Abstract
All picornaviruses have a protein, VPg, covalently linked to the 5′-ends of their genomes. Uridylylated VPg (VPg-pUpU) is thought to serve as the protein primer for RNA synthesis. VPg-pUpU can be produced in vitro by the viral polymerase, 3Dpol, in a reaction in which a single adenylate residue of a stem-loop structure, termed oriI, templates processive incorporation of UMP into VPg by using a “slide-back” mechanism. This reaction is greatly stimulated by viral precursor protein 3CD or its processed derivative, 3C; both contain RNA-binding and protease activities. We show that the 3C domain encodes specificity for oriI, and the 3D domain enhances the overall affinity for oriI. Thus, 3C(D) stimulation exhibits an RNA length dependence. By using a minimal system to evaluate the mechanism of VPg uridylylation, we show that the active complex contains polymerase, oriI, and 3C(D) at stoichiometry of 1:1:2. Dimerization of 3C(D) is supported by physical and structural data. Polymerase recruitment to and retention in this complex require a protein-protein interaction between the polymerase and 3C(D). Physical and functional data for this interaction are provided for three picornaviruses. VPg association with this complex is weak, suggesting that formation of a complex containing all necessary components of the reaction is rate-limiting for the reaction. We suggest that assembly of this complex in vivo would be facilitated by use of precursor proteins instead of processed proteins. These data provide a glimpse into the organization of the ribonucleoprotein complex that catalyzes this key step in picornavirus genome replication.
Picornaviruses are the etiologic agent of numerous diseases of medical and veterinary importance. Poliomyelitis, the common cold, summer flu, hepatitis, and foot-and-mouth disease can all be caused by picornaviruses (1). These viruses have a single-stranded RNA genome of positive polarity that is on the order of 7500 nt3 in length (1). A protein, VPg (virion protein genome-linked), is covalently linked to the 5′-end of the viral genome, the so-called plus-strand, and a poly(rA) tail is present at the 3′-end (1). Genome replication occurs in a process that uses the plus-strand as a template for minus-strand synthesis, which, in turn, is used as a template for production of an excess of plus-strands (2). Initiation of both plus- and minus-strand RNA synthesis is thought to be primed by a uridylylated form of VPg, VPg-pUpU (2).
Several years ago, Paul and Wimmer made the paradigm-shifting observation that an RNA stem-loop structure in the 2C-coding region of the poliovirus (PV) genome was capable of templating production of VPg-pUpU (3, 4) much more efficiently than the poly(rA) tail (5). Since this time, it has become clear that all picornaviruses appear to use a similar strategy for production of VP-pUpU (6–12). All picornaviruses have a cis-acting RNA element capable of templating the production of VPg-pUpU, although the position of this element in the genome varies. This element has been termed oriI (2). Although it is generally accepted that oriI is essential for genome replication (6–8, 13–15), some suggest that it is only required for production of primers for plus-strand synthesis (16–18), whereas others suggest that oriI-derived primers are required for both plus- and minus-strand synthesis (12).
The VPg uridylylation reaction can be mimicked in vitro by using purified components: the viral RNA-dependent RNA polymerase (3Dpol); the VPg peptide; (bio)synthetic oriI RNA; UTP; and Mg2+ or Mn2+ (3, 4, 6, 11, 12). The reaction is also stimulated by a viral precursor protein, 3CD. Protein 3CD has both RNA-binding and protease activities, but only the RNA-binding activity is required for stimulation of the VPg uridylylation reaction (3, 4). We have shown that the fully processed viral protein 3C also stimulates VPg uridylylation (19). The mechanism for 3C(D) stimulation is not known. Our previous studies of PV support a model in which 3C(D) binds to oriI and recruits polymerase to oriI (19). Moreover, we suggested that this recruitment was mediated by an interaction between the thumb subdomain of 3Dpol and some undefined subdomain of 3C (19). This model was based on the identification of 3Dpol derivatives that contained wild-type polymerase activity on primed RNA templates and retained basal VPg uridylylation activity that could not be stimulated by 3C(D) (19). Recently, the suggestion was made that our interpretation of a 3Dpol-3C(D) interaction could also be explained as an allosteric effect unique to the 3Dpol derivatives studied (20).
In this report, we describe the establishment of a minimal VPg uridylylation system for PV that we use to evaluate the composition, stoichiometry, and functional and structural organization of the active VPg uridylylation complex. Consistent with previous studies (19), PV polymerase is recruited to and retained in the VPg uridylylation complex by a direct, physical interaction with 3C(D) bound to oriI. This conclusion is the same for two other picornaviruses: coxsackievirus B3 (CVB3) and human rhinovirus type 14 (HRV14). Targeting of 3C(D) to oriI is mediated solely by the 3C subdomain, the 3D subdomain serves only to increase affinity on oriI and, in doing so, increases the observed specificity of 3CD relative to 3C when oriI is placed in the context of longer RNA sequence. Protein 3C(D) binds to oriI at a 2:1 stoichiometry; 3C dimerizes in solution in the presence and absence of oriI. One of the 3C subdomains of 3C(D) binds to oriI contacting the stem in a position near the loop that would facilitate appropriate positioning of the polymerase. 3C binds to both single- and double-stranded RNA. In the context of oriI, affinity for the two single strands of the stem individually is higher than that for the annealed stem, suggesting an isomerization step after 3C(D)-oriI complex formation. Finally, association of VPg with this complex is weak and is probably rate-limiting for uridylylation. This study provides the most complete view to date of assembly and organization of the picornavirus VPg uridylylation complex.
EXPERIMENTAL PROCEDURES
Materials
Deep Vent DNA polymerase and restriction enzymes were from New England Biolabs, Inc.; shrimp alkaline phosphatase was from USB; T4 DNA ligase was from Invitrogen; Difco-NZCYM was from BD Biosciences; QIAEX beads were from Qiagen; RNase A was from Sigma; Ultrapure UTP solution was from GE Healthcare; [α-32P]UTP (6000 Ci/mmol) was from PerkinElmer Life Sciences; synthetic PV and HRV14 VPg peptides were purchased from Alpha Diagnostic International (San Antonio, TX); synthetic CVB3 VPg was a gift from Willem Melchers; all other reagents and apparatuses were available through Fisher or VWR or as indicated.
Construction of Expression Plasmids
The CVB3 3C coding region was amplified by using oligonucleotides 1–4 (Table 1 lists all oligonucleotides used in this study; oligonucleotides were from Invitrogen or Integrated DNA Technologies, Inc.) to perform overlap-extension PCR using the Knowlton CVB3/H3 cDNA (21) as template. The 3C-coding region was cloned into the pET26Ub plasmid (22) using SacII and EcoRI sites to give the pET26Ub-CVB3-3C-C147G-CHIS plasmid. The HRV16 3C-coding region was amplified by using oligonucleotides 5–8 to perform overlap-extension PCR using the HRV16 cDNA (23) as template. The amplified fragment was cloned into the pET26Ub-CHIS plasmid (19) using the SacII and BamHI sites to give the pET26Ub-HRV16-3C-CHIS plasmid. Cloning of the CVB3 3D is described by van Ooij et al. (12); cloning of HRV14 3C and 3D will be described by Shen et al.4 PV 3C without a His tag was made by PCR amplification using oligonucleotides 17 and 18 and pET26Ub-3C-C147G-CHIS (19) as template. The amplified fragment was cloned into pET26Ub (22). DNA sequencing at the Penn State Nucleic Acid Facility was used to verify the integrity of the above clones.
TABLE 1.
Oligonucleotides used in this study
| Number | Name | Sequence |
|---|---|---|
| 1 | CVB3-3CD-SacII-f | 5′-GCG ACT AGT CCG CGG TGG AGG TCC TGC ATT TGA ATT TGC-3′ |
| 2 | CVB3-3C-Chis-EcoRI-r | 5′-GCG GAA TTC GCG GCC GCT TAC TAA TGG TGG TGA TGG TGG TGA CCA GAG GAT CCT TGT TCA TCA TTG AAA TAG TG-3′ |
| 3 | CVB3-3C-C147G-f | 5′-ACG AGA GCA GGT CAA GGT GGC GGA GTA CTC ATG-3′ |
| 4 | CVB3-3C-C147G-r | 5′-CAT GAG TAC TCC GCC ACC TTG ACC TGC TCT CGT-3′ |
| 5 | HRV16-3C-SacII-f | 5′-GCG CCG CGG TGG AGG TCC AGA AGA AGA ATT TGG AAT G-3′ |
| 6 | HRV16-3C-BamHI-r | 5′-GCG GGA TCC TTG TTG TTC AGT GAA GTA TGA TCT CAA TAG-3′ |
| 7 | HRV16-3C-C147G-f | 5′-ACA AAA TCT GGG TAT GGT GGT GGT GTG TTA TAC-3′ |
| 8 | HRV16-3C-C147G-r | 5′-GTA TAA CAC ACC ACC ACC ATA CCC AGA TTT TGT-3′ |
| 9 | BamHI-T7–51ntCRE-f | 5′-CGG GAT CCT AAT ACG ACT CAC TAT AGG GAC AAC TAC ATA CAG TTC A-3′ |
| 10 | EcoRI-NcoI-51ntCRE-r | 5′-GGA ATT CCC ATG GCA AAC ATA CTG GTT CAA T-3′ |
| 11 | BamHI-T7-CVB3/H3-CRE-f | 5′-CGG GAT CCT AAT ACG ACT CAC TAT AGG GAG TAA TTA CAT ACA GTT C-3′ |
| 12 | EcoRI-XhoI-CVB3/H3-CRE-r | 5′-GGC GAA TTC CTC GAG CAG ACA TAC AGG C-3′ |
| 13 | PV-3C-SacII-f | 5′-GCG GAA TTC CTC CGC GGT GGA GGA CCA GGG TTC GAT T-3′ |
| 14 | 3C-Authentic-Cterm-r | 5′-GCG GAA TTC GTT TAA ACT TAC TAT TGA CTC TGA GTG AAG TA-3′ |
Restriction sites are shown in boldface type; codons containing nucleotide changes are italicized. The T7 promoter is underlined.
Expression and Purification of 3C Proteins
All of the 3C proteins were expressed using the ubiquitin fusion system described previously for 3Dpol (22). BL21(DE3)pCG1 cells were transformed with either the pET26Ub-PV-3C-CHIS, pET26Ub-CVB3-3C-CHIS, pET26Ub-HRV14-3C-CHIS, or pET26Ub-HRV16-3C-CHIS plasmids and plated (10%) onto NZCYM plates containing kanamycin at 25 μg/ml (K25), chloramphenicol at 20 μg/ml (C20), and dextrose at 0.4%. Ten colonies were then used to seed 100 ml of NZCYM medium supplemented with K25, C20, and dextrose at 0.1%. The culture was grown at 37 °C until to an A600 of 1. Cells were chilled to 25 °C and induced by the addition of isopropyl-β-D-thiogalactopyranoside to a final concentration of 500 μM. Cells were grown for 4 h at 25 °C and harvested. Cell pellets were weighed and stored at −80 °C. Frozen cell pellets were thawed on ice and suspended in lysis buffer (100 mM potassium phosphate, pH 8.0, 20% glycerol, 10 mM 2-mercaptoethanol (β-ME), 5.6 μg/ml pepstatin A, 4 μg/ml leupeptin) at a concentration of 4 ml of lysis buffer/g of cell pellet. Cells were homogenized using a Dounce homogenizer; cells were lysed by passing through a French pressure cell at a pressure per square inch of 20,000. Phenylmethylsulfonyl fluoride and Nonidet P-40 were added after lysis to final concentrations of 2 mM and 0.1% (v/v), respectively. The lysates were clarified by centrifugation in a Beckman JA-30.50 Ti rotor for 30 min at 24,000 rpm at 4 °C. The clarified lysate was passed over Ni2+-nitrilotriacetic acid spin columns (Qiagen). The columns were equilibrated prior to loading and washed after loading with buffer A (50 mM HEPES, pH 7.5, 20% glycerol, 10 mM β-ME, and 0.1% Nonidet P-40) containing 500 mM NaCl according to the manufacturer’s protocol. Protein was eluted in two 100-μl fractions using buffer A containing 500 mM NaCl and 500 mM imidazole. Protein concentration was determined by using the Bio-Rad protein assay. Conductivities of the fractions were measured, and the fractions were then aliquoted and stored at −80 °C.
Purification of PV 3D and PV 3CD-His was described previously (19). Purification of CVB3 3D was described by van Ooij et al. (12); purification of HRV14 3D will be described by Shen et al.4; Purified PV 3C containing an authentic carboxyl terminus was prepared by Dr. Jamie Arnold as a part of another study.5
Cloning and Transcription of oriIs
The region corresponding to the 51-nt oriI presented in Fig. 3 was amplified from an expression vector encoding the 2C gene6 using oligonucleotides 9 and 10. The purified 51-nt oriI PCR product was linearized using NcoI, and a transcription reaction (40 mM HEPES, pH 7.5, 32 mM magnesium acetate, 40 mM dithiothreitol, 2 mM spermidine, 28 mM NTPs, 25 μg/ml linearized template, and 25 μg/ml T7 RNA polymerase) was performed at 37 °C for 2 h. Magnesium pyrophosphate was removed by centrifugation for 2 min. The supernatant was treated with RQ1DNase (1 unit/μg of template; Promega) for 30 min to remove the template. The RNA was spun through a Micropure-EZ centrifugal filter (Amicon Bioseparations; Millipore) to remove any protein from the RNA according to the manufacturer’s instructions. Two G25 Sephadex (Sigma) spin columns were then used to remove any free nucleotides. The RNA was precipitated with ammonium acetate and suspended in TE (10 mM Tris·HCl, pH 8.0, 1 mM EDTA), and RNA concentration was calculated by measuring absorbance at 260 nm. The extinction coefficient was calculated for the 51-nt oriI plus three guanosine residues that are transcribed at the 5′-end of oriI to be 0.627000 μM−1·cm−1 (24). These extra nucleotides do not alter the fold of the 51-nt oriI as predicted by the mfold RNA folding server (available on the World Wide Web at www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi) (25, 26).
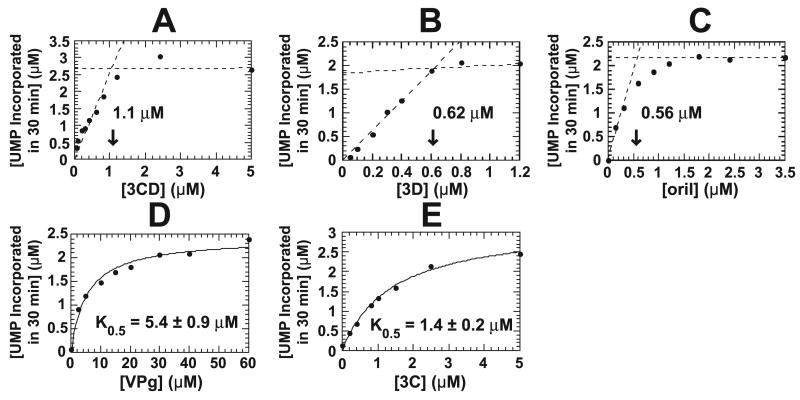
FIGURE 3. The active uridylylation complex contains one 3Dpol and two 3C(D) molecules per 29-nt RNA.
Shown is product formed in reactions in which protein and nucleic acid components were titrated individually. Reactions were performed in buffer (50 mM HEPES, pH 7.5, 5 mM manganese acetate, 10 mM β-ME, 10% glycerol, 10 μM UTP, and 0.04 μM [α-32P]UTP (6000 Ci/mmol)) at 30 °C for 30 min using 3D, 3CD, and 29-nt oriI at 1 μM and VPg at 50 μM unless they were being titrated. A, titration of 3CD. Lines defining the two phases of the curve were defined by linear regression, and the point of intersection was obtained algebraically. The intersection is 1.1 μM. B, titration of 3Dpol. The data were analyzed as described for 3CD titration. The intersection is 0.62 μM. C, titration of oriI. The data were analyzed as described for 3CD titration. The intersection is 0.56 μM. D, titration of VPg. The data were fit to a hyperbola. The K0.5 value is 5.4 ± 0.9 μM. E, titration of 3C. The data were fit to a hyperbola. The K0.5 value is 1.4 ± 0.2 μM.
The region corresponding to the CVB3 oriI was amplified using oligonucleotides 11 and 12 and the Knowlton CVB3/H3 cDNA as template. The purified oriI PCR product was linearized using XhoI and RNA transcribed as described above. The concentration was measured as described above using the calculated extinction coefficient (0.665700 μM−1·cm−1) that includes two guanosine residues at the 5′-end and a guanosine and adenosine at the 3′-end, a consequence of the XhoI digestion. These extra nucleotides do not alter the fold of the CVB3/H3 oriI as predicted by mfold.
The cloning of the HRV14 97-nt oriI is described elsewhere4; cloning of the PV 61-nt oriI was described previously (19). Transcription and determination of the concentration for the HRV14 97-nt oriI and the PV 61-nt oriI were done as described above.
RNA transcripts from the PV subgenomic replicon, pRLucRA (27, 28), were generated after linearization with ApaI. Transcription reactions, typically 20 μl, consisted of 350 mM HEPES, pH 7.5, 32 mM magnesium acetate, 40 mM dithiothreitol, 2 mM spermidine, 28 mM NTPs, 0.5 μg of template, and 0.5 μg of T7 RNA polymerase. Reactions were incubated at 37 °C for 3 h followed by removal of magnesium pyrophosphate. DNase treatment with RQ1DNase (Promega) was used to remove the template; lithium chloride precipitation of the RNA was used to remove unincorporated nucleotides. RNA concentration was calculated by measuring absorbance at 260 nm, assuming that an A260 of 1 was equivalent to 40 μg/ml. RNA transcripts of the PV 2C gene were generated after linearization of pET26Ub-2C plasmid with HindIII. Transcription reactions and RNA quantification were performed as described above.
The 29-, 22-, and 14-nt oriIs presented in Fig. 3 were chemically synthesized by Dharmacon, Inc. (Lafayette, CO). Each RNA was deprotected prior to use in the VPg uridylylation assays. Deprotection was done by suspension of the RNA in 500 mM acetic acid and incubation at 65 °C for 15 min, followed by the addition of an equal volume of 660 mM Tris, pH 8.0, and incubation at 65 °C for 15 min. Concentrations were measured as described above. The extinction coefficients used for each are 0.378500 μM−1·cm−1 (29 nt), 0.261100 μM−1·cm−1 (22 nt), and 0.166400 μM−1·cm−1 (14 nt).
Footprint Analysis Using an Iodine Cleavage Assay
oriI containing phosphorothioated ATP (ATPαS) and a 5′-OH to be used for footprinting by the iodine cleavage assay was obtained by performing a 1-ml transcription reaction (40 mM HEPES, pH 7.5, 32 mM magnesium acetate, 40 mM dithiothreitol, 2 mM spermidine, 12 mM NTPs, 0.3 mM ATPαS (10% of the total ATP concentration), 8 mM guanosine, 25 μg/ml linearized template, and 25 μg/ml T7 RNA polymerase) at 37 °C for 2 h. Magnesium pyrophosphate was removed by centrifugation for 2 min. The supernatant was treated with RQ1DNase (1 unit/μg of template; Promega) for 30 min to remove the template; two phenol/chloroform extractions followed by a chloroform extraction were performed to deproteinate the RNA. Next, the RNA was precipitated with ammonium acetate and washed with 70% ethanol, and the pellet was suspended in 50% formamide. This provided the starting material for gel purification. The entire volume was loaded onto a 10% acrylamide, 50% formamide gel (18 cm × 24 cm × 2 mm). The gel was run at 25 mA for ~4 h (the band corresponding to the full-length oriI had migrated to the middle of the gel by this time, as determined by the migration of bromphenol blue and xylene cyanol indicator dyes). The oriI band was excised from the gel by using UV-shadowing using a TLC plate with a fluorescent indicator (PEI Cellulose F; EM Science). The gel piece was cut into tiny squares and placed in an Elutrap electrophoresis chamber (Schleicher & Schuell). The eluted RNA was precipitated with ammonium acetate, washed with 70% ethanol, and suspended in TE. This was then passed over two Sephadex G-25 (Sigma) spin columns. The gel-purified RNA was radiolabeled using [γ-32P]ATP and polynucleotide kinase (New England Biolabs, Inc.). The labeled RNA was gel-purified again following the same procedure described above. RNA quality was assessed by 15% denaturing PAGE. RNA concentration was calculated by measuring absorbance at 260 nm. The extinction coefficient was calculated for the 61-nt oriI plus three guanidine residues that are transcribed at the 5′-end of oriI (0.749400 μM−1·cm−1) as described previously (19).
The 5′-end-labeled phosphorothioate oriI (3.3 μM) was incubated at 80 °C for 5 min in folding buffer (166 mM HEPES, pH 7.5, 66.6 mM NaCl) and then gradually cooled to room temperature for 1 h. At 55 °C, MgCl2 (16.6 mM) was added. The cleavage reaction proceeded as follows. The folded RNA was incubated with varying concentrations of PV 3C-His and incubated at 30 °C for 5 min. To initiate cleavage, 2 μl of 500 μM iodine dissolved in ethanol was added. After incubation at 30 °C for 2 min, the cleavage was quenched with 2 μl of 20 mM dithiothreitol. The final reaction mixture contained 50 mM HEPES, pH 7.5, 5 mM MgCl2, 20 mM NaCl, 1 μM RNA, 0–10 μM protein, 100 μM iodine, and 20% ethanol.
An alkaline hydrolysis ladder was generated by incubating the RNA in 50 mM sodium bicarbonate and 3 mM EDTA, pH 8.0, at 90 °C for 5 min. The samples were placed on ice to quench the cleavage reaction.
Prior to analysis, an equal volume of 100% formamide was added to each sample. The RNA was denatured at 90 °C for 5 min and placed on ice. The cleavage products were separated on a 15% denaturing polyacrylamide gel run at constant power of 90 W. The gel was visualized by using a Typhoon 8600 scanner in the storage phosphor mode and quantified by using ImageQuant software.
Fluorescence Polarization Assay
Experiments were performed using a Beacon fluorescence polarization system (GE Healthcare) as described previously (29). Briefly, PV 3C-His (0–100 μM) was mixed with the indicated 3′-fluorescein-labeled RNA (0.1 nM) in binding buffer (1 mM HEPES, pH 8.0). NaCl (10 mM) was included in the binding buffer when indicated in the figure legend. Protein-RNA complexes were incubated for 30 s at 25 °C. Binding of PV 3C was measured by the change in polarization. All steps were performed in reduced light.
VPg Uridylylation Assays
VPg uridylylation reactions were performed essentially as described previously (19) with slight modifications. Typically, reaction mixtures for each virus VPg uridylylation system contained 3Dpol, 3C-His, or 3CD-His as indicated, oriI template as described, and VPg peptide in reaction buffer (50 mM HEPES, pH 7.5, 10% glycerol, 5 mM magnesium or manganese acetate, 10 mM β-ME, 10 μM UTP, and 0.04 μM [α-32P]UTP (6000 Ci/mmol)). All reactions were adjusted to a final NaCl concentration of 30 mM. Concentrations of the components are indicated in each figure legend. All components were diluted to working concentrations immediately prior to use. Reactions were incubated at 30 °C for the indicated times and quenched with an equal volume of 100 mM EDTA in 75% formamide containing 0.05% bromphenol blue dye. Quenched reactions (5 μl) were analyzed by using Tris-Tricine SDS-polyacrylamide gel electrophoresis. Gels contained 15% acrylamide and 0.4% bisacrylamide. The cathode buffer (upper chamber) contained 0.1 M Tris base, 0.1 M Tricine, and 0.1% (w/v) SDS; the anode buffer contained 0.2 M Tris·Cl, pH 8.9. Gels were run at 80 W (for a 33 × 39-cm gel) for 2 h. Products were visualized by using a Typhoon 8600 scanner in the storage phosphor mode and quantified by using ImageQuant version 5.0 software.
Cross-linking by Using Glutaraldehyde and Western Blot Analysis
Cross-linking reactions were performed as follows. 3D (1 μM) and 3C-His (or 3C without a His tag or no 3C, as indicated) (1 μM) were incubated in reaction buffer (50 mM HEPES, pH 7.5, 10% glycerol, 5 mM magnesium acetate, 10 mM β-ME, 10 μM UTP, 60 μM ZnCl2) for 15 min at 30 °C. Glutaraldehyde was then added to a final concentration of 1 mM, and the reactions were incubated at 37 °C for 1 min. All reactions were adjusted to a final NaCl concentration of 30 mM. All components were diluted to working concentrations immediately prior to use. Glutaraldehyde, purchased from Sigma, was provided in 1-ml ampules; this was aliquoted and stored at −20 °C in small, single use volumes. Aliquots were used within 2 weeks to ensure maximal cross-linking efficiency of the glutaraldehyde, which is sensitive to oxidation. Reactions were then quenched by the addition of an equal volume of 2× SDS-PAGE dye (225 mM Tris, pH 6.8, 5% SDS, 50% glycerol, 5% β-mercaptoethanol, and 0.05% bromphenol blue) containing 100 mM glycine. Quenched reactions (5 μl) were analyzed by Western blot analysis. Proteins were separated by using 8 or 12.5% SDS-PAGE; transfer to nitrocellulose membrane (Osmonics, Inc. and/or GE Healthcare) was performed using the Genie transfer unit from Idea Scientific Company (Minneapolis, MN) for 50 min at 24 V in transfer buffer (25 mM Tris-glycine, 3 mM SDS, 20% (v/v) methanol (pH 8.3)); dry milk (5%, w/v) in TBS-T (20 mM Tris, pH 7.6, 137 mM NaCl, 0.1% Tween 20) was used to block the membrane (the blocking and probing steps were performed at room temperature for 1 h). Polyclonal antisera produced in rabbits against purified recombinant proteins PV 3D, CVB3/H3 3D, HRV14 3D, and PV 3C by Covance Research Products, Inc. (Denver, PA) were used to probe for the 3C-3D cross-linked heterodimer. Antisera were capable of detecting at least 100 pg of purified protein at 1:1000 dilutions in TBS-T (1:1000 for the anti-rabbit horseradish peroxidase secondary antibody).
PV 3D (PA 473) antiserum was used at a 1:80,000 dilution in TBS-T; CVB3 3D (PA 700) and HRV14 3D (PA 722) antisera were used at 1:8000 dilutions in TBS-T; PV 3C (PA 484) antiserum was used at a dilution of 1:40,000 in TBS-T. Either horseradish peroxidase- or alkaline phosphatase-conjugated goat anti-rabbit antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:2000 dilution in TBS-T was used as the secondary antibody. ECL (with horseradish peroxidase) and ECF (with alkaline phosphatase) detection systems (GE Healthcare) were used to visualize the Western blots. The Typhoon 8600 scanner was used in the fluorescence mode for the ECF system, and Eastman Kodak Co. BioMax MR film was used for the ECL system.
RESULTS
PV Protein 3CD Exhibits Enhanced Specificity for oriI When Compared with Protein 3C
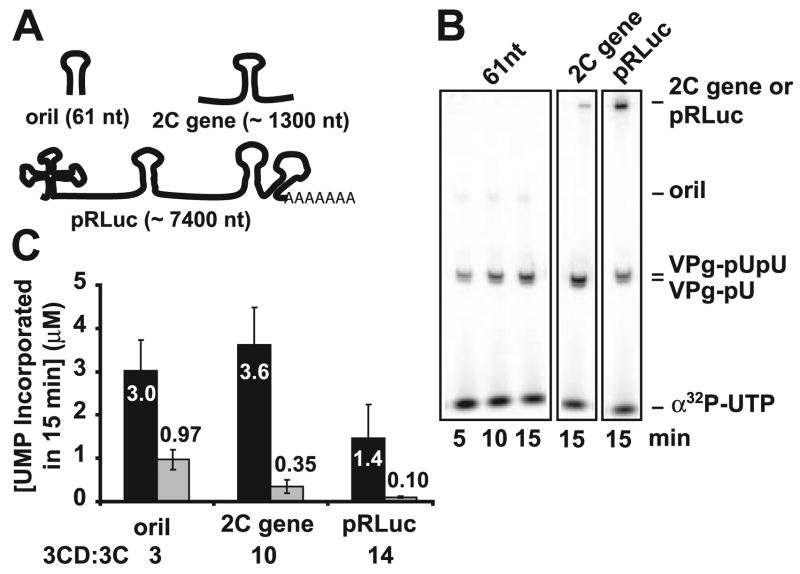
Our previous studies suggested that 3C could substitute for 3CD in VPg uridylylation reactions that employed a 61-nt oriI template (19). These data were reproduced here (Fig. 1). Reactions produced VPg-pU(pU) (Fig. 1B). The observed reaction rate is constant for at least 45 min (data not shown). Product formed in 15 min in reactions containing 3CD or 3C is shown in Fig. 1C. As before, only a 2-fold difference was observed when 3CD or 3C was present at equivalent concentrations. Increasing the concentration of 3C removed this difference, as reported previously (19). The capacity for 3C to substitute for 3CD may not occur in all cases. It is possible that oriI-containing templates employed may alter the capacity of 3C to substitute for 3CD. Because oriI exists in the context of the full-length genome in vivo, we asked whether the presence of additional “competitor” RNA supplied by increasing the length of the RNA would impact the stimulatory activity of 3CD or 3C. Two RNAs were employed. The first spanned the 2C-coding sequence in the PV genome (Fig. 1A); the second was the full-length subgenomic replicon RNA (27, 28). Both templates supported VPg uridylylation, yielding VPg-pUpU as the primary product (Fig. 1B). In contrast to results obtained by using the 61-nt oriI RNA, the ratio of 3CD-stimulated VPg uridylylation to 3C-stimulated VPg uridylylation increased as the RNA length was increased (Fig. 1C). This difference reflected a systematic decrease in the effectiveness of 3C stimulation by using the longer RNAs (Fig. 1C, gray bars). These results are consistent with 3C containing the primary determinant for stimulation of the VPg uridylylation reaction; however, the 3D subdomain of 3CD increases the affinity and/or specificity of 3C binding to oriI in the context of “competitor” RNA.
FIGURE 1. PV protein 3CD exhibits enhanced specificity for oriI when compared with protein 3C.
A, oriI-containing templates employed: 61-nt stem-loop, the 2C gene (~1300-nt RNA), and the PV subgenomic replicon (~6600-nt RNA). B, analysis of the products of VPg uridylylation. Shown is the phosphor image of a Tris-Tricine gel used to resolve the products of the VPg uridylylation reaction templated by the various oriI RNAs. Indicated are the positions of free UTP, the VPg-pU(pU), and the RNAs. C, RNA length dependence of 3C(D) stimulation. VPg uridylylation reactions were performed using 3Dpol (1 μM), 3C or 3CD (1 μM), and the indicated RNAs (1 μM for the 61-nt oriI and 0.1 μM for the 2C RNA and pRLuc) in reaction buffer (50 mM HEPES, pH 7.5, 5 mM magnesium acetate, 10 mM β-ME, 10% glycerol, 50 μM VPg, 10 μM UTP, and 0.04 μM [α-32P]UTP (6000 Ci/mmol)) at 30 °C. Shown is the amount of UMP incorporated in 15 min when stimulated by either PV 3CD (black bars) or PV 3C (gray bars). The average of at least two independent experiments is reported along with the corresponding S.D. values. The ratio of 3CD to 3C stimulation is indicated for each RNA.
A 29-nt oriI Element and Protein 3C Are Sufficient to Direct Specific and Efficient VPg Uridylylation in Vitro
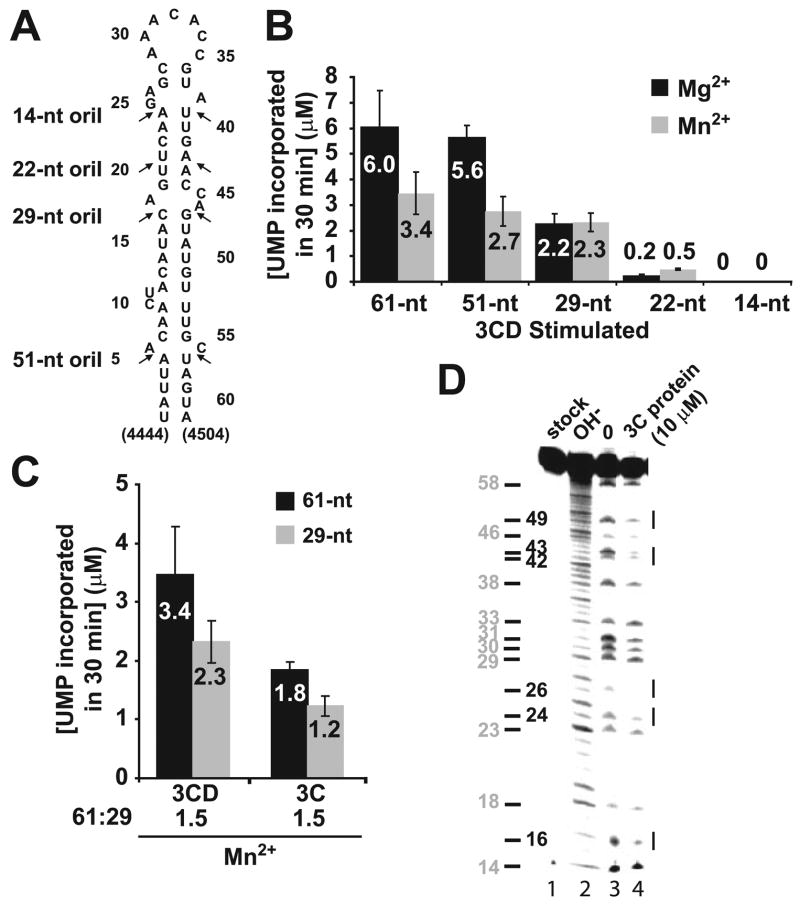
Given the capacity of competitor RNA to interfere with the assembly of the VPg uridylylation complex in vitro, interrogation of the kinetics and mechanism of the VPg uridylylation reaction would be facilitated by using a minimal oriI element. In order to define the minimal template for VPg uridylylation, the 61-nt oriI element (nucleotides 4444–4504 of the PV genome) was truncated, primarily at the bulges, to yield RNAs that were 51, 29, 22, or 14 nt in length (Fig. 2A). It should be noted that although only single, stem-loop structures having the same base pairings as the full-length RNA were predicted by mfold for the truncated oriIs, formation of unpredicted structures remains a possibility.
FIGURE 2. A 29-nt oriI element and protein 3C are sufficient to direct specific and efficient VPg uridylylation in vitro.
A, sites of oriI truncation. PV oriIs containing stems of various lengths as indicated were produced by using in vitro transcription (61 and 51 nt) or chemical synthesis (29, 22, and 14 nt). B, a 29-nt oriI is sufficient for VPg uridylylation in vitro. Shown is product formed in reactions containing oriI templates of varying length (1 μM) in the presence of Mg2+ (black bars) or Mn2+ (gray bars) as the cofactor for PV 3Dpol. Reactions were stimulated using 3CD (1 μM) in the presence of 3D (1 μM) and the indicated RNAs (1 μM). The average of at least two independent experiments is reported along with the corresponding S.D. values. C, 3C is sufficient for VPg uridylylation in vitro. Shown is product formed in reactions containing the 61-nt (black bars) or 29-nt oriI (gray bars) stimulated by using either 3CD or 3C. The average of at least two independent experiments is reported along with the corresponding S.D. values. D, footprint of 3C on oriI. Shown is iodine cleavage of AMPαS-substituted 61-nt oriI. Cleavage products were analyzed by denaturing PAGE. Shown is the phosphor image of the gel. Lane 1, untreated oriI; lane 2, oriI cleaved by alkaline hydrolysis; lane 3, iodine-treated oriI in the absence of 3C; lane 4, iodine-treated RNA in the presence of 3C (10 μM). Sites of 3C binding are indicated by lines to the right of the gel. The position of the adenosine residues is indicated to the left of the gel.
In the presence of Mg2+, truncation of oriI RNA to 29 nt caused an approximately 2-fold reduction in uridylylation activity (Fig. 2B). The 22-nt oriI RNA, however, caused a 15-fold reduction in activity (Fig. 2B). The 14-nt RNA was essentially inactive (Fig. 2B). The truncations will clearly have a large effect on the stability of the stem. Because Mn2+ binds to the phosphodiester backbone more tightly than Mg2+, stability of the stem should be enhanced by using Mn2+ as the divalent cation in the reaction (30). The use of Mn2+ causes a 3-fold reduction in the efficiency of uridylylation relative to Mg2+ (compare black bars with gray bars for 61-nt RNA in Fig. 2B) that may be due to a decrease in the nucleotidyl transfer rate of the polymerase (31). However, no change in uridylylation activity was observed in the presence of Mn2+ in going from 61 to 29 nt, suggesting that the decrease observed in the presence of Mg2+ was due to the stability of the stem.
In the presence of Mn2+, a condition in which the stability of the stem should be the same for the 61- and 29-nt RNAs, 3C functioned as well as 3CD, based upon the ratio of activity on the 61-nt RNA to that on the 29-nt RNA (Fig. 2C). The approximately 2-fold reduction in the observed activity of 3C relative to 3CD reflects the use of equivalent concentrations of both proteins and a reduced apparent dissociation constant of 3CD relative to 3C (discussed below).
Interestingly, footprint analysis by using iodine cleavage of phophorothioate-substituted oriI and 3C shows protection of oriI (i.e. 3C binding) mainly in the upper portion of the RNA stem, corresponding to the truncation made for the 29-nt oriI (compare the protected residues, labeled explicitly in Fig. 2D, with the oriI truncations shown in Fig. 2A). Together, these data show that a 29-nt oriI fragment and 3C are sufficient for uridylylation in vitro, consistent with genetic studies performed previously (32).
The Active Uridylylation Complex Contains One 3Dpol and Two 3C(D) Molecules per 29-nt RNA
Evaluation of the VPg uridylylation reaction by using the 29-nt oriI fragment in the presence of Mn2+ permits interrogation of the optimal stoichiometry of the components in the reaction. Intuitively, it is easy to imagine that one molecule of each component (RNA, polymerase, 3C(D) and VPg) would be required for assembly of an active complex. In this experiment, each component was titrated under conditions in which 3Dpol, 3CD, and/or oriI were present at a final concentration of 1 μM. VPg was present at a final concentration of 50 μM, since previous titrations with a 61-nt oriI RNA in the presence of Mg2+ showed a K0.5 value of 7 ± 2 μM (19).
Titrations of 3CD, 3D, and the 29-nt oriI element are shown in Fig. 3, A–C, respectively. Under the conditions employed, stoichiometric binding rather than hyperbolic binding was observed, suggesting that the dissociation constant for these components in the active complex is less than 0.2 μM. Maximum VPg-pU(pU) formation observed with the 3CD titration (3.1 μM) was higher than observed for 3D (2.1 μM) or oriI (2.2 μM), suggesting that more 3CD was required in the active uridylylation complex than either 3D or oriI (Fig. 3, A–C). The points of intersection were 1.1 μM for 3CD, 0.62 μM for 3D, and 0.56 μM for oriI (Fig. 3, A–C, indicated by the arrows). These data are consistent with a stoichiometry of 3CD/3D/oriI of 2:1:1. Stability of VPg within this complex was weak, since the K0.5 value was 5.4 ± 0.9 μM (Fig. 3D).
Titration of 3C showed hyperbolic binding, yielding a K0.5 value of 1.4 ± 0.2 μM (Fig. 3E). Therefore, 3C binding to oriI is at least 7-fold weaker than 3CD. Importantly, the extrapolated end point of the 3C reaction (3.2 ± 0.1 μM) was equivalent to that observed for 3CD in Fig. 3A. This result argues that the 3D domain of 3CD acts indirectly. If the 3D domain of 3CD participated directly, then the end point of the reaction containing 3C should never reach that of a reaction containing 3CD. Because the end point of 3C is equivalent to that of 3CD, it is likely that in the active 3C-3Dpol-oriI complex, two molecules of 3C are present.
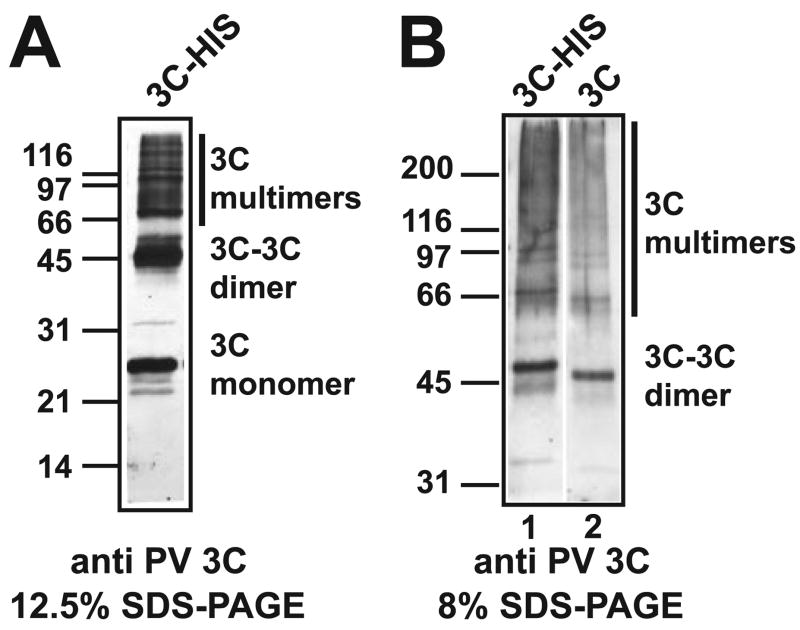
PV 3C Can Form Dimers
Whether or not 3C has the capacity to multimerize is not clear. Yeast two-hybrid data and cross-linking experiments performed in the past suggested that 3C does not multimerize (33). However, structural data for PV 3C reveal a very specific interface that predicts the formation of dimers in solution (34). In order to evaluate this possibility, we performed glutaraldehyde cross-linking experiments with purified PV 3C protein in vitro. The initial experiments employed a His-tagged derivative. Monomers, dimers, and multimeric forms of 3C were observed (Fig. 4A). In order to confirm that the observed cross-linking was not due to the His tag, authentic 3C protein was prepared, and the capacity for this protein to multimerize was evaluated and compared directly with the His-tagged 3C protein. As shown in Fig. 4B, authentic 3C formed clear dimers and multimers as well.
FIGURE 4. PV 3C can form dimers.
A, cross-linking of His-tagged 3C. PV 3C containing a carboxyl-terminal His tag (1 μM) was incubated in reaction buffer at 30 °C for 15 min. Glutaraldehyde (1 mM) was added, and the reaction was incubated at 37 °C for 1 min, quenched with 2× SDS-PAGE dye, and analyzed by using 12.5% SDS-PAGE followed by Western blot analysis. B, cross-linking of “authentic” 3C. The experiment was performed as described above. An 8% gel was employed to resolve the multimers. Lane 1, 3C-His; lane 2, “authentic” 3C.
3C Binds Single- and Double-stranded RNA
The footprint of 3C on oriI showed greater protection in the double-stranded rather than the single-stranded loop region of oriI (Fig. 2D). Because iodine cleaves phosphorothioates in both single- and double-stranded RNA equivalently, this technique does not permit conclusions to be reached regarding changes in nucleic acid structure induced by 3C binding. To date, only qualitative data have been obtained regarding 3C(D) binding to oriI (32). As a starting point, we performed fluorescence polarization experiments in order to determine if 3C could bind to single-stranded RNA. We determined the Kd values for PV 3C binding to 3′-fluorescein labeled oligo(rU) RNAs of varying length (rU5, rU7, rU8, rU9, rU10, rU12, rU15, and rU20). The data for rU10 in the presence of 0.1 or 10 mM NaCl are shown in Fig. 5A. PV 3C bound to all single-stranded RNAs evaluated; the minimal site size was in the 9–10-nt range (Table 2). 3C binding to rU10 was quite salt-sensitive, since the Kd value increased by 10-fold in going from 0.1 to 10 mM NaCl (Fig. 5A and Table 2). The salt sensitivity was not unique to oligo(rU) RNA, since a GC-containing, 9-nt RNA showed this effect as well (Table 2). 3C also bound to double-stranded RNA (Table 2). In this case, 3C binding to double-stranded RNA was on the order of 10-fold tighter than binding to single-stranded RNA of equivalent length (Table 2). 3C binding to double-stranded RNA was also salt-sensitive (Table 2).
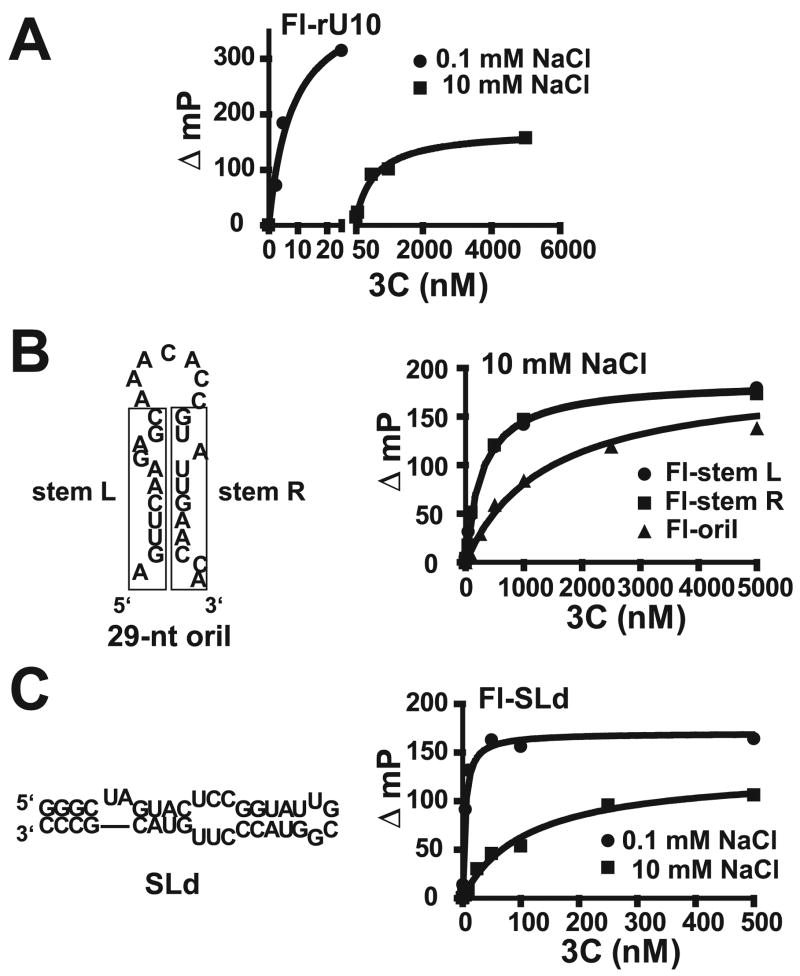
FIGURE 5. RNA binding by PV 3C measured by using fluorescence polarization.
A, 3C binds single-stranded RNA and is sensitive to salt concentration. 3′-Fluorescein-labeled rU10 (FL-rU10) RNA was titrated with 3C in the presence of 0.1 and 10 mM NaCl. Binding was detected as a change in fluorescence polarization (mP) in a Beacon instrument. The data were fit to a hyperbola. B, 3C binds to single-stranded oriI RNA fragments better than to intact oriI. The intact oriI employed is shown. The rectangles on the left and right define the RNA sequences of “stemL” and “stemR” single-stranded RNAs, respectively. All RNAs were labeled at the 3′-end with fluorescein. Experiments were performed as described above in the presence of 10 mM NaCl. C, 3C binding to SLd is tight. The 5′-end of PV RNA contains a cis-acting element (oriL) that contains a high affinity binding site for 3CD termed SLd. The sequence and secondary structure are shown on the left. SLd RNA was labeled at the 3′-end with fluorescein. Experiments were performed as described above in the presence of 0.1 and 10 mM NaCl.
TABLE 2.
3C RNA binding measured by using fluorescence polarization
| RNA | NaCl | Kd |
|---|---|---|
| mm | nm | |
| rU5 | 0.1 | 1300 ± 300 |
| rU7 | 0.1 | 120 ± 20 |
| rU8 | 0.1 | 43 ± 5 |
| rU9 | 0.1 | 19 ± 4 |
| rU10 | 0.1 | 6 ± 2 |
| 10 | 550 ± 100 | |
| rU12 | 0.1 | 9 ± 1 |
| rU15 | 0.1 | 9 ± 1 |
| rU20 | 0.1 | 7 ± 1 |
| GCCGCCCGG | 0.1 | 56 ± 10 |
| 10 | 4590 ± 1400 | |
| GCCGCCCGG CGGCGGGCC | 0.1 | 18 ± 4 |
| 10 | 480 ± 100 | |
| oriI (29 nt) | 0.1 | 14 ± 4 |
| 10 | 1400 ± 200 | |
| AGUUCAAGAGC (left stem) | 0.1 | 2 ± 1 |
| 10 | 290 ± 50 | |
| GUAUUGAACCA (right stem) | 0.1 | 2 ± 1 |
| 10 | 290 ± 50 | |
| SLd | 0.1 | 4 ± 2 |
| 10 | 100 ± 10 |
The capacity for 3C to bind to both single- and double-stranded RNA prevented us from reaching conclusions about binding to cis-acting elements like oriI (Fig. 5B) or stem-loop D (SLd) (Fig. 5C), both of which contain single- and double-stranded RNA. SLd is an extensively studied stem-loop structure residing in the 5′ cloverleaf and serves as a positive control (27, 35–42). Binding to oriI was salt-sensitive (Table 2). In the presence of 10 mM NaCl, the observed Kd value was 1.4 ± 0.2 μM (Table 2), a value identical to that inferred from the 3C concentration dependence of the VPg uridylylation reaction (Fig. 3E). However, this Kd value was 2-fold higher than observed for a random sequence of double-stranded RNA (Table 2). We separated the two strands of the oriI stem into the single-stranded components (indicated as “left stem” and “right stem” in Fig. 5B and Table 2). Interestingly, binding of 3C to each of these single-stranded RNAs was tighter than binding to the intact RNA (Fig. 5B and Table 2), suggesting that 3C may unwind the stem in some postbinding, isomerization step. In contrast, 3C binding to SLd was exceptionally tight, even in the presence of salt, consistent with the capacity of investigators to observe this interaction by gel mobility shift analysis among other methods (Fig. 5C and Table 2) (27, 35).
Functional Interaction between Picornavirus 3C and 3D Proteins
We suggested previously that an interaction between 3C and 3Dpol is required for VPg uridylylation (19). This conclusion was based on a study in which Arg residues 455 and 456 of 3Dpol were changed simultaneously to Ala, Ser, or Asp. VPg uridylylation catalyzed by these 3Dpol derivatives was not stimulated by 3CD to levels observed for WT 3Dpol. Trivial explanations for the observed uridylylation phenotype, such as differences in the linearity of reactions and K0.5 values for UTP, were ruled out (19). Importantly, none of these mutants supported PV replication in tissue culture (19).
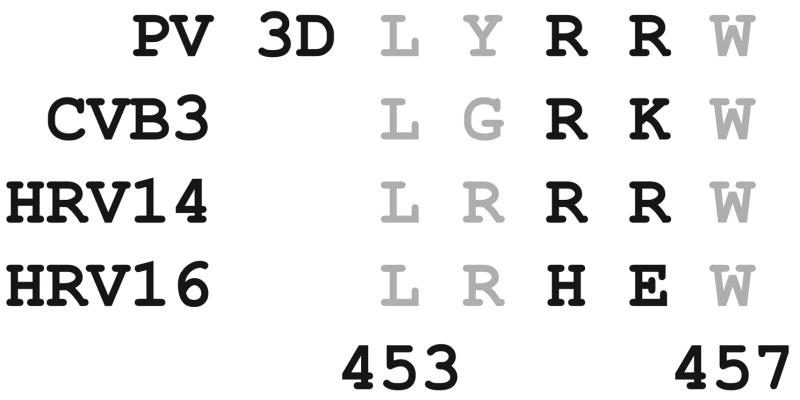
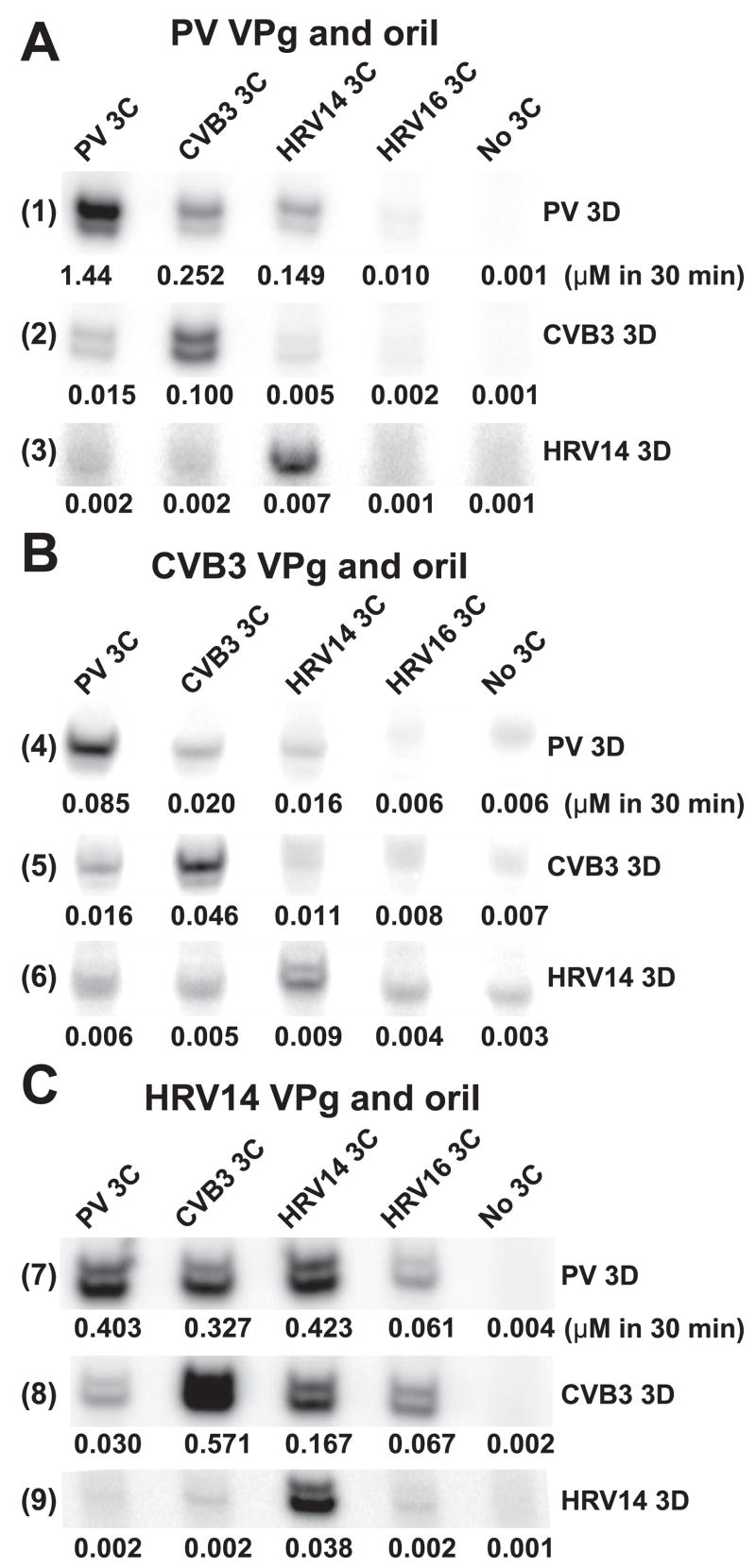
In order to obtain additional evidence for the existence of a 3C-3Dpol interaction and to probe the universality of this interaction among picornaviruses, we evaluated the 3Dpol interaction surface of all of the picornaviruses to identify viruses that encoded polymerases with changes at or near positions 455 and 456. If an interaction exists between 3C and 3D, then 3C proteins from viruses with a 3Dpol thumb substantially different from PV 3Dpol would not be expected to stimulate PV 3Dpol-catalyzed VPg uridylylation as well as PV 3C. As shown in Fig. 6, 3Dpol sequences from CVB3 and HRV14 and -16 show clear differences relative to PV in the region of the thumb spanning residues 453–457. We established in vitro uridylylation systems for CVB3 and HRV14. The sequences for VPg and oriI and the purified proteins employed are shown in Fig. 7. In all cases, maximal VPg uridylylation activity of the polymerase was observed when the cognate 3C was utilized regardless of the origin of the VPg and oriI (Fig. 8). For example, CVB3 3D was stimulated by CVB3 3C to a greater extent than by any of the other 3C proteins (Fig. 8B, row 5) even when the oriI and VPg were from PV (Fig. 8A, row 2) or from HRV14 (Fig. 8C, row 8). Similar observations were made for the HRV14 3D and 3C pair (Fig. 8, rows 3, 6, and 9) and the PV 3D and 3C pair (Fig. 8, rows 1, 4, and 7). We predicted that CVB3 and HRV14 3C proteins would substitute for PV 3C better than HRV16 3C, because the 3Dpol enzymes from CVB3 and HRV14 only contained one substantial difference relative to PV 3Dpol, whereas HRV16 3Dpol contained three differences. This prediction was supported by the data (Fig. 8). These data are all consistent with our previous findings that an important functional link exists between 3Dpol and 3C, perhaps a physical interaction.
FIGURE 6. Comparison of the thumb subdomains of the 3Dpol enzymes from PV, CVB3, HRV14, and HRV16.
The 3Dpols of PV, CVB3, HRV14, and HRV16 show variations in their thumb subdomains. Arg455 and Arg456 (shown in black) are hypothesized to be involved in an interaction with the surface of 3C (19).
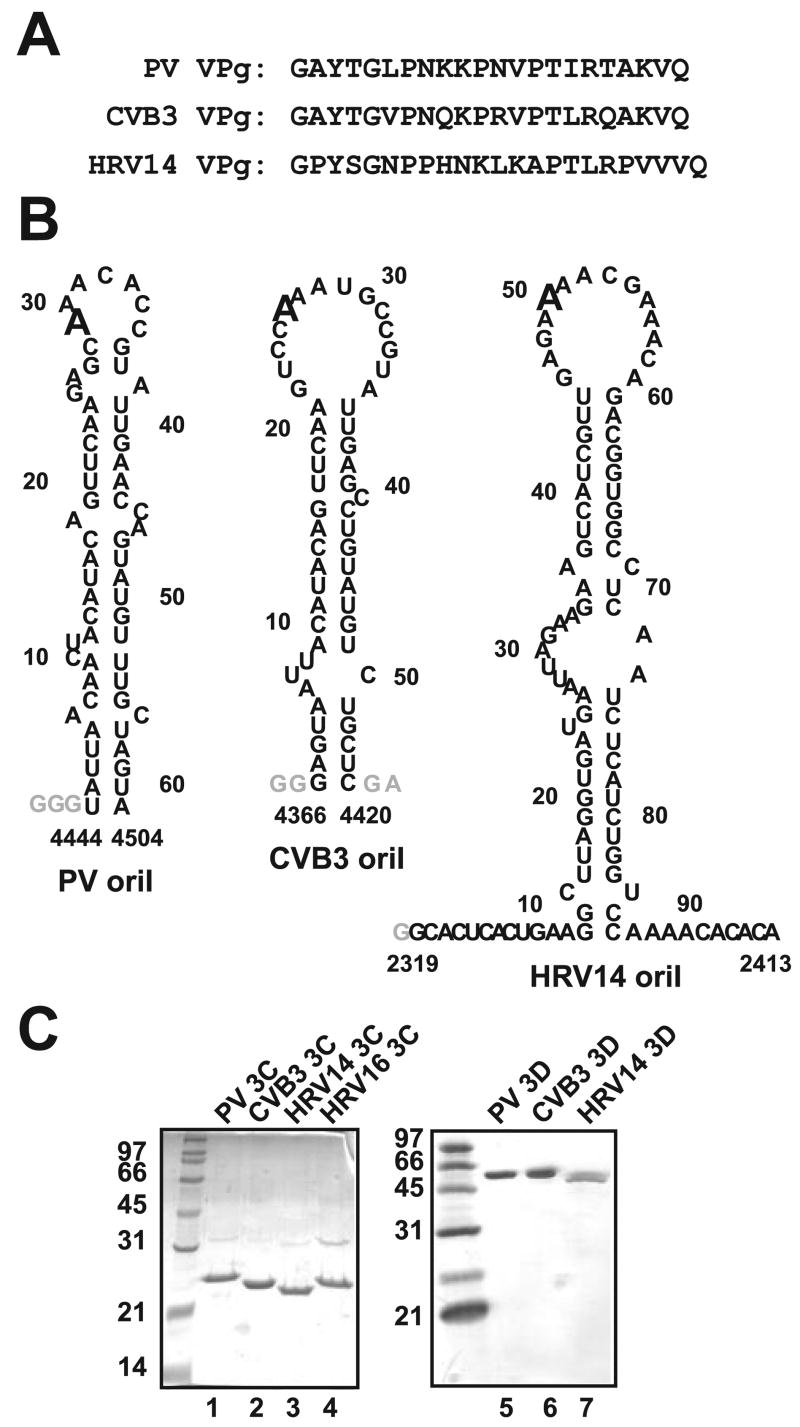
FIGURE 7. VPg uridylylation components employed in this study.
A, VPg peptides. Only five of the 22–23 amino acids are strictly conserved. One of these is Tyr-3, the nucleophile employed for the first nucleotidyl transfer reaction. B, oriIs. Although structural differences are apparent among the three stem-loop RNAs, the oriIs of enteroviruses and rhinoviruses have been shown to be functionally exchangeable (3, 8, 10, 32), suggesting that structure is not the primary determinant for recognition by the initiation complex. The templating “A” located in the loop is in an enlarged font. C, purified 3C and 3Dpol proteins. SDS-PAGE analysis of 1.5 μg of each 3C and 1 μg of each 3D is shown (left and right polyacrylamide gels, respectively).
FIGURE 8. Evidence for a 3C-3Dpol interaction: Cognate 3C-3Dpol pairs are more efficient at VPg uridylylation independent of the origin of the VPg and oriI.
VPg uridylylation reactions contained cognate VPg-oriI pairs from PV (A), CVB3 (B), or HRV14 (C). The source of 3C and 3Dpol is as indicated. Shown is a representative phosphor image of the VPg-pU(pU) products. Quantifications of UMP incorporation are indicated under each lane (μM VPg-pU(pU) in 30 min). The S.E. value of the measurement did not exceed 20%. Reactions were performed in buffer (50 mM HEPES, pH 7.5, 5 mM magnesium acetate, 10 mM β-ME, 10% glycerol, 10 μM UTP, and 0.04 μM [α-32P]UTP (6000 Ci/mmol)) at 30 °C for 30 min using 3D (1 μM for PV and CVB3, 10 μM for HRV14), 3C (1 μM), VPg (10 μM), and oriI (1 μM).
Physical Interaction between Picornavirus 3C and 3D Proteins
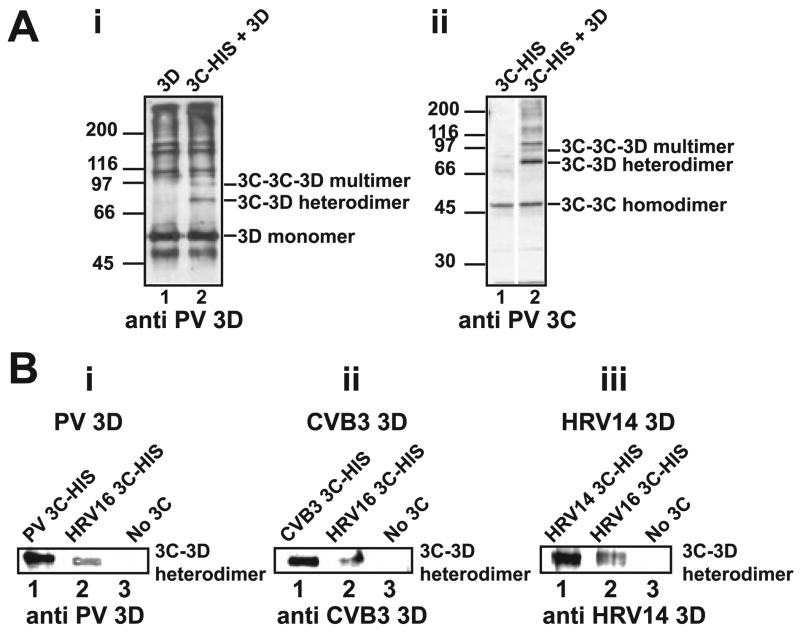
In order to demonstrate a physical interaction between PV 3C and 3Dpol proteins, we performed glutaraldehyde cross-linking experiments. After cross-linking, cross-linked species were resolved by SDS-PAGE and identified by Western blotting. Inter- and intramolecular cross-links were observed for purified PV 3Dpol (Fig. 9, Ai, lane 1). Cross-linking of PV 3Dpol (52 kDa) in the presence of PV 3C (21 kDa) produced two unique cross-linked bands that were recognized by the PV 3Dpol antibody and migrated at a position consistent with the molecular weight of cross-linked 3C-3Dpol and 3C-3C-3Dpol (Fig. 9, Ai, lane 2). By performing the analogous experiment using a PV 3C antibody (Fig. 9, Aii, lane 2), we demonstrated that these cross-linked species contained 3C. Cross-linking efficiency of PV 3Dpol to 3C proteins correlated with the capacity of the 3C proteins to stimulate VPg uridylylation (compare Fig. 9, Bi with Fig. 8A).
FIGURE 9. Evidence for a 3C-3Dpol interaction: Physical interaction between 3C-3Dpol pairs determined by cross-linking correlates with VPg uridylylation efficiency.
Physical interaction between 3C and 3Dpol proteins was probed by glutaraldehyde cross-linking experiments as described under “Experimental Procedures.” A, interaction between PV 3C and 3Dpol. i, cross-linking reactions contained 3Dpol alone (lane 1) or in the presence of 3C-His (lane 2). Cross-linked products were resolved by SDS-PAGE and identified by Western blotting using an antibody against PV 3D. ii, exactly as described in i using an antibody against PV 3C for detection. B, 3C-3Dpol cross-linking efficiency correlates with VPg uridylylation efficiency. A 3C-3Dpol heterodimer can be trapped by cross-linking for PV (i), CVB3 (ii), and HRV14 (iii). Experiments were performed as described above, and the cross-linked product was detected by using the indicated 3Dpol antibody. Shown is the band corresponding to the 3C-3Dpol cross-linked species. Lane 1, cognate 3C-His; lane 2, HRV16 3C-His; lane 3, no 3C. In each case, the use of HRV16 3C-His shows reduced cross-linking, consistent with the inability of this 3C protein to support robust uridylylation for these 3Dpol proteins (Fig. 8).
The 3C-3Dpol interaction observed for PV was also observed for CVB3 (Fig. 9, Bii, lane 1) and HRV14 (Fig. 9, Biii, lane 1). In addition, the efficiency of cross-linking for these proteins correlated with uridylylation efficiency (compare 9B, ii and iii, with Fig. 8, B and C). Together, these results provide compelling evidence for a physical interaction between a 3Dpol monomer and a 3C dimer that is required for VPg uridylylation.
DISCUSSION
Picornaviruses are positive-stranded RNA viruses with a single-stranded genome. This family of viruses contains numerous human and veterinary pathogens. Picornavirus genomic (plus-strand) and antigenomic (minus-strand) RNAs contain a protein (VPg) linked to the 5′-end. Attachment is thought to occur by using uridylylated VPg (VPg-pUpU) as primer for the virus-encoded RNA-dependent RNA polymerase (3Dpol), an enzyme that it is quite inefficient at initiation of RNA synthesis de novo (30). VPg-pUpU can be produced by the polymerase by using a genome-specific RNA stem-loop structure (oriI) (3, 4) or a poly(rA) tail (5). oriI-templated uridylylation of VPg is much more efficient than observed for poly(rA)-templated uridylylation reactions (3, 4). Substantial genetic and biochemical evidence exists supporting the use of oriI for VPg uridylylation during picornavirus genome replication (3, 4, 6, 8, 11, 12). However, there is some debate regarding the use of VPg-pUpU produced by using oriI for both plus- and minus-strand synthesis (12, 16–18).
Model for Assembly and Organization of the VPg Uridylylation Ribonucleoprotein Complex
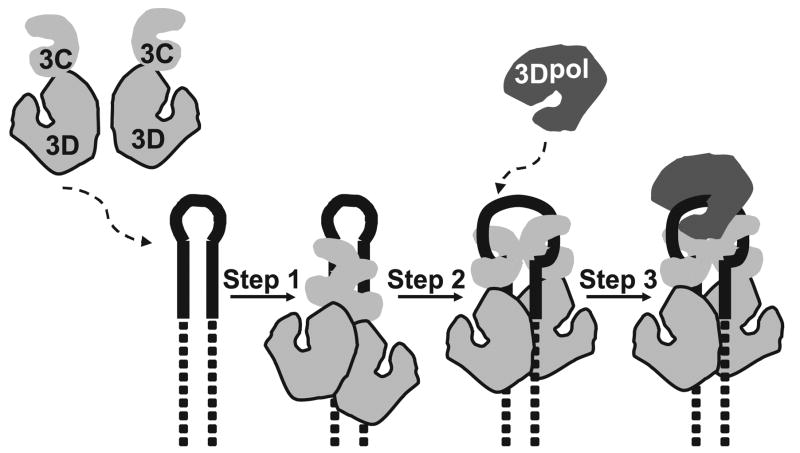
The VPg uridylylation reaction has been reconstituted in vitro from purified components (3, 4). However, the structural organization, corresponding assembly mechanism, and functional roles for some factors are largely unknown (2). We provided a glimpse into the structural organization several years ago by revealing an interaction between protein 3C(D) and 3Dpol that was required for VPg uridylylation, leading to the model proposed by Pathak et al. (19). The experiments reported here are consistent with the model for VPg uridylylation presented in Fig 10. A dimer of 3C(D) binds to the stem of oriI in the vicinity of the loop directed by 3C binding to the intact, double-stranded stem. This complex isomerizes to separate the strands of the oriI stem, opening the loop to facilitate binding of 3Dpol. 3Dpol is then recruited to and retained on the “open” loop by a protein-protein interaction with one or both subunits of the 3C subdomain of 3CD. Association of VPg with this complex is weak and could either be prebound to the open loop, enter with 3Dpol, or enter after.
FIGURE 10. Assembly and organization of the VPg ribonucleoprotein complex.
Step 1, two 3CD molecules bind to oriI with the 3C domains contacting the upper stem (solid lines) and the 3D domains contacting the lower stem (dashed lines). Step 2, the 3C dimer opens the RNA stem by forming a more stable interaction with single strands forming the stem. Step 3, 3Dpol is recruited to and retained in this complex by a physical interaction between the back of the thumb subdomain of 3Dpol and a surface of one or both 3C subdomains of 3CD.
Specificity Determinants of oriI
oriI has three domains: a loop and upper stem (solid lines in Fig. 10) and a lower stem (dotted lines in Fig. 10). The loop contains adenylate residues critical for templating uridylylation and facilitating processive uridylylation by using a slide-back mechanism (6, 9). The upper stem contains determinants for uridylylation, since mutations in this region reduce uridylylation efficiency (Fig. 2B) (8, 32). Loss- and gain-of-function mutations for uridylylation map to this region (10). An oriI chimera that contains the upper stem from HRV-14 and the lower stem from PV functions in the context of the HRV-14 genome (10). The reciprocal chimera does not support replication (10). These data are consistent with the upper stem providing the specificity for binding to proteins required for uridylylation and the lower stem serving a more structural role. For example, the number of mispairings leading to misfolded oriI structures would be reduced by adding the lower stem. In addition, the presentation of the stem in the context of the structured genome may be enhanced.
3C(D) Binding to oriI
Our capacity to establish a minimal system for uridylylation that was quite robust by employing only a 29-nt oriI and 3C (Fig. 2B) suggested that 3C interacted with the upper stem. This interaction could be demonstrated directly by footprinting (Fig. 2D). These data are consistent with filter binding data that correlated integrity of the upper stem with 3CD binding efficiency (32). Importantly, binding of 3C to the stem does not have a substantial affect on the accessibility of the loop (Fig. 2D). On minimal templates, 3C and 3CD function essentially equivalently (Fig. 1C); however, on longer templates, 3CD is superior (Fig. 1C). This difference probably reflects the capacity for 3CD to bind more tightly to oriI than 3C (Fig. 3). We conclude that 3C determines specificity and 3D enhances affinity. Consistent with this observation is the finding that mutations in 3C can change oriI specificity (10).
Titration experiments suggested that two molecules of 3CD were required per molecule of oriI and 3Dpol for maximal uridylylation (Fig. 3). Dimers of 3C could be observed in solution (Fig. 4). 3CD was not evaluated in this way, because it is well known that the 3D domain promotes multimerization in solution, confounding interpretation of any observed cross-linking (43). In support of 3C dimerization is the observation that PV 3C crystallizes as a dimer (34). The inability of the 3C dimer to be scored by using the yeast two-hybrid system or cross-linking in the past is unclear (33). We conclude that a dimer of 3C(D) binds to the upper stem of oriI.
Isomerization of the 3C(D)-oriI Complex
All of the picornavirus cis-acting elements are structured (e.g. Figs. 1A and 5C). However, these elements have single-stranded regions, double-stranded regions, and bulges as well as tertiary structures (2). As shown in Table 2, 3C binds to both single- and double-stranded RNA. The minimum site size for maximal binding to single-stranded RNA is 10 nt (Table 2). RNA binding affinity is quite salt-sensitive (Table 2) and correlates quite well with the salt sensitivity of the VPg uridylylation reaction (data not shown). Interestingly, binding of 3C to the intact oriI (Table 2) was weaker than binding to the single strands that form the stem (compare oriI with left stem and right stem in Table 2). We speculate that, after the initial binding event, 3C captures each of the individual strands, effectively opening the duplex and extending the loop. This aspect of the model is intellectually satisfying, because it provides a mechanism for loading the “loop” into the 3Dpol-template-binding site in an extended conformation. The solution structure for a minimal oriI from HRV-14 reveals no obvious mechanism for interaction of the oriI loop with 3Dpol (44). This aspect of the model must be explored more rigorously in the future.
Recruitment of 3Dpol to the Isomerized 3C(D)-oriI Complex and Retention Therein
We have obtained reverse genetic evidence for an interaction between 3Dpol and 3C that is required for VP uridylylation (19). By changing Arg residues at positions 455 and 456 on the back of the thumb of PV 3Dpol (Fig. 6) to Ala, Asp, or Ser, 3Dpol derivatives were constructed whose basal VPg uridylylation activity was unaffected relative to wild-type 3Dpol, but these derivatives could not be stimulated by 3C or 3CD at all (19). Sequence analysis shows that these two residues exhibit variability across the Picornavirus family (Fig. 6). In all cases evaluated, VPg uridylylation reactions performed by using cognate pairs of 3C and 3Dpol worked substantially better than those employing noncognate pairs (Fig. 8). The nature of the 3C-3Dpol pair was even more important than the origin of either VPg or oriI (Fig. 8). The sequence of the 3Dpol thumb (Fig. 6) was a major predictor of the capacity for a 3C to stimulate (Fig. 8), consistent with our hypothesis that the back of the thumb interacts with 3C (19). Interestingly, the ratio of VPg-pUpU to VPg-pU formed by the 3D and 3C pairs varied. The relationship between uridylylation processivity and genome replication efficiency is unclear but is currently under investigation.
The argument that a 3C-3Dpol interaction existed was supported by cross-linking (Fig. 9, A and B). Interestingly, in addition to observing a 3C-3Dpol species, a 3C-3C-3Dpol species was also observed (Fig. 9A, i and ii), further supporting a 3C dimer in the form of the oriI complex that recruits 3Dpol. The efficiency of 3C-3Dpol cross-linking correlates directly with the capacity of a 3C to stimulate 3Dpol-catalyzed VPg uridylylation (Fig. 9B, i–iii). It is worth noting that the crystal structure for PV 3CD has been solved (45). Interactions between 3C and 3D subdomains are observed that are required for VPg uridylylation. We conclude that both recruitment of 3Dpol to the VPg uridylylation complex and retention of 3Dpol therein are mediated by a direct, physical interaction between 3C and 3Dpol.
It has been suggested recently that our original interpretation of a 3C-3Dpol interaction should have been interpreted as allele-specific allosteric effect on the 3Dpol active site (20). These data refute this possibility; in this regard, the conclusions from that study are incorrect.
Association of VPg with the Uridylylation Complex Is Weak
VPg must associate with the final complex in order for VPg uridylylation to occur. VPg binds very weakly to the uridylylation complex (Fig. 3D). To what VPg binds is unclear. A crystal structure has been solved for a picornavirus 3Dpol-VPg complex; VPg binds to the RNA-binding site of 3Dpol in an extended conformation with numerous specific interactions. Therefore, it is possible that it is a 3Dpol-VPg complex that is recruited to oriI. We have also observed that binding of a VPg-3C fusion protein to oriI increases the stability by 10-fold relative to 3C alone, and this VPg-3C fusion protein can be uridylylated by 3Dpol. Therefore, it is possible that VPg binds to oriI in the open conformation followed by 3Dpol binding, a possibility favored by us that will be tested in the future.
Implications on Genome Replication in Vivo
Although the in vitro VPg uridylylation reaction described here predicts biological phenotypes (4, 8, 19), the reaction kinetics are too slow to support genome replication in vivo. Initiation events should not occur any slower than 0.1/s (46), a rate constant at least 50-fold faster than observed here (Fig. 3). Given the number of components that need to associate and dissociation constants in the micromolar range, formation of a complex containing all of the components is probably inefficient, and the use of stoichiometric amounts of VPg would probably make VPg association rate-limiting. Because VPg (also known as 3B), 3C(D), and 3Dpol originate from the same precursor protein (P3) in vivo, it is possible that P3 or 3BCD associates with oriI followed by processing concomitant with or after uridylylation, substantially increasing the overall efficiency of the process in vivo. Pathak (47) has shown that the PV VPg precursor, 3BC, serves as both the VPg donor and 3Dpol recruitment factor. Nayak et al. reached similar conclusions for foot-and-mouth disease virus (48). Production of P3 and various P3 processing intermediates is now feasible and will permit evaluation of these precursors as well (47).7
Acknowledgments
We thank Miaoqing Shen, Christian Castro, Mark J. M. van Ooij and Willem J. G. Melchers for providing us with reagents and/or communicating unpublished results. We thank Victoria Korneeva, Aniko Paul and Eric Smidansky for useful comments on the manuscript.
Footnotes
This work was supported, in part, by NIAID, National Institutes of Health, Grant AI053531 (to C. E. C.).
The abbreviations used are: nt, nucleotide(s); PV, poliovirus; CVB3, coxsackievirus B3; HRV14, human rhinovirus type 14; HRV16, human rhinovirus type 16; SLd, stem-loop d; β-ME, β-mercaptoethanol; ATPαS, adenosine 5′-O-(1-thiotriphosphate); Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
M. Shen, Q. Wang, Y. Yang, H. B. Pathak, J. J. Arnold, S. M. Lemon, and C. E. Cameron, manuscript in preparation.
J. J. Arnold and C. E. Cameron, unpublished results.
C. E. Cameron, unpublished results.
H. B. Pathak and C. E. Cameron, unpublished results.
References
- 1.Racaniello VR. In: Fields Virology. 4. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Vol. 1. Lippincott-Raven Publishers; Philadelphia, PA: 2001. pp. 685–722. [Google Scholar]
- 2.Paul AV. In: Molecular Biology of Picornaviruses. 1. Semler BL, Wimmer E, editors. American Society for Microbiology Press; Washington, D. C: 2002. pp. 227–246. [Google Scholar]
- 3.Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. J Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rieder E, Paul AV, Kim DW, van Boom JH, Wimmer E. J Virol. 2000;74:10371–10380. doi: 10.1128/jvi.74.22.10371-10380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul AV, van Boom JH, Filippov D, Wimmer E. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 6.Gerber K, Wimmer E, Paul AV. J Virol. 2001;75:10979–10990. doi: 10.1128/JVI.75.22.10979-10990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason PW, Bezborodova SV, Henry TM. J Virol. 2002;76:9686–9694. doi: 10.1128/JVI.76.19.9686-9694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Rijnbrand R, McKnight KL, Wimmer E, Paul A, Martin A, Lemon SM. J Virol. 2002;76:7485–7494. doi: 10.1128/JVI.76.15.7485-7494.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul AV, Yin J, Mugavero J, Rieder E, Liu Y, Wimmer E. J Biol Chem. 2003;278:43951–43960. doi: 10.1074/jbc.M307441200. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Rijnbrand R, Watowich S, Lemon SM. J Biol Chem. 2004;279:12659–12667. doi: 10.1074/jbc.M312992200. [DOI] [PubMed] [Google Scholar]
- 11.Nayak A, Goodfellow IG, Belsham GJ. J Virol. 2005;79:7698–7706. doi: 10.1128/JVI.79.12.7698-7706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Ooij MJ, Vogt DA, Paul A, Castro C, Kuijpers J, van Kuppeveld FJ, Cameron CE, Wimmer E, Andino R, Melchers WJ. J Gen Virol. 2006;87:103–113. doi: 10.1099/vir.0.81297-0. [DOI] [PubMed] [Google Scholar]
- 13.McKnight KL, Lemon SM. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobert PE, Escriou N, Ruelle J, Michiels T. Proc Natl Acad Sci U S A. 1999;96:11560–11565. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond JW, Barclay W, Evans DJ. J Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray KE, Barton DJ. J Virol. 2003;77:4739–4750. doi: 10.1128/JVI.77.8.4739-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morasco BJ, Sharma N, Parilla J, Flanegan JB. J Virol. 2003;77:5136–5144. doi: 10.1128/JVI.77.9.5136-5144.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodfellow IG, Polacek C, Andino R, Evans DJ. J Gen Virol. 2003;84:2359–2363. doi: 10.1099/vir.0.19132-0. [DOI] [PubMed] [Google Scholar]
- 19.Pathak HB, Ghosh SK, Roberts AW, Sharma SD, Yoder JD, Arnold JJ, Gohara DW, Barton DJ, Paul AV, Cameron CE. J Biol Chem. 2002;277:31551–31562. doi: 10.1074/jbc.M204408200. [DOI] [PubMed] [Google Scholar]
- 20.Boerner JE, Lyle JM, Daijogo S, Semler BL, Schultz SC, Kirkegaard K, Richards OC. J Virol. 2005;79:7803–7811. doi: 10.1128/JVI.79.12.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knowlton KU, Jeon ES, Berkley N, Wessely R, Huber S. J Virol. 1996;70:7811–7818. doi: 10.1128/jvi.70.11.7811-7818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gohara DW, Ha CS, Kumar S, Ghosh B, Arnold JJ, Wisniewski TJ, Cameron CE. Protein Expression Purif. 1999;17:128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
- 23.Cheney IW, Naim S, Shim JH, Reinhardt M, Pai B, Wu JZ, Hong Z, Zhong W. J Virol. 2003;77:7434–7443. doi: 10.1128/JVI.77.13.7434-7443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll SS, Benseler F, Olsen DB. Methods Enzymol. 1996;275:365–382. doi: 10.1016/s0076-6879(96)75022-x. [DOI] [PubMed] [Google Scholar]
- 25.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathews DH, Sabina J, Zuker M, Turner DH. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 27.Andino R, Rieckhof GE, Achacoso PL, Baltimore D. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herold J, Andino R. J Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. J Biol Chem. 2005;280:36417–36428. doi: 10.1074/jbc.M508175200. [DOI] [PubMed] [Google Scholar]
- 30.Arnold JJ, Ghosh SK, Cameron CE. J Biol Chem. 1999;274:37060–37069. doi: 10.1074/jbc.274.52.37060. [DOI] [PubMed] [Google Scholar]
- 31.Arnold JJ, Gohara DW, Cameron CE. Biochemistry. 2004;43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin J, Paul AV, Wimmer E, Rieder E. J Virol. 2003;77:5152–5166. doi: 10.1128/JVI.77.9.5152-5166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. J Virol. 1998;72:6732–6741. doi: 10.1128/jvi.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosimann SC, Cherney MM, Sia S, Plotch S, James MN. J Mol Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 35.Andino R, Rieckhof GE, Baltimore D. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 36.Dildine SL, Semler BL. J Virol. 1992;66:4364–4376. doi: 10.1128/jvi.66.7.4364-4376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohll JB, Percy N, Ley R, Evans DJ, Almond JW, Barclay WS. J Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 39.Rieder E, Xiang W, Paul A, Wimmer E. J Gen Virol. 2003;84:2203–2216. doi: 10.1099/vir.0.19013-0. [DOI] [PubMed] [Google Scholar]
- 40.Ohlenschlager O, Wohnert J, Bucci E, Seitz S, Hafner S, Ramachandran R, Zell R, Gorlach M. Structure. 2004;12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Du Z, Yu J, Ulyanov NB, Andino R, James TL. Biochemistry. 2004;43:11959–11972. doi: 10.1021/bi048973p. [DOI] [PubMed] [Google Scholar]
- 42.Headey SJ, Huang H, Claridge JK, Soares GA, Dutta K, Schwalbe M, Yang D, Pascal SM. RNA. 2007;13:351–360. doi: 10.1261/rna.313707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen JL, Long AM, Schultz SC. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 44.Thiviyanathan V, Yang Y, Kaluarachchi K, Rijnbrand R, Gorenstein DG, Lemon SM. Proc Natl Acad Sci U S A. 2004;101:12688–12693. doi: 10.1073/pnas.0403079101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, Cameron CE, Hogle JM. J Virol. 2007;81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnold JJ, Cameron CE. J Biol Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 47.Pathak HB. Ph.D. thesis. Pennsylvania State University; University Park, PA: 2006. Towards a Unified Model for Initiation of the First Step of Picornavirus Genome Replication; pp. 173–215. [Google Scholar]
- 48.Nayak A, Goodfellow IG, Woolaway KE, Birtley J, Curry S, Belsham GJ. J Virol. 2006;80:9865–9875. doi: 10.1128/JVI.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]