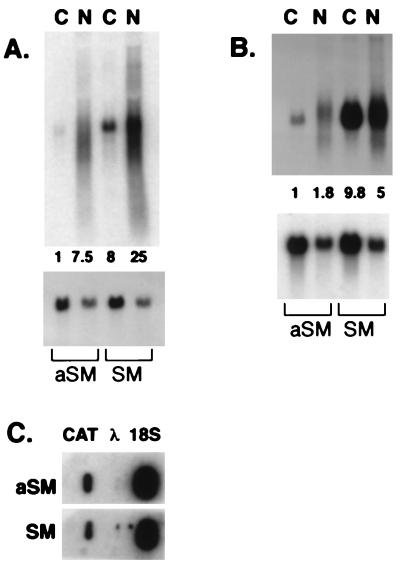
As shown in Fig. 3a, cotransfection of BJAB cells with SM and the hGH plasmid pXGH5 led to almost complete inhibition of hGH secretion. Northern analysis of nuclear and cytoplasmic RNA from aSM-transfected cells demonstrated that hGH mRNA is easily detected in both nuclear and cytoplasmic fractions (Fig. 3b). In contrast, there was no detectable cytoplasmic hGH mRNA in SM-transfected cells. The amount of nuclear hGH mRNA also was greatly diminished, although detectable. The blots were stripped and hybridized with a glyceraldehyde-3-phosphate dehydrogenase probe to verify equal RNA loading (Fig. 3c). These data are consistent with SM reducing nuclear pre-mRNA stability. Although blockade of nucleo-cytoplasmic transport also could explain the absence of cytoplasmic hGH mRNA, such a block should lead to increased levels of nuclear mRNA unless mRNA stability were concomitantly decreased. Regardless, it is clear that the inhibition of hGH expression is caused by an effect of SM at the RNA level rather than on protein translation.
Figure 3.
Effect of SM on expression of hGH protein and RNA. (A) BJAB cells were transfected with pXGH5 and either SM (shaded bar) or aSM (open bar) and growth hormone in the supernatant measured 48 hr after transfection by radioimmunoassay. (B) RNA from cells transfected in A was blotted and hybridized with labeled hGH probe. N, nuclear; C, cytoplasmic. (C) Membrane from B was stripped and probed with labeled glyceraldehyde-3-phosphate dehydrogenase cDNA. (D) Nuclear transcripts from cells transfected with pXGH5 and SM or aSM were hybridized to immobilized cDNAs for GH and 18S RNA and bacteriophage λ DNA.
Selective inhibition by SM of intron-containing genes strongly suggests that the effect is post-transcriptional. To rule out a direct inhibitory effect on transcriptional initiation, nuclear run-on assays were performed to compare the rate of transcript initiation in the presence of SM and aSM. SM did not inhibit the rate of hGH transcript initiation (Fig. 3d). Hybridization to hGH cDNA and 18S rRNA cDNA (as an internal control) were quantitated. The ratio of hGH to 18S RNA-specific counts was 31% in the presence of SM and 25% in the presence of aSM.
Transactivation by SM is Dependent on the 3′ UTR.
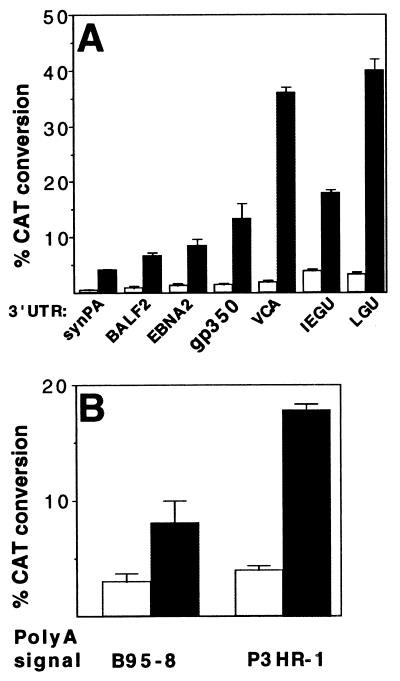
The mechanism of SM transactivation is controversial, with studies reporting either no change in the levels of the target mRNA (6) or an increase in the levels of cytoplasmic mRNA (11). ICP27 is reported to interact with the 3′ UTR and affect processing of target mRNAs (27–29). To determine whether SM exerts its trans-activating effect by a similar interaction, we constructed a series of reporter plasmids which differ in the 35 nucleotides immediately following the consensus hexanucleotide polyadenylation signal, AAUAAA. Although a strict consensus sequence has not been identified, cleavage and polyadenylation is dependent on the sequence immediately following the canonical hexanucleotide (30). Thus, a reporter plasmid without such a 3′ UTR, synPA, expresses CAT poorly (ref. 12 and Fig. 4a). Insertion of 3′ UTRs from various HSV or EBV genes led to increased baseline CAT expression (Fig. 4a). However, the different 3′ UTRs conferred varying amounts of responsiveness to SM transactivation. The most active EBV 3′ UTRs were those from the late lytic genes gp350 and the major VCA. Cotransfection of SM with these reporter plasmids led to a 10- and 18-fold increase in CAT activity, respectively. Interestingly, the 3′ UTRs from two lytic HSV genes [ICP27 (IEGU) and UL 38 (LGU)] led to high levels of transactivation. The baseline activity of the reporter plasmids (with cotransfected aSM) did not differ significantly, suggesting that the 3′ UTRs differ in their ability to interact with SM.
Figure 4.
Effect of 3′ UTR and polyadenylation signal on transactivation by SM. (A) CAT assays on lysates from cells transfected with SM (shaded bars) or aSM (open bars) and a series of reporter plasmids varying in the 3′ UTR sequence. The gene from which each UTR is derived is shown below the panel. (B) CAT assays on lysates from cells transfected with SM (shaded bars) or aSM (open bars) and CAT reporter plasmids with polyadenylation signals from B95–8 or P3HR-1 strains of EBV.
The EBV DNA polymerase gene of the P3HR-1 virus has been reported to possess a noncanonical polyadenylation signal AUUAAA, whereas the polymerase gene of the B95–8 prototype has a deletion in this region without a clearly defined polyadenylation signal (16). To determine whether SM could transactivate expression of genes with such atypical 3′-processing signals, we constructed analogous reporter constructs containing the putative P3HR-1 and B95–8 polyadenylation signals and the 60 downstream nucleotides. Results from cotransfection of these plasmids with SM or aSM are shown in Fig. 4b. Although baseline CAT expression from these plasmids is less than that from those containing canonical polyadenylation signals, SM nevertheless transactivates CAT expression ≈4-fold. These data indicate that the SM effect does not require the AAUAAA sequence for activity and may act on a variety of intron-less genes, albeit at differing levels.
SM Increases Levels of Both Cytoplasmic and Nuclear Target mRNAs and Enhances Nucleo-Cytoplasmic mRNA Transport.
Previous studies on the mechanism of transactivation by SM have led to differing conclusions. Most authors have reported that there is either little or no effect of SM on mRNA levels (3, 6). A more recent report concluded that SM leads to increased cytoplasmic levels of target mRNAs (11). To determine whether SM acts at the post-transcriptional level to increase mRNA stability, nucleo-cytoplasmic transport, or protein translation, we compared the levels of nuclear and cytoplasmic CAT mRNA in cells cotransfected with an intron-less CAT reporter plasmid (pCMV-CAT) and either SM or aSM. As shown in Fig. 5a, SM expression led to markedly increased levels of both nuclear and cytoplasmic CAT mRNA as compared with control Cytoplasmic CAT mRNA was increased 8-fold by SM, and nuclear CAT mRNA was increased 3-fold in the presence of SM. These data indicate that SM leads to greater accumulation of nuclear and cytoplasmic CAT mRNA. Although a direct effect on nuclear export also may be present, it is clear that the increased levels of nuclear mRNA in the presence of SM are caused by either enhanced stability or transcription. To eliminate the possibility that SM directly enhances transcription, nuclear run-on assays were performed with nuclei from cells transfected with CMV-CAT and SM or aSM. The rate of CAT transcription initiation was not affected by SM. The CAT-specific transcripts were 3.64% and 3.79% of 18S RNA transcripts in the presence of SM and aSM, respectively (Fig. 5c).
Figure 5.
Effect of SM on RNA levels of an intron-less gene. (A) RNA from BJAB cells transfected with aSM or SM and pCMV-CAT separated into nuclear (N) and cytoplasmic (C), fractions and probed with CAT (Top) or with glyceraldehyde-3-phosphate dehydrogenase cDNA (Bottom). Relative levels of CAT RNA-specific cpm in each lane are shown below CAT in A and B. (B) PolyA RNA from cells used in A probed with labeled CAT (Top) or glyceraldehyde-3-phosphate dehydrogenase (Bottom), cDNA. (C) Labeled nuclear transcripts from SM- or aSM-transfected cell nuclei were hybridized to immobilized 18SrRNA cDNA, CAT, or bacteriophage λ DNA.
To evaluate whether SM directly facilitates transport of mRNA, we measured the nuclear and cytoplasmic levels of polyadenylated (polyA) CAT mRNA in cells transfected with SM and compared them with levels in cells transfected with aSM (Fig. 5b). SM led to an absolute increase in both cytoplasmic (9.8-fold), and nuclear (2.8-fold), polyA CAT RNA levels. In the presence of aSM, the amount of cytoplasmic polyA CAT RNA was only 55% of the nuclear polyA CAT RNA. In contrast, in the presence of SM, the amount of cytoplasmic polyA CAT RNA was almost twice that found in the nucleus, suggesting that SM facilitates nuclear mRNA export. These data demonstrate that SM exerts its activating effect at a post-transcriptional level. The SM-mediated increase in protein expression correlated with an increase in cytoplasmic mRNA. The increase in cytoplasmic RNA was accompanied by greater accumulation of non-polyA and polyA CAT RNAs in the nucleus.
SM Has Stimulatory and Inhibitory Effects on EBV Gene Expression.
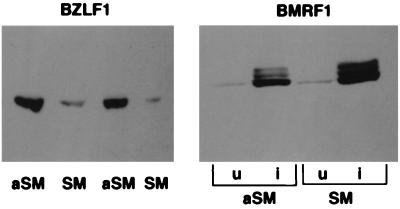
Previous studies of SM have used a variety of bacterial and eukaryotic genes but not EBV genes in reporter constructs (3–11). SM activity has been reported to be gene-dependent (10), and the only gene on which SM has previously been shown to have any effect is CAT. Because we wished to determine whether SM had an inhibitory effect on spliced EBV genes and a stimulatory effect on unspliced genes, we examined the effect of SM on expression of the intron-containing immediate-early gene BZLF1 and the unspliced early gene BMRF1. BJAB cells were transfected with either aSM or SM and SVNaeZ. B95–8 cells were transfected with either aSM or SM and EBV replication induced with phorbol 12-tetradecanoate 13-acetate. Forty-eight hours after induction or transfection, cells were harvested for analysis by immunoblotting. The results demonstrate that SM has a marked inhibitory effect on expression of cotransfected BZLF1 (Fig. 6 Left). In contrast, SM enhances the expression of BMRF1 produced during EBV reactivation (Fig. 6 Right).
Figure 6.
Effect of SM on expression of EBV genes. Lysates from BJAB cells transfected with BZLF1 and SM or aSM were immunoblotted with anti-BZLF1 mAb (Left). Lysates from B95–8 cells transfected with SM or aSM and untreated (u) or treated (i) with phorbol 12-tetradecanoate 13-acetate to induce EBV replication, immunoblotted with anti-BMRF1 mAb (Right).
ssRNA Binding by SM-GST Fusion Proteins.
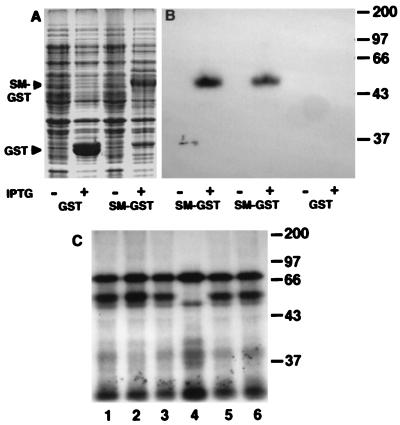
A mechanism by which SM can enhance processing, stability, and transport of mRNA could involve direct binding to ssRNA. A fusion protein containing the 281 NH2-terminal amino acids of SM fused in frame to GST was overexpressed in E. coli (SM-GST). Bacterial lysates containing SM-GST or GST alone were electrophoresed and transferred to polyvinylidene difluoride membrane. The expected molecular mass of this fusion is approximately 57 kDa. A protein of the appropriate size was seen as the major product only in lysates of isopropyl β-d-thiogalactoside-induced cells containing SM-GST (Fig. 7a). After denaturation of the blotted proteins in 8 M urea and renaturation, membranes were probed with 32P-labeled, in vitro-synthesized RNA corresponding to the sense strand of a 3′ UTR highly responsive to SM (LGU). As shown in Fig. 7b, the RNA probe does not bind to GST alone, nor does it show reactivity with lysates from uninduced bacteria. However, in lysates from induced cells that carry SM-GST, the fusion protein binds the RNA probe. To confirm RNA-binding by SM-GST and to determine whether it bound RNA in solution, RNA-protein crosslinking experiments were performed. SM-GST lysates or GST lysates were incubated in vitro with radiolabeled 3′ UTR probe and UV-crosslinked. After treatment with RNase A to remove unbound probe, reaction mixtures were analyzed by SDS/PAGE (Fig. 7c). Two labeled proteins were observed in all the bacterial lysates and presumably represent bacterial proteins capable of binding the ssRNA probe. However, in reaction mixtures containing SM-GST, an additional protein of ≈57 kDa also was detected. These experiments demonstrate that the N-terminal 281 amino acids of SM are sufficient to bind single-stranded RNA. To determine whether SM-GST bound to other 3′ UTR containing RNA probes, we also constructed and used probes from the synPA and VCA plasmids. These probes also bound immobilized SM-GST with equal affinity (data not shown).
Figure 7.
RNA binding by SM-GST fusion proteins. (A) Bacterial cultures carrying either GST or SM-GST fusion plasmids were uninduced (−) or induced with isopropyl β-d-thiogalactoside (+) to produce fusion proteins. Lysates were separated by SDS/PAGE and stained with Coomassie blue. Arrows denote position of induced GST and SM-GST. Molecular masses in kilodaltons are shown at right. (B) Bacterial lysates from cells containing two independently derived SM-GST fusion proteins and GST control were subjected to SDS/PAGE, transferred to polyvinylidene difluoride membrane, and probed with radioactively labeled RNA. Bacterial cultures were uninduced (−) or induced with isopropyl β-d-thiogalactoside (+) to produce fusion proteins. (C) Lysates from bacteria expressing SM-GST fusions (lanes 1–3, 5–6) or GST alone (lane 4), were crosslinked to radiolabeled RNA probe, subjected to SDS/PAGE and autoradiographed.
DISCUSSION
In this study, we demonstrate that the SM gene product of EBV has two distinct activities, enhancement of intron-less gene expression and inhibition of intron-containing gene expression. Both activities are shown to be mediated at the post-transcriptional level. We show that the activating function is dependent on the sequence of the 3′ UTR of the RNA transcript of the target gene. Finally, we find that recombinant SM protein is capable of binding RNA in vitro. Expression of intron-less genes is inherently inefficient (31). Whereas the vast majority of eukaryotic cell genes contain multiple exons, many viral genes, including many EBV lytic phase genes, are encoded as a single ORF (32). The findings of this study suggest that SM directly interacts with mRNA and has the potential to differentially regulate gene expression of both cell and EBV genes based on the structure of their nascent RNA transcripts.
Previous investigations have shown that SM trans-activates gene expression in a gene-dependent manner, but the mechanism for such selectivity was not further explored (6, 10). The data presented in this report suggest several explanations for the gene specificity of SM. First, it is clear that the presence of introns in the transcript leads to inhibition rather than activation by SM. Our finding that the 3′ UTR of the mRNA transcript has an effect on responsiveness to SM suggests several mechanisms for the gene specificity of SM trans-activation. A likely gene-specific determinant of sensitivity to SM activation is the inherent stability of a given RNA, which is affected by the 3′ UTR sequence. Expression of relatively unstable RNAs is more likely to be enhanced by SM. Such a hypothesis is supported by the effect of SM on the accumulation of cytoplasmic CAT mRNA, which is known to be highly labile (33). Other potential gene-dependent factors include the ability of the RNA sequence to bind cellular 3′-processing factors and perhaps SM itself. Thus intron-less genes with particularly inefficient processing signals may be preferentially activated by SM whereas those with poor SM-binding RNAs would not. SM has a highly arginine-rich region from amino acids 151–184, which may serve as an RNA-binding motif. The in vitro RNA binding we have detected, however, does not appear to depend on the 3′ UTR sequence. Such sequence-independent binding is demonstrated by other RNA-binding proteins, including ICP27 (13, 34), which also has gene-specific effects dependent on the 3′ UTR sequence of the target gene (12).
We have described a novel property of SM, its ability to inhibit expression of spliced mRNA transcripts. SM is therefore similar to ICP27 of HSV, which is involved in shutoff of splicing and host protein synthesis (25, 26). SM homologues are found in several other herpes viruses, but they vary considerably in sequence and function (12, 29, 35–39). This inhibitory function of SM is of particular interest because all of the genes expressed during EBV latency are spliced (for review, see ref. 32). In particular, the latent nuclear genes (EBNAs) are extensively spliced and derived from multicistronic transcripts. SM expression shortly after lytic reactivation would therefore be expected to lead to a decrease in accumulation of all latent gene mRNAs. Diversion of the cellular processing machinery away from latent EBV and cell transcripts likely plays an important role in the switch from latent to lytic infection. Selective repression of spliced genes also is likely to play a role in regulating the sequential expression of lytic genes. For example, the immediate early BZLF1 gene, whose expression directly activates expression of BMLF1 and other early genes, is spliced (40). SM would therefore be expected to down-regulate BZLF1, while enhancing expression of unspliced early and late genes later in the lytic cascade. That SM plays such a role is supported by our finding that transfection of SM leads to repression of BZLF1 and activation of BMRF1, the gene encoding EBV DNA polymerase accessory protein. A model in which SM binds pre-mRNA at the 3′ terminus is consistent with the negative effect of SM on intron-containing transcripts. Such binding may inhibit interaction of the spliceosome with the polyadenylation complex, which is required for proper excision of the terminal intron (41). Thus SM may lead to incompletely spliced RNAs, which are known to be retained and degraded in the nucleus rather than transported to the cytoplasm for translation (42).
It is well documented that the presence of introns facilitates both post-transcriptional processing and cytoplasmic accumulation of mature mRNA. The possible basis for these effects has become evident from recent studies on the mechanisms of polyadenylation and nuclear export. Splicing and polyadenylation are closely linked, and various components of the spliceosome and 5′ cap have been shown to interact with the polyadenylation complex (for review, see ref. 43). For example, the U1 snRNP-A protein interacts directly with the 160-kDa subunit of cleavage-polyadenylation specificity factor and increases polyadenylation efficiency. Similarly, mutation of splicing signals inhibits polyadenylation (44). Because polyadenylation is critical to the stability of mRNAs (45), a lack of introns in certain genes poses special problems. The SM protein may act to enhance stability of such mRNAs either by binding and directly stabilizing pre-mRNA or by facilitating the assembly of the polyadenylation complex.
Nuclear export of all types of RNA appears to be an active process dependent on binding to specific proteins that carry a nuclear export signal (NES), (reviewed in ref. 46). The NES allows interaction with nuclear export receptors, which facilitate exit via nuclear pores. Each class of RNA appears to have an uniquely saturable pathway characterized by interactions with different NES-containing proteins, export receptors, and nucleoporins (47, 48). Proteins involved in processing pre-mRNA may mediate association with the appropriate export receptors. For example, the cap-binding complex, CBP20 and CBP80, potentially contains an NES (48). Similarly, hnRNPA1, which complexes with pre-mRNA and mRNA, has an NES allowing rapid export (49, 50). Thus, RNAs on which spliceosome formation does not occur may not have access to the same export pathways available to spliced mRNAs. Our finding that the SM protein of EBV binds RNA and stimulates RNA export suggests that it compensates for the lack of spliceosome assembly on certain EBV transcripts. SM may act as a bridge between RNAs and one of the family of nuclear export receptors, thereby targeting EBV transcripts to a specific cellular export pathway. Such a role is played by the HIV rev protein in facilitating export of unspliced, intron-containing HIV RNAs which would otherwise be retained in the nucleus. HIV rev contains an NES, binds unspliced HIV RNA, and directs it to a cellular pathway used for 5S and U1 snRNA export (51–53). Whether SM contains such a nuclear export signal and interacts with one or more nuclear export receptors is currently under investigation.
Acknowledgments
This work was supported in part by the John Sealy Memorial Endowment Fund for Biomedical Research.
ABBREVIATIONS
- EBV
Epstein–Barr virus
- SM
nuclear protein BS-MLF1
- CAT
chloramphenicol acetyltransferase
- GH
growth hormone
- hGH
human GH
- GST
glutathione-S-transferase
- CMV
cytomegalovirus
- NES
nuclear export signal
- UTR
untranslated region
- VCA
viral capsid antigen
- polyA
polyadenylated
- HSV
herpes simplex virus
References
- 1.Cho M S, Jeang K T, Hayward S D. J Virol. 1985;56:852–859. doi: 10.1128/jvi.56.3.852-859.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sample J, Lancz G, Nonoyama M. J Virol. 1986;57:145–154. doi: 10.1128/jvi.57.1.145-154.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chevalier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong K-M, Levine A J. J Virol. 1986;60:149–156. doi: 10.1128/jvi.60.1.149-156.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman P M, O’Hare P, Hayward G S, Hayward S D. J Virol. 1986;60:140–148. doi: 10.1128/jvi.60.1.140-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenney S, Kamine J, Holley-Guthrie E, Mar E C, Lin J C, Markovitz D, Pagano J. J Virol. 1989;63:3870–3877. doi: 10.1128/jvi.63.9.3870-3877.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenney S, Kamine J, Markovitz D, Fenrick R, Pagano J. Proc Natl Acad Sci USA. 1988;85:1652–1656. doi: 10.1073/pnas.85.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buisson M, Manet E, Trescol-Biemont M C, Gruffat H, Durand B, Sergeant A. J Virol. 1989;63:5276–5284. doi: 10.1128/jvi.63.12.5276-5284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chavrier P, Gruffat H, Chevalier-Greco A, Buisson M, Sergeant A. J Virol. 1989;63:607–614. doi: 10.1128/jvi.63.2.607-614.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markovitz D M, Kenney S, Kamine J, Smith M S, Davis M, Huang E-S. Virology. 1989;173:750–754. doi: 10.1016/0042-6822(89)90591-6. [DOI] [PubMed] [Google Scholar]
- 11.Cook I D, Shanahan F, Farrell P J. Virology. 1994;205:217–227. doi: 10.1006/viro.1994.1637. [DOI] [PubMed] [Google Scholar]
- 12.Sandri-Goldin R M, Mendoza G E. Genes Dev. 1992;63:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 13.Mears E, Rice S A. J Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baer R, Bankier A T, Biggin M D. Nature (London) 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 15.Farrell P. Adv Viral Oncol. 1989;8:103–132. [Google Scholar]
- 16.Furnari F B, Adams M D, Pagano J S. Proc Natl Acad Sci USA. 1993;90:378–382. doi: 10.1073/pnas.90.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorman C G, Moffat L F, Howard B H. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selden R F, Howie K B, Rowe M E, Goodman H G, Moore D D. Mol Cell Biol. 1986;6:3173–3179. doi: 10.1128/mcb.6.9.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaminathan S, Tomkinson B, Kieff E. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menezes J, Liebold W, Klein G, Clements G. Biomedicine (Paris) 1975;22:276–284. [PubMed] [Google Scholar]
- 21.Gorman C M, Moffat L F, Howard B H. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 23.Swaminathan S. Virology. 1996;217:532–541. doi: 10.1006/viro.1996.0148. [DOI] [PubMed] [Google Scholar]
- 24.St. Johnston D, Brown N H, Gall J G, Jantsch M. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy W R, Sandri-Goldin R M. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardwicke M A, Sandri-Goldin R M. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown C R, Nakamura M S, Mosca J D, Hayward G S, Straus S E, Perera L P. J Virol. 1995;69:7187–7195. doi: 10.1128/jvi.69.11.7187-7195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLauchlan J, Phelan A, Loney C, Sandri-Goldin R M, Clements J B. J Virol. 1992;66:6939–6945. doi: 10.1128/jvi.66.12.6939-6945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGregor F, Phelan A, Dunlop J, Clements J B. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDevitt M A, Imperiale M J, Nevins J R. Cell. 1984;37:993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- 31.Huang M T F, Gorman C M. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieff E. In: Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 1889–1910. [Google Scholar]
- 33.Thompson J F, Hayes L S, Lloyd D B. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- 34.Ingram A, Phelan A, Dunlop J, Clements J B. J Gen Virol. 1996;77:1847–1851. doi: 10.1099/0022-1317-77-8-1847. [DOI] [PubMed] [Google Scholar]
- 35.Albrecht J C, Nicholas J, Biller D, Cameron K R, Beisinger B C N, Wittman S, Craxton M A, Coleman H. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chee M, Barrell B. Trends Genet. 1990;6:86–91. doi: 10.1016/0168-9525(90)90099-r. [DOI] [PubMed] [Google Scholar]
- 37.Davison A J, Taylor P. J Gen Virol. 1987;68:1067–1079. doi: 10.1099/0022-1317-68-4-1067. [DOI] [PubMed] [Google Scholar]
- 38.Winkler M, Rice S A, Stamminger T. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perera L P, Kaushal S, Kinchington P R, Mosca J D, Hayward G S, Straus S E. J Virol. 1994;68:2468–2477. doi: 10.1128/jvi.68.4.2468-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manet E, Gruffat H, Trescol-Biemont M C, Moreno N, Chambard P, Giot J F, Sergeant A. EMBO J. 1989;8:1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niwa M, Berget S. Genes Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 42.Legrain P, Rosbash M. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 43.Colgan D F, Manley J L. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 44.Niwa M, Rose S D, Berget S M. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 45.Decker C, Parker R. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 46.Ullman K S, Powers M A, Forbes D J. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 47.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 49.Pinol-Roma S, Dreyfuss G. Nature (London) 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 50.Michael W M, Choi M, Dreyfuss G. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 51.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 52.Meyer B E, Malim M H. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 53.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]