Abstract
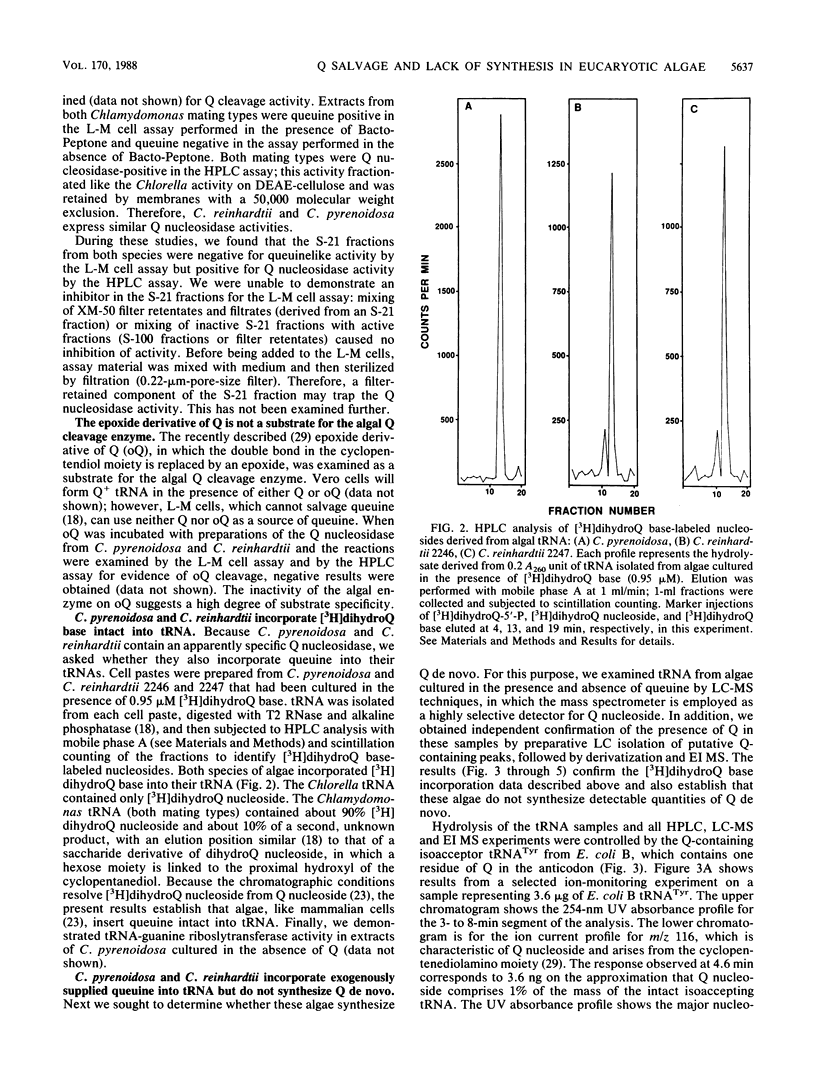
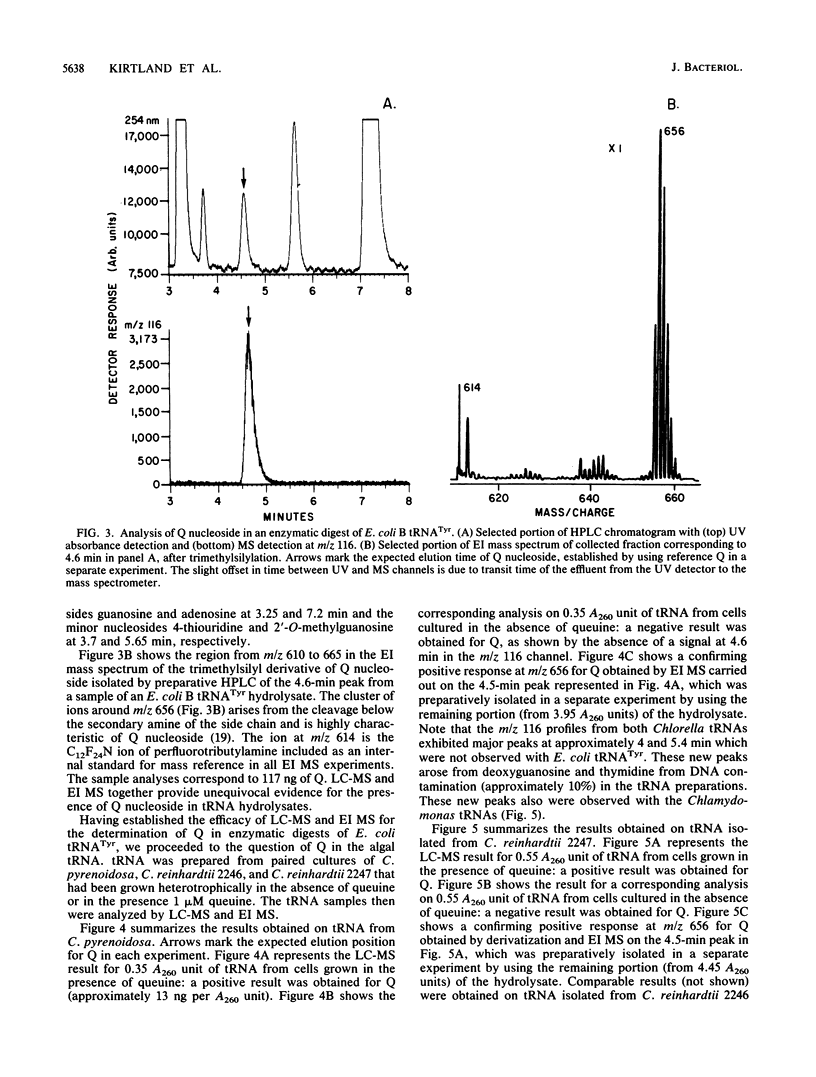
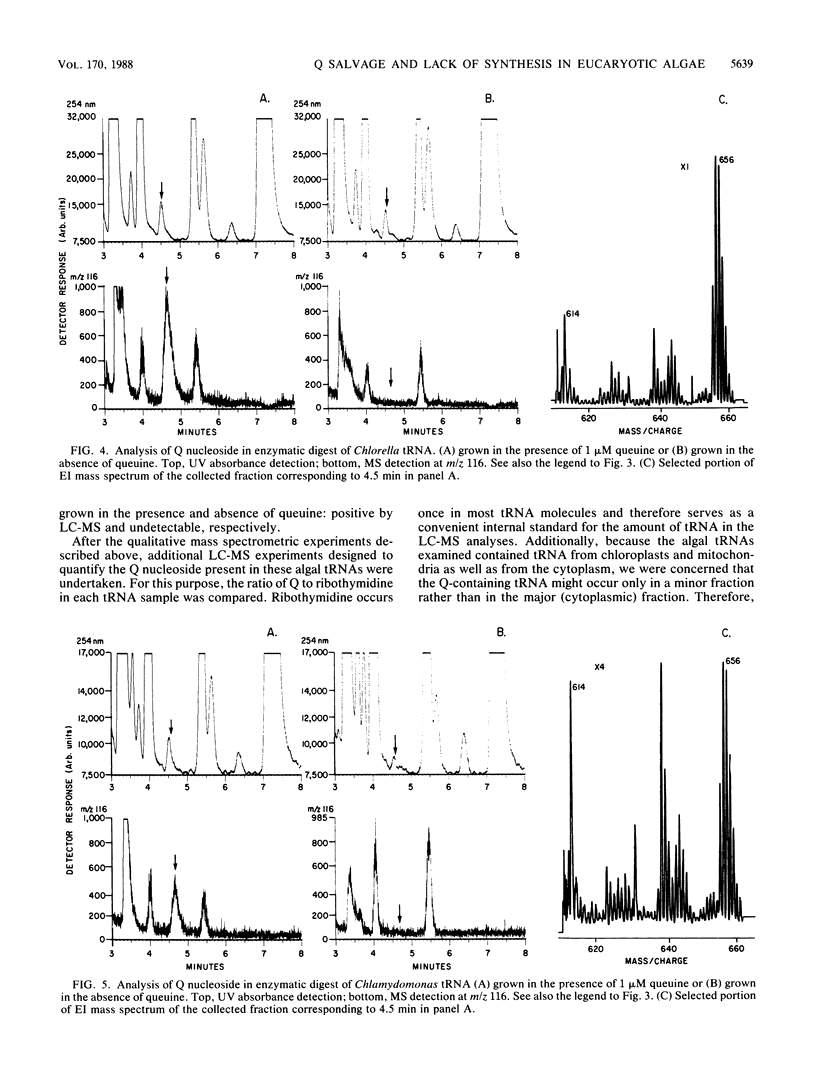
Partially purified extracts from Chlorella pyrenoidosa and Chlamydomonas reinhardtii catalyze the cleavage of queuosine (Q), a modified 7-deazaguanine nucleoside found exclusively in the first position of the anticodon of certain tRNAs, to queuine, the base of Q. This is the first report of an enzyme that specifically cleaves a 7-deazapurine riboside. Guanosine is not a substrate for this activity, nor is the epoxide a derivative of Q. We also establish that both algae can incorporate exogenously supplied queuine into their tRNA but lack Q-containing tRNA when cultivated in the absence of queuine, indicating that they are unable to synthesize Q de novo. Although no physiological function for Q has been identified in these algae, Q cleavage to queuine would enable algae to generate queuine from exogenous Q in the wild and also to salvage (and recycle) queuine from intracellular tRNA degraded during the normal turnover process. In mammalian cells, queuine salvage occurs by the specific cleavage of queuine from Q-5'-phosphate. The present data also support the hypothesis that plants, like animals, cannot synthesize Q de novo.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beier H., Barciszewska M., Krupp G., Mitnacht R., Gross H. J. UAG readthrough during TMV RNA translation: isolation and sequence of two tRNAs with suppressor activity from tobacco plants. EMBO J. 1984 Feb;3(2):351–356. doi: 10.1002/j.1460-2075.1984.tb01810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H., Barciszewska M., Sickinger H. D. The molecular basis for the differential translation of TMV RNA in tobacco protoplasts and wheat germ extracts. EMBO J. 1984 May;3(5):1091–1096. doi: 10.1002/j.1460-2075.1984.tb01934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Connick M., Ames B. N. Complete analysis of tRNA-modified nucleosides by high-performance liquid chromatography: the 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal Biochem. 1983 Feb 15;129(1):1–13. doi: 10.1016/0003-2697(83)90044-1. [DOI] [PubMed] [Google Scholar]
- Crain P. F., McCloskey J. A. Analysis of modified bases in DNA by stable isotope dilution gas chromatography-mass spectrometry: 5-methylcytosine. Anal Biochem. 1983 Jul 1;132(1):124–131. doi: 10.1016/0003-2697(83)90434-7. [DOI] [PubMed] [Google Scholar]
- Crain P. F., Sethi S. K., Katze J. R., McCloskey J. A. Structure of an amniotic fluid component, 7-(4,5-cis-dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanine (queuine), a substrate for tRNA: guanine transglycosylase. J Biol Chem. 1980 Sep 25;255(18):8405–8407. [PubMed] [Google Scholar]
- Deflaun M. F., Paul J. H., Davis D. Simplified method for dissolved DNA determination in aquatic environments. Appl Environ Microbiol. 1986 Oct;52(4):654–659. doi: 10.1128/aem.52.4.654-659.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn D. B., Trigg M. D. Minor nucleotides in tRNA from blue-green algae [proceedings]. Biochem Soc Trans. 1980 Feb;8(1):87–88. doi: 10.1042/bst0080087. [DOI] [PubMed] [Google Scholar]
- Edmonds C. G., Vestal M. L., McCloskey J. A. Thermospray liquid chromatography-mass spectrometry of nucleosides and of enzymatic hydrolysates of nucleic acids. Nucleic Acids Res. 1985 Nov 25;13(22):8197–8206. doi: 10.1093/nar/13.22.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas W. R. Effect of diet on the queuosine family of tRNAs of germ-free mice. J Biol Chem. 1980 Jul 25;255(14):6832–6835. [PubMed] [Google Scholar]
- Green G. A., Jones D. S. The nucleotide sequences of a cytoplasmic and a chloroplast tRNATyr from Scenedesmus obliquus. Nucleic Acids Res. 1985 Mar 11;13(5):1659–1663. doi: 10.1093/nar/13.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guranowski A. Purine catabolism in plants : purification and some properties of inosine nucleosidase from yellow lupin (lupinus luteus L.) seeds. Plant Physiol. 1982 Aug;70(2):344–349. doi: 10.1104/pp.70.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gündüz U., Katze J. R. Queuine salvage in mammalian cells. Evidence that queuine is generated from queuosine 5'-phosphate. J Biol Chem. 1984 Jan 25;259(2):1110–1113. [PubMed] [Google Scholar]
- Gündüz U., Katze J. R. Salvage of the nucleic acid base queuine from queuine-containing TRNA by animal cells. Biochem Biophys Res Commun. 1982 Nov 16;109(1):159–167. doi: 10.1016/0006-291x(82)91579-0. [DOI] [PubMed] [Google Scholar]
- Kasai H., Oashi Z., Harada F., Nishimura S., Oppenheimer N. J., Crain P. F., Liehr J. G., von Minden D. L., McCloskey J. A. Structure of the modified nucleoside Q isolated from Escherichia coli transfer ribonucleic acid. 7-(4,5-cis-Dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanosine. Biochemistry. 1975 Sep 23;14(19):4198–4208. doi: 10.1021/bi00690a008. [DOI] [PubMed] [Google Scholar]
- Katze J. R., Basile B., McCloskey J. A. Queuine, a modified base incorporated posttranscriptionally into eukaryotic transfer RNA: wide distribution in nature. Science. 1982 Apr 2;216(4541):55–56. doi: 10.1126/science.7063869. [DOI] [PubMed] [Google Scholar]
- Katze J. R., Farkas W. R. A factor in serum and amniotic fluid is a substrate for the tRNA-modifying enzyme tRNA-guanine transferase. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3271–3275. doi: 10.1073/pnas.76.7.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze J. R., Gündüz U., Smith D. L., Cheng C. S., McCloskey J. A. Evidence that the nucleic acid base queuine is incorporated intact into tRNA by animal cells. Biochemistry. 1984 Mar 13;23(6):1171–1176. doi: 10.1021/bi00301a022. [DOI] [PubMed] [Google Scholar]
- Katze J. R. Q-factor: a serum component required for the appearance of nucleoside Q in tRNA in tissue culture. Biochem Biophys Res Commun. 1978 Sep 29;84(2):527–535. doi: 10.1016/0006-291x(78)90201-2. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Structure, biosynthesis, and function of queuosine in transfer RNA. Prog Nucleic Acid Res Mol Biol. 1983;28:49–73. doi: 10.1016/s0079-6603(08)60082-3. [DOI] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., DeFlaun M. F. Dynamics of extracellular DNA in the marine environment. Appl Environ Microbiol. 1987 Jan;53(1):170–179. doi: 10.1128/aem.53.1.170-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillipson D. W., Edmonds C. G., Crain P. F., Smith D. L., Davis D. R., McCloskey J. A. Isolation and structure elucidation of an epoxide derivative of the hypermodified nucleoside queuosine from Escherichia coli transfer RNA. J Biol Chem. 1987 Mar 15;262(8):3462–3471. [PubMed] [Google Scholar]
- Reyniers J. P., Pleasants J. R., Wostmann B. S., Katze J. R., Farkas W. R. Administration of exogenous queuine is essential for the biosynthesis of the queuosine-containing transfer RNAs in the mouse. J Biol Chem. 1981 Nov 25;256(22):11591–11594. [PubMed] [Google Scholar]
- Sheen M. R., Martin H. F., Parks R. E., Jr The interaction of the nucleoside analogues, formycins A and B, with xanthine oxidase and hepatic aldehyde oxidase. Mol Pharmacol. 1970 May;6(3):255–265. [PubMed] [Google Scholar]
- Shindo-Okada N., Okada N., Ohgi T., Goto T., Nishimura S. Transfer ribonucleic acid guanine transglycosylase isolated from rat liver. Biochemistry. 1980 Jan 22;19(2):395–400. doi: 10.1021/bi00543a023. [DOI] [PubMed] [Google Scholar]


