Abstract
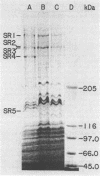
The syrA and syrB genes involved in syringomycin production in Pseudomonas syringae pv. syringae B301D were identified from an EcoRI-pLAFR3 cosmid library and then physically and functionally analyzed in relation to plant pathogenicity. Homologous recombination of the genes required for syringomycin production from cosmids pGX183 (syrA) and pGX56 (syrB), respectively, introduced into nontoxigenic (Tox-) Tn5 mutants W4S2545 and W4S770 resulted in the concomitant restoration of toxin production and full virulence. The disease indices of the Tox+ strains obtained by recombination of the cloned, homologous DNA into the corresponding Tn5 mutant were essentially equivalent to that of strain B301D-R and significantly higher than those of W4S2545 and W4S770. A 12-kilobase (kb) EcoRI fragment from pGX183 was subcloned (i.e., pGX15) and found to contain the sequences necessary for syringomycin production. A map of pGX15 prepared by a combination of restriction endonuclease digestions and Tn5 mutagenesis showed that the syrA sequence was 2.3 to 2.8 kb. Marker exchange of syrA::Tn5 from pGX15 into B301D-R yielded nonpathogenic phenotypes, indicating that syrA is a regulatory gene since it is necessary for both syringomycin production and pathogenicity. The 4.9-kb EcoRI fragment from pGX56 was subcloned (i.e., pGX4) and shown to carry the syrB sequence which was 2.4 to 3.3 kb. Sodium dodecyl sulfate-polyacrylamide gel electrophoretic analysis of protein extracts from B301D-R associated five proteins, ranging from approximately 130,000 to approximately 470,000 in molecular weight, with syringomycin production. The syrA and syrB genes were required for the formation of proteins SR4 (approximately 350,000) and SR5 (approximately 130,000), which are believed to be components of the syringomycin synthetase complex.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baxter-Gabbard K. L. A simple method for the large-scale preparation of sucrose gradients. FEBS Lett. 1972 Jan 15;20(1):117–119. doi: 10.1016/0014-5793(72)80031-0. [DOI] [PubMed] [Google Scholar]
- Bidwai A. P., Takemoto J. Y. Bacterial phytotoxin, syringomycin, induces a protein kinase-mediated phosphorylation of red beet plasma membrane polypeptides. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6755–6759. doi: 10.1073/pnas.84.19.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppels D. A. Generation and Characterization of Tn5 Insertion Mutations in Pseudomonas syringae pv. tomato. Appl Environ Microbiol. 1986 Feb;51(2):323–327. doi: 10.1128/aem.51.2.323-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis P. B., Pearson C. K. Characterization of density gradients prepared by freezing and thawing a sucrose solution. Anal Biochem. 1978 Nov;91(1):343–349. doi: 10.1016/0003-2697(78)90848-5. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. C., Cody Y. S., Proebsting E. L., Radamaker G. K., Spotts R. A. Distribution, population dynamics, and characteristics of ice nucleation-active bacteria in deciduous fruit tree orchards. Appl Environ Microbiol. 1983 Dec;46(6):1370–1379. doi: 10.1128/aem.46.6.1370-1379.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. C. Regulation of syringomycin synthesis in Pseudomonas syringae pv. syringae and defined conditions for its production. J Appl Bacteriol. 1985 Feb;58(2):167–174. doi: 10.1111/j.1365-2672.1985.tb01444.x. [DOI] [PubMed] [Google Scholar]
- Jones G. H., Hopwood D. A. Molecular cloning and expression of the phenoxazinone synthase gene from Streptomyces antibioticus. J Biol Chem. 1984 Nov 25;259(22):14151–14157. [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kleinkauf H., von Döhren H. Biosynthesis of peptide antibiotics. Annu Rev Microbiol. 1987;41:259–289. doi: 10.1146/annurev.mi.41.100187.001355. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindgren P. B., Peet R. C., Panopoulos N. J. Gene cluster of Pseudomonas syringae pv. "phaseolicola" controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol. 1986 Nov;168(2):512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morgan M. K., Chatterjee A. K. Genetic organization and regulation of proteins associated with production of syringotoxin by Pseudomonas syringae pv. syringae. J Bacteriol. 1988 Dec;170(12):5689–5697. doi: 10.1128/jb.170.12.5689-5697.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. K., Chatterjee A. K. Isolation and characterization of Tn5 insertion mutants of Pseudomonas syringae pv. syringae altered in the production of the peptide phytotoxin syringotoxin. J Bacteriol. 1985 Oct;164(1):14–18. doi: 10.1128/jb.164.1.14-18.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepold F., Anderson D., Mills D. Cloning determinants of pathogenesis from Pseudomonas syringae pathovar syringae. Proc Natl Acad Sci U S A. 1985 Jan;82(2):406–410. doi: 10.1073/pnas.82.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet R. C., Lindgren P. B., Willis D. K., Panopoulos N. J. Identification and cloning of genes involved in phaseolotoxin production by Pseudomonas syringae pv. "phaseolicola". J Bacteriol. 1986 Jun;166(3):1096–1105. doi: 10.1128/jb.166.3.1096-1105.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Selvaraj G., Iyer V. N. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol. 1983 Dec;156(3):1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Measurement of DNA length by gel electrophoresis. Anal Biochem. 1979 Dec;100(2):319–323. doi: 10.1016/0003-2697(79)90235-5. [DOI] [PubMed] [Google Scholar]
- Vidaver A. K. Synthetic and complex media for the rapid detection of fluorescence of phytopathogenic pseudomonads: effect of the carbon source. Appl Microbiol. 1967 Nov;15(6):1523–1524. doi: 10.1128/am.15.6.1523-1524.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. W., Gross D. C. Evaluation of the Role of Syringomycin in Plant Pathogenesis by Using Tn5 Mutants of Pseudomonas syringae pv. syringae Defective in Syringomycin Production. Appl Environ Microbiol. 1988 Jun;54(6):1345–1353. doi: 10.1128/aem.54.6.1345-1353.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]