Abstract
Background
Although asthma is highly prevalent among certain Hispanic subgroups, genetic determinants of asthma and asthma‐related traits have not been conclusively identified in Hispanic populations. A study was undertaken to identify genomic regions containing susceptibility loci for pulmonary function and bronchodilator responsiveness (BDR) in Costa Ricans.
Methods
Eight extended pedigrees were ascertained through schoolchildren with asthma in the Central Valley of Costa Rica. Short tandem repeat (STR) markers were genotyped throughout the genome at an average spacing of 8.2 cM. Multipoint variance component linkage analyses of forced expiratory volume in 1 second (FEV1) and FEV1/ forced vital capacity (FVC; both pre‐bronchodilator and post‐bronchodilator) and BDR were performed in these eight families (pre‐bronchodilator spirometry, n = 640; post‐bronchodilator spirometry and BDR, n = 624). Nine additional STR markers were genotyped on chromosome 7. Secondary analyses were repeated after stratification by cigarette smoking.
Results
Among all subjects, the highest logarithm of the odds of linkage (LOD) score for FEV1 (post‐bronchodilator) was found on chromosome 7q34–35 (LOD = 2.45, including the additional markers). The highest LOD scores for FEV1/FVC (pre‐bronchodilator) and BDR were found on chromosomes 2q (LOD = 1.53) and 9p (LOD = 1.53), respectively. Among former and current smokers there was near‐significant evidence of linkage to FEV1/FVC (post‐bronchodilator) on chromosome 5p (LOD = 3.27) and suggestive evidence of linkage to FEV1 on chromosomes 3q (pre‐bronchodilator, LOD = 2.74) and 4q (post‐bronchodilator, LOD = 2.66).
Conclusions
In eight families of children with asthma in Costa Rica, there is suggestive evidence of linkage to FEV1 on chromosome 7q34–35. In these families, FEV1/FVC may be influenced by an interaction between cigarette smoking and a locus (loci) on chromosome 5p.
Spirometric measurements of pulmonary function are important markers of asthma severity and critical intermediate phenotypes for asthma research. Several groups have identified genomic regions linked to pulmonary function measurements (such as forced expiratory volume in 1 second (FEV1) and FEV1/ forced vital capacity (FVC) ratio) in families ascertained through probands with asthma.1,2,3 The prevalence and severity of asthma are markedly variable among different Hispanic populations living in the US and Latin America, and these differences may have an underlying genetic component.4 In the Collaborative Study on the Genetics of Asthma,5 genome‐wide linkage analysis of asthma and some of its intermediate phenotypes (eg, total serum IgE) were performed in Hispanic families from New Mexico. However, there has been no genome‐wide linkage analysis of pulmonary function phenotypes in any Hispanic population.
Bronchodilators, specifically β2‐adrenergic receptor agonists, are the most widely prescribed drugs in the treatment of asthma. There is a substantial inter‐individual variation in the response to inhaled β2‐agonists, and bronchodilator responsiveness (BDR) has been shown to aggregate in families,6 consistent with a genetic component to the variation in BDR. Although multiple studies have examined genetic linkage for airway responsiveness to bronchoconstrictor agents or pre‐bronchodilator and post‐bronchodilator spirometry measurements, a genome‐wide linkage analysis of BDR in families with asthma has not been published. The only reported linkage analysis for BDR was in families of probands with severe, early‐onset chronic obstructive disease (COPD).7
In this study, we performed genome‐wide linkage analyses of pulmonary function measurements and BDR in families of children with asthma in Costa Rica, a nation with a high prevalence of childhood asthma.8 Most Costa Ricans live in the Central Valley, where there is a genetically isolated population of predominantly mixed Spanish and Amerindian origin.9 Extensive genealogical records can be used to track the rapid expansion of this population from approximately 4000 founding individuals registered in the census of 169710 to about 2.85 million current residents of the Central Valley. The unique characteristics of the population of the Central Valley make it ideal for studies of the genetics of asthma and/or its intermediate phenotypes.
Methods
Study subjects
Seven probands were recruited through an ongoing study of children with asthma; an eighth proband was recruited from phase II of the International Study of Asthma and Allergies in Childhood in Costa Rica.11 Eligible probands were 6–12 years old and had physician‐diagnosed asthma, ⩾2 respiratory symptoms (cough, wheeze, or dyspnoea) or asthma attacks in the previous year, airway hyper‐responsiveness (provocative dose of methacholine causing a 20% decline in FEV1 (PD20) ⩽8.58 μmol), ⩾1 sibling with physician‐diagnosed asthma, and ⩾6 great‐grandparents born in the Central Valley of Costa Rica. All first‐degree and second‐degree relatives of the proband (⩾6 years old) were invited to participate. Families were further extended by including first‐degree relatives of individuals with asthma.
All adults gave written informed consent; parental consent was obtained for participating children, who also gave written assent. The study was approved by the Institutional Review Boards of the Hospital Nacional de Niños, San José, Costa Rica and Brigham and Women's Hospital (BWH), Boston, Massachusetts.
Phenotypic assessment
Each study participant completed a questionnaire and spirometry and gave a blood sample for DNA extraction. The study questionnaire was modified from that used by the Collaborative Study on the Genetics of Asthma12 and translated into Spanish. Two versions of the questionnaire were used: one for adolescents and adults (>12 years old) and one for children (⩽12 years old). Pack‐years of cigarette smoking were calculated by multiplying the number of years of smoking by the average number of cigarettes per day, divided by 20 to convert to packs. Children (⩽12 years old) were assumed to be non‐smokers.
Spirometry was performed using a Survey Tach Spirometer (Warren E Collins, Braintree, Massachusetts) according to the American Thoracic Society recommendations.13 Height was measured to the nearest half‐inch. Spirometry was performed while subjects were seated and wearing a noseclip. Up to eight attempts were made to obtain three acceptable flow‐volume loops. Participants were asked to refrain from use of short‐acting bronchodilator drugs for at least 4 hours before testing. Spirometry was repeated 15 min after the administration of 200 μg (two puffs) of albuterol through a spacer. The best FEV1 and FVC were selected for both pre‐bronchodilator and post‐bronchodilator spirometry.
Genotyping
DNA was extracted from blood samples using Puregene Kits (Gentra Systems). A panel of 380 short tandem repeat (STR) markers was genotyped by the Genome Quebec Innovation Centre using Applied Biosystems (Foster City, California) 3700 and 3730 analysers on 671 family members. This marker panel is a modified version of the Cooperative Human Linkage Center Human Screening set/V.614 that also includes selected Genethon markers.15 Marker locations were based on the deCODE map.16 Markers were located at an average spacing of 8.2 cM.
An additional nine STR markers on chromosome 7 were genotyped at Brigham and Women's Hospital: D7S2452, D7S2533, D7S500, D7S509, D7S495, D7S2505, D7S3044 and D7S2511. Primer sequences available in the National Center for Biotechnology Information's UniSTS database (http://www.ncbi.nlm.nih.gov/) were used to design assays. Fluorescent‐labelled and unlabelled primers were obtained from Invitrogen (Carlsbad, California) and Applied Biosystems. PCR product sizes were assessed on an Applied Biosystems 3100 instrument. GeneScan and GeneMapper V.3.7 software were used to assist with genotype determination, and calls were manually reviewed.
Pedigree relationships were assessed by Relpair17; four subjects that did not match reported relationships were excluded from analysis. Mendelian inconsistencies at individual markers for the remaining 667 individuals were assessed using PedCheck.18
Statistical analysis
BDR was defined as: 1) BDRbase = change in FEV1 as a percentage of the baseline FEV1; 2) BDRpred = change in FEV1 as a percentage of predicted FEV1; and 3) BDRabs = absolute change in FEV1 (ml).7,19 The three BDR measurements were normally distributed after log10 transformation.
Heritability estimation and multipoint linkage analysis of FEV1, FEV1/FVC and BDR measurements were performed by a variance component approach in Sequential Oligogenic Linkage Analysis Routines (SOLAR), which uses identity‐by‐descent sharing to estimate the additive genetic variance due to a chromosomal region.20 Results are expressed as logarithm of the odds of linkage (LOD) scores (logarithm [base 10] of the odds of linkage versus no linkage). Covariates included age, sex, height, weight, smoking status (ever vs never) and pack‐years of cigarette smoking, including quadratic terms for continuous variables. Significant covariates (p<0.05) were included in the linkage models. Only 19 subjects (3%) were using anti‐inflammatory drugs for asthma, so drug usage was not included as a covariate. Multipoint identity‐by‐descent matrices were estimated by a Markov‐Chain Monte Carlo algorithm implemented in the Loki program.21 Because of potential misclassification of chronic obstructive pulmonary disease (COPD) as asthma, secondary analyses were performed in non‐smokers only by removing the phenotype measurements of current and former smokers, and in smokers‐only by removing the phenotypes of non‐smokers.
Log10 transformed BDR and raw FEV1 measurements had acceptable kurtosis after adjustment for covariates in the final variance component models. Because of residual kurtosis (>3), FEV1/FVC measurements were analysed by the t‐distribution in SOLAR. Empirical p values for the multipoint LOD scores were estimated by comparing the observed LOD scores with the empirical distribution of LOD scores resulting from 100 000 simulations in SOLAR.
One thousand simulations were run in SOLAR to assess our statistical power to detect linkage (LOD scores ranging from ⩾1 to ⩾3) to a biallelic locus influencing a quantitative trait with heritability ranging from 10% to 25% in the Costa Rican pedigrees. These simulations assumed that the trait of interest was influenced by a single quantitative trait locus and that fully informative marker data were available for study subjects.
Results
Study subjects
Of the 667 members of the eight participating families, 640 and 624 had spirometric measurements of lung function before and after administration of albuterol, respectively (table 1). There was marked variability among participating families in family size, percentage of former and current smokers and percentage of patients with asthma. As expected in individuals with asthma and their relatives with and without asthma, average values of FEV1 and FEV1/FVC were within the normal range. However, post‐bronchodilator values were improved.
Table 1 Characteristics of families of children with asthma.
| Family | Individuals | Former and current smokers* n (%) | Asthma† n (%) | Mean (SD) FEV1, % predicted‡ | Mean (SD) FEV1/FVC, % predicted‡ | Mean (SD) bronchodilator response¶** | ||
|---|---|---|---|---|---|---|---|---|
| Pre‐BD§ | Post‐BD¶ | Pre‐BD§ | Post‐BD¶ | |||||
| 1 | 34 | 8 (23.5) | 12 (35.3) | 96.1 (16.8) | 101.1 (13.5) | 94.5 (8.7) | 97.2 (8.7) | 6.6 (12.1) |
| 2 | 18 | 4 (23.5) | 7 (38.9) | 103.9 (24.0) | 109.2 (27.4) | 97.8 (6.9) | 101.2 (7.0) | 6.1 (10.7) |
| 3 | 224 | 37 (16.5) | 56 (25.1) | 96.6 (14.8) | 100.3 (14.5) | 95.8 (7.0) | 98.7 (7.1) | 4.3 (7.2) |
| 4 | 97 | 17 (17.5) | 16 (16.5) | 102.6 (17.4) | 107.0 (17.0) | 96.9 (8.0) | 99.7 (7.8) | 5.0 (7.3) |
| 5 | 8 | 5 (62.5) | 2 (28.6) | 100.1 (13.2) | 102.5 (13.4) | 98.1 (7.7) | 101.4 (5.5) | 2.6 (6.8) |
| 6 | 107 | 10 (9.4) | 8 (7.5) | 99.7 (16.4) | 102.7 (16.0) | 98.5 (6.7) | 101.0 (5.7) | 3.1 (7.3) |
| 7 | 23 | 9 (39.1) | 5 (21.7) | 103.9 (12.4) | 108.8 (11.4) | 98.1 (5.6) | 101.4 (4.5) | 5.0 (5.3) |
| 8 | 129 | 12 (9.3) | 24 (18.6) | 96.8 (13.9) | 100.0 (13.1) | 96.9 (8.7) | 99.3 (7.5) | 3.4 (7.7) |
| All | 640 | 102 (16.0) | 130 (20.4) | 98.5 (15.8) | 102.3 (15.4) | 96.8 (7.6) | 99.5 (7.2) | 4.2 (7.7) |
FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; BD, bronchodilator.
*n = 638 individuals with complete data on smoking.
†Physician‐diagnosed asthma and wheezing within the past year. n = 637 subjects with complete data.
‡Based on prediction equations for Mexican‐Americans in Hankinson et al.22
§n = 640.
¶n = 624. Sixteen individuals did not have post‐bronchodilator spirometry.
**As a percentage of baseline FEV1 (BDRbase).
Analysis of FEV1
After adjustment for significant covariates (table 2), narrow‐sense heritability (h2N, the proportion of phenotypic variance explained by genetic factors) for pre‐bronchodilator FEV1 was 24.0% (SD 6.8%; p = 6×10−6). Heritability was similar for post‐bronchodilator measurements of FEV1 (h2N = 23.0% (SD 6.5%), p = 3×10−6).
Table 2 Genome‐wide linkage analysis for FEV1 and FEV1/FVC in all subjects, non‐smokers and smokers only.
| Phenotype | Bronchodilator | Subjects | Covariates | Chromosome | cM | LOD | p Value |
|---|---|---|---|---|---|---|---|
| FEV1 | Pre | All | Age, age2, ht, ht2, wt2, gender, smoker | 6 | 127 | 1.63 | 0.004 |
| Non‐smokers | Age, age2, ht, ht2, wt2, gender | 9 | 153 | 1.09 | 0.014 | ||
| Smokers | Age, age2, ht, gender | 3 | 241 | 2.74 | <0.001 | ||
| Post | All | Age, age2, ht, ht2, wt2, gender | 7 | 150 | 2.13 | 0.002 | |
| Non‐smokers | Age, age2, ht, ht2, wt2, gender | 7 | 150 | 1.73 | 0.003 | ||
| Smokers | Age, age2, ht, gender | 4 | 138 | 2.66 | <0.001 | ||
| FEV1/FVC | Pre | All | Age | 2 | 245 | 1.53 | 0.004 |
| Non‐smokers | Age, ht | 4 | 95 | 1.50 | 0.004 | ||
| Smokers | Age, age2 | 9 | 72 | 1.52 | 0.002 | ||
| Post | All | Age, age2, ht, wt | 7 | 74 | 1.40 | 0.007 | |
| Non‐smokers | Age, ht, wt | 18 | 117 | 1.46 | 0.006 | ||
| Smokers | Age, age2, packs | 5 | 49 | 3.27 | <0.001 |
FEV1, forced expiratory volume in one second; FVC, forced vital capacity; ht, height; smoker, ever smoker; packs, pack‐years; wt, weight.
Age2, ht2 and wt2 represent the quadratic terms for age, height and weight, respectively.
The maximum LOD score for each analysis is reported. Full results are available in supplementary tables 1 and 2.
Table 2 lists the highest LOD scores from the genome‐wide linkage analyses of FEV1 in all subjects, in non‐smokers and in smokers only; full results are available in Supplementary table 1 (available online at http://thorax.bmj.com/supplemental). In the genome‐wide linkage analysis of pre‐bronchodilator FEV1 in all subjects, the highest LOD score (1.63 at 127 cM) was found on chromosome 6q. Four additional genomic regions (chromosomes 4q, 6p, 7p and 16q) showed modest evidence of linkage (LOD >1) to pre‐bronchodilator FEV1. After excluding the phenotypic data of former and current smokers from the analysis, there was only modest evidence of linkage to pre‐bronchodilator FEV1 on chromosomes 4q and 9q. In smokers‐only, the highest LOD score was on chromosome 3q (LOD = 2.74 at 241 cM). Chromosomes 4q and 8p also showed suggestive evidence of linkage.
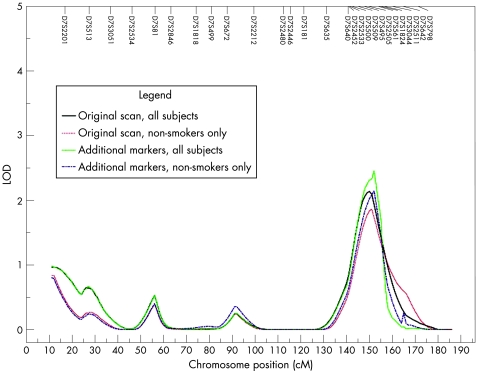
In the genome‐wide linkage analysis of post‐bronchodilator FEV1 in all subjects, there was suggestive evidence of linkage to chromosome 7q (LOD = 2.13 at 150 cM)23 and modest evidence of linkage (LOD >1) to chromosomes 1p, 2p, 15q and 16q. Nine additional STR markers were genotyped on chromosome 7q, the region with the highest LOD score in the analyses of all subjects. With the additional markers, the LOD score for post‐bronchodilator FEV1 increased to 2.45 (at 152 cM) in all subjects (fig 1). Among non‐smokers only, chromosomes 1p, 7q and 14q showed modest evidence of linkage to post‐bronchodilator FEV1. After inclusion of additional markers on chromosome 7q, there was suggestive evidence of linkage (LOD = 2.14) in non‐smokers. Chromosomes 2p, 4q (LOD = 2.66 at 138 cM), 8p and 14q all showed suggestive evidence of linkage in smokers only.
Figure 1 Linkage analysis on chromosome 7 for post‐bronchodilator FEV1 in all subjects and in non‐smokers only using short tandem repeat (STR) markers from the initial genome scan and including nine additional STR markers.
Analysis of FEV1/FVC
The estimated heritability was 19.9% (SD 7.8%; p = 2×10−4) for pre‐bronchodilator values of FEV1/FVC ratio and 15.4% (SD 6.6%; p = 0.001) for post‐bronchodilator values. Table 2 and Supplementary table 2 (available online at http://thorax.bmj.com/supplemental) show the results of the genome‐wide linkage analysis of FEV1/FVC. In the genome‐wide linkage analysis of pre‐bronchodilator FEV1/FVC in all subjects, the highest LOD score was on chromosome 2q (LOD = 1.53 at 245 cM). There was also modest evidence of linkage to pre‐bronchodilator FEV1/FVC on chromosomes 4q and 7q. Among non‐smokers there was modest evidence of linkage to pre‐bronchodilator FEV1/FVC on chromosomes 2p, 3q, 4q (LOD = 1.50 at 95 cM), 6p, 7q and 13q. In smokers only the highest LOD score was on chromosome 9q (LOD = 1.52 at 72 cM).
For post‐bronchodilator measurements of FEV1/FVC, the highest LOD score in all subjects was on chromosome 7p (LOD = 1.40 at 74 cM). There was also modest evidence of linkage to post‐bronchodilator FEV1/FVC on chromosomes 1q, 3p, 4q and 20q. Among non‐smokers there was modest evidence of linkage to post‐bronchodilator FEV1/FVC on chromosomes 2p, 3p, 4q, 5q, 6p, 7p and 18q. In smokers only the LOD score of 3.27 on chromosome 5p (49 cM) approached genome‐wide significant evidence of linkage.23
Analysis of bronchodilator responsiveness
The three measures of bronchodilator responsiveness (log10 transformed) were significantly heritable (BDRbase: h2N = 10.5% (SD 6.1%), p = 0.01; BDRpred: h2N = 10.4% (SD 6.1%), p = 0.01; BDRabs: h2N = 8.0% (SD 5.6%), p = 0.04), although the heritability estimates were lower than those of the pulmonary function measures. No covariates were significant in the variance component models of the three BDR outcomes in all subjects and in non‐smokers. In the genome‐wide linkage analysis of BDRbase (log10 transformed), the highest LOD score was found on chromosome 4q (LOD = 1.26 at 70 cM; table 3 and supplementary table 3 (available online at http://thorax.bmj.com/supplemental)). A LOD score >1 was found in this location in the analysis of BDRpred. In the analyses of BDRpred and BDRabs, the highest LOD scores were found on chromosome 9p at 49 cM (BDRpred: LOD = 1.25, BDRabs: LOD = 1.53). Despite the reduced sample size, the LOD scores were increased on chromosomes 4q and 9p in analyses restricted to non‐smokers only. In non‐smokers only, a region on chromosome 10p had LOD scores >1 for all three BDR phenotypes. In smokers, regions on chromosomes 12q and 16q had LOD scores >1 for all three BDR measures.
Table 3 Genome‐wide linkage analysis for bronchodilator responsiveness in all subjects, non‐smokers and smokers only.
| Phenotype* | Subjects | Covariates | Chromosome | cM | LOD | p Value |
|---|---|---|---|---|---|---|
| BDRbase | All | None | 4 | 70 | 1.26 | 0.008 |
| Non‐smokers | None | 10 | 31 | 1.59 | 0.002 | |
| Smokers | Gender | 16 | 66 | 1.31 | 0.019 | |
| BDRpred | All | None | 9 | 49 | 1.25 | 0.007 |
| Non‐smokers | None | 10 | 31 | 1.69 | 0.002 | |
| Smokers | Gender | 16 | 66 | 1.22 | 0.018 | |
| BDRabs | All | None | 9 | 49 | 1.53 | 0.003 |
| Non‐smokers | None | 9 | 49 | 1.72 | <0.001 | |
| Smokers | Gender | 16 | 66 | 1.60 | 0.012 |
The maximum LOD score for each analysis is reported. Full results are available in supplementary table 3.
*Definitions of BDR phenotypes7:
BDRbase = (FEV1post‐bronchodilator‐FEV1pre‐bronchodilator)/FEV1pre‐bronchodilator ×100%.
BDRpred = (FEV1post‐bronchodilator‐FEV1pre‐bronchodilator)/FEV1predicted ×100%.
BDRabs = (FEV1post‐bronchodilator‐FEV1pre‐bronchodilator).
All three BDR variables were log10 transformed for analysis.
Statistical power
Our statistical power to detect a LOD score ⩾1 was 42% for traits with h2N = 10% (eg, BDRbase) and 98% for traits with h2N = 25% (eg, pre‐bronchodilator FEV1). For LOD scores ⩾2, our power ranged from 13.6% for traits with h2N = 10% to 87% for traits with h2N = 25%. Finally, our power to detect a LOD score ⩾3 ranged from 4% for traits with h2N = 10% to 67% for traits with h2N = 25%.
Discussion
In families of schoolchildren with asthma in Costa Rica, we found significant genetic contributions (heritability) to inter‐individual variation in measures of pulmonary function and BDR. The heritability estimates of FEV1 were similar in magnitude to previous studies,24 but the heritabilities of FEV1/FVC and BDR were lower than have been reported.7 Because of low heritability, our study had limited statistical power to detect linkage to BDR. However, we had adequate statistical power to detect and found genome‐wide suggestive evidence of linkage23 to post‐bronchodilator FEV1, on chromosome 7q34–35, which was improved after inclusion of additional STR markers.
Although no significant or suggestive evidence of linkage was found in the other genome‐wide linkage analyses in all subjects, we uncovered potential regions of interest, including chromosome 9p for BDR and chromosome 2q for FEV1/FVC. Despite a small number of smokers and correspondingly limited power, the evidence for linkage for FEV1/FVC (post‐bronchodilator) on chromosome 5p approached genome‐wide significance.
Several authors have reported genome‐wide linkage analyses for spirometric measures of pulmonary function in families ascertained through probands with asthma; however, the present study is the only genome‐wide linkage analysis of FEV1 and FEV1/FVC in a Hispanic population. In a study of 2551 members of 533 families in China, Xu et al1 found the strongest evidence for linkage to FEV1 on chromosomes 10p and 22q. In 591 individuals in 202 Australian families, Ferreira et al2 showed suggestive evidence of linkage to FEV1 on chromosomes 5q, 8p, 12q, 17q and 20q and to FEV1/FVC on chromosomes 4q, 9q and 12q. Postma et al3 showed different regions of linkage to FEV1 in genome‐wide analyses of all subjects, smokers and non‐smokers among 1183 members of 200 Dutch families. They reported genome‐wide significant evidence of linkage to FEV1/FVC (both pre‐bronchodilator and post‐bronchodilator) on chromosome 2q.
There have been no previous reports of suggestive or significant evidence of linkage to FEV1 on chromosome 7q. However, the genome‐wide linkage analyses of pulmonary function described above were completed in populations of European and Asian descent. A unique aspect of our study is that we were able to recruit large extended pedigrees of children with asthma because of detailed genealogical records and low migration out of the relatively genetically isolated population of the Central Valley of Costa Rica. Our statistical power to detect linkage to lung function measures may have been increased by inclusion of extended pedigrees (which offer more power for linkage analysis of quantitative traits than sib‐pair studies with the same sample size)25 and by founder effects leading to relatively few susceptibility genes for asthma‐related traits in Costa Rica. On the other hand, genetic heterogeneity in determinants of lung function among the Spanish and Amerindian founders of the population of the Central Valley may have hindered our statistical power. Although the characteristics of the Costa Rican population may limit the generalisability of our results, it should be noted that the G protein coupled receptor‐154 (GPR154) was first identified as a potential asthma‐susceptibility gene in a genetically isolated population in Finland26 and then shown to be relevant in other European nations.27,28 Thus, some of our results may be relevant across ethnic groups and others may be more relevant to Costa Ricans and other Hispanic groups of predominant Spanish and Amerindian ancestry.
Genome‐wide linkage analyses of pulmonary function have also been performed in families from the general population24,29,30,31 and in families of probands with severe, early‐onset COPD.32 Several of the regions of interest (LOD ⩾1.5) in our study are similar to the findings in those studies (table 4), suggesting that some genomic regions are likely to contain genetic variants that influence pulmonary function in normal individuals, in patients with COPD and in those with asthma. Some of these regions may be relevant across different ethnic groups as well. In our analysis, the highest LOD score for FEV1/FVC was found on chromosome 2q. This region overlaps the linkage peaks for FEV1/FVC in families from the general population in Utah31 and in families from the Boston Early‐Onset COPD Study.32 The FEV1/FVC linkage found in Dutch asthma families is also located on chromosome 2q, but closer to the centromere.3
Table 4 Overlapping regions of linkage for pulmonary function phenotypes.
| Chromosome | Present study | Previous studies | |||
|---|---|---|---|---|---|
| Phenotype | LOD score (cM) | Author, year | Phenotype/Population | LOD score (cM) | |
| 2 | FEV1/FVC, pre‐BD | 1.53 (245) | Palmer, 20037 | FEV1/FVC, post‐BD/Early‐onset COPD | 4.42 (222) |
| Malhotra, 200331 | FEV1/FVC, pre‐BD/General population | 2.36 (216–251)* | |||
| Postma, 20053 | FEV1/FVC, pre‐BD/Asthma | 4.92 (195) | |||
| 4 | FEV1/FVC, pre‐BD | 1.50 (95) | Wilk, 200330 | FEV1/FVC, pre‐BD/General population | 1.64 (89.3) |
| Ferreira, 20052 | FEV1/FVC, pre‐BD/Asthma | 2.07† (94) | |||
| 7 | FEV1, pre‐BD | 1.53 (11) | Bouzigon, 200433 | FEV1 % predicted, pre‐BD/Asthma | 2.17 (17.7) |
Chromosomal regions with LOD score ⩾1.5 in the present study (all subjects) and in at least one previous study are shown.
*1−LOD drop support interval.
†−log10 (p Value).
The highest LOD score in any of the genome scans was found for FEV1/FVC (post‐bronchodilator) on chromosome 5p13 in an analysis limited to former and current smokers. Despite the limited sample size, the LOD score of 3.27 approached genome‐wide significance,23 possibly identifying a locus (or loci) for smoking‐related air flow obstruction in families with a genetic predisposition to asthma, consistent with the Dutch hypothesis, which proposes a common origin for asthma and COPD. Several cadherin genes (CDH‐6, 9 and 10) are located on chromosome 5p13. E‐cadherin (CDH1), another member of the cadherin family, is a cell adhesion molecule involved in epithelial permeability in allergic asthma.34 The importance of other cadherin genes in asthma and COPD is unknown.
Although other studies of asthma have analysed both pre‐bronchodilator and post‐bronchodilator spirometry, the only previous genome‐wide linkage analysis of BDR as a distinct phenotype is from the Boston Early‐Onset COPD Study.7 An analysis limited to chromosome 12q examined BDR in families from the Childhood Asthma Management Program Study35; the present study is the first reported genome‐wide linkage analysis of BDR in families of subjects with asthma. Similar to the Boston Early‐Onset COPD Study, we found significant heritability of BDR, although the heritability estimates in our asthma families (h2N range 8.0–10.5% for the three BDR measures) are lower than those found in the COPD families (h2N range 10.1–26.3%). As in our study, no significant linkage for BDR measurements was found in the Boston Early‐Onset COPD Study, although regions on chromosomes 3q and 4q had LOD scores of ⩾1.5.
The ability to detect linkage to measures of BDR may be limited by the day‐to‐day variability in BDR that is inherent in asthma. It is not clear which definition of BDR is most useful in genetic studies, so we used three commonly accepted measures.7 The fact that many of the regions with LOD scores >1 were similar across the analyses of two or all three BDR variables implies that these three definitions are likely to reflect the same underlying phenotype. The concordance of results is not perfect, as some regions were found in only one of the analyses of the BDR variables. The heritability estimates for the BDR measures were lower than those of the spirometric traits, although all heritabilities were statistically significant. This may also reduce the ability to detect significant linkage for BDR.
Additionally, as all subjects did not have post‐bronchodilator spirometry, the power in the analyses of post‐bronchodilator traits and BDR may be reduced. However, this power reduction should be minimal, as only 16 subjects did not complete the post‐bronchodilator measurements. Most patients with asthma (especially children) will have pulmonary function test values within the normal range; our study is no exception (table 1). The limited phenotype range may also limit power in genetic studies. Despite this, we were able to find suggestive evidence for linkage to FEV1.
Post‐bronchodilator spirometry is less likely to have substantial day‐to‐day variability in asthma, as the post‐bronchodilator values generally reflect underlying lung function.7 In our analysis, the strongest linkage evidence was for post‐bronchodilator FEV1. However, pre‐bronchodilator spirometry may be more variable within an individual subject and may be more reflective of asthma symptoms and severity. Because the analyses of pre‐bronchodilator and post‐bronchodilator spirometry may yield different information on lung development, asthma severity and asthma susceptibility, we chose to perform genome scans on both pre‐bronchodilator and post‐bronchodilator phenotypes.
Even though we did not find genome‐wide significant evidence of linkage for pulmonary function or BDR, the suggestive evidence of linkage to FEV1 on chromosome 7q warrants further investigation. Several plausible asthma candidate genes are located in this region, including the T cell receptor, β subunit (TRB@) and endothelial nitric oxide synthase (NOS3). Polymorphisms in NOS3 have been associated with asthma in some studies36,37 but not in others.38,39 As in any genetic analysis, our findings may be due to chance or to causal genetic variants. Although our results were adjusted for multiple testing in the setting of a genome‐wide linkage analysis of a single phenotype, we did not adjust for testing of multiple traits because of correlation among lung function phenotypes. Because current methods for association studies cannot be used in a small number of extended pedigrees, we plan to assess our findings further by testing for an association between variants in candidate genes on chromosome 7q34–35 and FEV1 in nuclear families of children with asthma in Costa Rica.
Supplementary tables 1, 2 and 3 available online at http://thorax.bmj.com/supplemental
Supplementary Material
Acknowledgements
We thank the participating families for their enthusiastic cooperation, the members of our field team in Costa Rica (Ligia Sanabria, Jenny Vega, Marvin Corrales, Adriana Gonzalez, Raquel Boza, Joaquín Acuña, Laura Rojas, Ana Castillo, Gabriela Ivankovich, Marcia Solano, Herminia Solano) and the staff at McGill University and the Genome Quebec Innovation Centre (Genevieve Geneau, Alexandre Belisle, Corinne Darmon‐Zwaig, Frederick Robidoux, David Roquis and Yannick Renaud) and the Channing Laboratory, Brigham and Women's Hospital (Benedict Bodota, Vimala Chompupong, and Elizabeth Bevilacqua).
Abbreviations
BDR - bronchodilator responsiveness
COPD - chronic obstructive pulmonary disease
FEV1 - forced expiratory volume in 1 second
FVC - forced vital capacity
LOD - logarithm of the odds of linkage
SOLAR - Sequential Oligogenic Linkage Analysis Routines
STR - short tandem repeat
Footnotes
Funding: This work was supported by US National Institutes of Health (NIH) grants HL66289, HL04370 and HL073373. Dr Hersh is supported by NIH grant HL080242 and a grant from the Alpha‐1 Foundation. The study sponsors had no role in study design, data collection, data analysis, manuscript preparation or submission.
Competing interests: Edwin K Silverman received grant support, consulting fees and honoraria from GlaxoSmithKline for studies of COPD genetics. He has received a speaker fee from Wyeth for a talk on COPD genetics and has also received honoraria from Bayer. None of the other authors declare any competing interests.
Supplementary tables 1, 2 and 3 available online at http://thorax.bmj.com/supplemental
References
- 1.Xu X, Fang Z, Wang B.et al A genomewide search for quantitative‐trait loci underlying asthma. Am J Hum Genet 2001691271–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira M A, O'Gorman L, Le Souef P.et al Robust estimation of experimentwise P values applied to a genome scan of multiple asthma traits identifies a new region of significant linkage on chromosome 20q13. Am J Hum Genet 2005771075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postma D S, Meyers D A, Jongepier H.et al Genomewide screen for pulmonary function in 200 families ascertained for asthma. Am J Respir Crit Care Med 2005172446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunninghake G M, Weiss S T, Celedon J C. Asthma in Hispanics. Am J Respir Crit Care Med 2006173143–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Collaborative Study on the Genetics of Asthma (CSGA) A genome‐wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 199715389–392. [DOI] [PubMed] [Google Scholar]
- 6.Niu T, Rogus J J, Chen C.et al Familial aggregation of bronchodilator response: a community‐based study. Am J Respir Crit Care Med 20001621833–1837. [DOI] [PubMed] [Google Scholar]
- 7.Palmer L J, Celedon J C, Chapman H A.et al Genome‐wide linkage analysis of bronchodilator responsiveness and post‐bronchodilator spirometric phenotypes in chronic obstructive pulmonary disease. Hum Mol Genet 2003121199–1210. [DOI] [PubMed] [Google Scholar]
- 8.The International Study of Asthma and Allergies in Childhood (ISAAC) Steering Committee Worldwide variations in the prevalence of asthma symptoms: the International Study of Asthma and Allergies in Childhood (ISAAC). Eur Respir J 199812315–335. [DOI] [PubMed] [Google Scholar]
- 9.Service S, DeYoung J, Karayiorgou M.et al Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome‐wide association studies. Nat Genet 200638556–560. [DOI] [PubMed] [Google Scholar]
- 10.Melendez C.Conquistadores y Pobladores: Origenes Historico‐Sociales de los Costarricenses. San Jose, Costa Rica: Editorial Universidad Estatal a Distancia, 1982
- 11.Celedon J C, Soto‐Quiros M E, Silverman E K.et al Risk factors for childhood asthma in Costa Rica. Chest 2001120785–790. [DOI] [PubMed] [Google Scholar]
- 12.Blumenthal M N, Banks‐Schlegel S, Bleecker E R.et al Collaborative studies on the genetics of asthma—National Heart, Lung and Blood Institute. Clin Exp Allergy 199525(Suppl 2)29–32. [DOI] [PubMed] [Google Scholar]
- 13.American Thoracic Society Standardization of spirometry, 1994 update. Am J Respir Crit Care Med 19951521107–1136. [DOI] [PubMed] [Google Scholar]
- 14.Dubovsky J, Sheffield V C, Duyk G M.et al Sets of short tandem repeat polymorphisms for efficient linkage screening of the human genome. Hum Mol Genet 19954449–452. [DOI] [PubMed] [Google Scholar]
- 15.Dib C, Faure S, Fizames C.et al A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 1996380152–154. [DOI] [PubMed] [Google Scholar]
- 16.Kong A, Gudbjartsson D F, Sainz J.et al A high‐resolution recombination map of the human genome. Nat Genet 200231241–247. [DOI] [PubMed] [Google Scholar]
- 17.Epstein M P, Duren W L, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet 2000671219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connell J R, Weeks D E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 199863259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Celedon J C, Speizer F E, Drazen J M.et al Bronchodilator responsiveness and serum total IgE levels in families of probands with severe early‐onset COPD. Eur Respir J 1999141009–1014. [DOI] [PubMed] [Google Scholar]
- 20.Almasy L, Blangero J. Multipoint quantitative‐trait linkage analysis in general pedigrees. Am J Hum Genet 1998621198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heath S C. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet 199761748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hankinson J L, Odencrantz J R, Fedan K B. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med 1999159179–187. [DOI] [PubMed] [Google Scholar]
- 23.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 199511241–247. [DOI] [PubMed] [Google Scholar]
- 24.Ober C, Abney M, McPeek M S. The genetic dissection of complex traits in a founder population. Am J Hum Genet 2001691068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams J T, Blangero J. Power of variance component linkage analysis to detect quantitative trait loci. Ann Hum Genet 199963545–563. [DOI] [PubMed] [Google Scholar]
- 26.Laitinen T, Polvi A, Rydman P.et al Characterization of a common susceptibility locus for asthma‐related traits. Science 2004304300–304. [DOI] [PubMed] [Google Scholar]
- 27.Melen E, Bruce S, Doekes G.et al Haplotypes of G protein‐coupled receptor 154 are associated with childhood allergy and asthma. Am J Respir Crit Care Med 20051711089–1095. [DOI] [PubMed] [Google Scholar]
- 28.Kormann M S, Carr D, Klopp N.et al G‐Protein‐coupled receptor polymorphisms are associated with asthma in a large German population. Am J Respir Crit Care Med 20051711358–1362. [DOI] [PubMed] [Google Scholar]
- 29.Joost O, Wilk J B, Cupples L A.et al Genetic loci influencing lung function: a genome‐wide scan in the Framingham Study. Am J Respir Crit Care Med 2002165795–799. [DOI] [PubMed] [Google Scholar]
- 30.Wilk J B, DeStefano A L, Arnett D K.et al A genome‐wide scan of pulmonary function measures in the National Heart, Lung, and Blood Institute Family Heart Study. Am J Respir Crit Care Med 20031671528–1533. [DOI] [PubMed] [Google Scholar]
- 31.Malhotra A, Peiffer A P, Ryujin D T.et al Further evidence for the role of genes on chromosome 2 and chromosome 5 in the inheritance of pulmonary function. Am J Respir Crit Care Med 2003168556–561. [DOI] [PubMed] [Google Scholar]
- 32.Silverman E K, Palmer L J, Mosley J D.et al Genomewide linkage analysis of quantitative spirometric phenotypes in severe early‐onset chronic obstructive pulmonary disease. Am J Hum Genet 2002701229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouzigon E, Dizier M H, Krahenbuhl C.et al Clustering patterns of LOD scores for asthma‐related phenotypes revealed by a genome‐wide screen in 295 French EGEA families. Hum Mol Genet 2004133103–3113. [DOI] [PubMed] [Google Scholar]
- 34.Goto Y, Uchida Y, Nomura A.et al Dislocation of E‐cadherin in the airway epithelium during an antigen‐induced asthmatic response. Am J Respir Cell Mol Biol 200023712–718. [DOI] [PubMed] [Google Scholar]
- 35.Raby B A, Silverman E K, Lazarus R.et al Chromosome 12q harbors multiple genetic loci related to asthma and asthma‐related phenotypes. Hum Mol Genet 2003121973–1979. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y C, Cheon K T, Lee H B.et al Gene polymorphisms of endothelial nitric oxide synthase and angiotensin‐converting enzyme in patients with asthma. Allergy 200055959–963. [DOI] [PubMed] [Google Scholar]
- 37.Yanamandra K, Boggs P B, Thurmon T F.et al Novel allele of the endothelial nitric oxide synthase gene polymorphism in Caucasian asthmatics. Biochem Biophys Res Commun 2005335545–549. [DOI] [PubMed] [Google Scholar]
- 38.Holla L I, Buckova D, Kuhrova V.et al Prevalence of endothelial nitric oxide synthase gene polymorphisms in patients with atopic asthma. Clin Exp Allergy 2002321193–1198. [DOI] [PubMed] [Google Scholar]
- 39.Leung T F, Liu E K, Tang N L.et al Nitric oxide synthase polymorphisms and asthma phenotypes in Chinese children. Clin Exp Allergy 2005351288–1294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.