Abstract
We report that interleukin 4 (IL-4) inhibits the propagation of non-syncytia-inducing and increases the propagation of syncytia-inducing HIV-1 isolates by two mechanisms. It differentially regulates the two major HIV-1 coreceptors, CCR5 and CXCR4, in human peripheral blood mononuclear cells, increasing CXCR4 and decreasing CCR5 expression in primary CD4(+) T-lymphocytes. In addition, IL-4 stimulates the expression of all HIV-1 isolates via a transcriptional activation mechanism. The combination of these effects results in increased propagation of CXCR4-using and inhibition of CCR5-using HIV-1 strains. IL-4 also activates HIV-1 expression in primary monocytes/macrophages but does not affect CCR5 expression. These results identify IL-4 as an important regulator of HIV-1 and suggest a critical role for this cytokine in the control of viral evolution and in the phenotypic switch from non-syncytia-inducing to syncytia-inducing, which leads to accelerated disease progression.
HIV-1 isolates have been classified as rapid/high or syncytia-inducing (SI) and slow/low or non-SI (NSI) (1–3) according to their biological phenotype. The main coreceptors for NSI and SI virus entry are the chemokine receptors CCR5 and CXCR4, respectively (4–9). NSI viruses are more likely to be transmitted and are found early after infection in most infected individuals. In contrast, SI viruses are usually present in the later stages of disease, and their appearance is associated with increased viral load and poor prognosis (10–13). A switch from NSI to SI has been demonstrated in a majority of HIV-1 infected individuals, but the mechanism regulating this process is poorly understood.
Although the loss of T lymphocytes with helper function is a central feature of HIV-1 infection, other signs of immune dysregulation, such as early loss of cellular responses to recall antigens, polyclonal B cell activation, and hypergammaglobulinemia are present in all HIV-1-infected individuals (14–16) and can be attributed to TH2 produced cytokines [interleukin (IL)-4, IL-5, and IL-13]. A switch to TH2 immune responses has been proposed to be important in AIDS pathogenesis (17), although a TH0 state or abnormal pattern of cytokine production have been also reported (18–20). Increased production of IL-4, the primary TH2-defining cytokine, has been demonstrated by intracellular staining (21, 22), in situ hybridization (23), and by increased frequency of T cell clones with TH2 phenotype from HIV-1-infected individuals (24–27). Higher virus replication correlated with increased IL-4 levels in lymph nodes from infected children (28).
To evaluate the role of TH2 cytokines on the propagation of NSI and SI HIV-1 isolates we studied their effects in peripheral blood mononuclear cells (PBMC) and in purified subpopulations under different conditions. We found that IL-4 has differential effects on the expression of CCR5 and CXCR4 and that it also stimulates intracellular pathways leading to increased HIV-1 production. The sum of these effects leads to the decrease of NSI and increase of SI expression in primary lymphocytes.
MATERIALS AND METHODS
Cells, Molecular Clones, and Viral Infections.
PBMC from healthy, HIV-1-seronegative blood donor were purified by Histopaque gradient centrifugation (Sigma). The purified cells were stimulated for 3 days with phytohemagglutinin (5 μg/ml).
CD4+ T lymphocytes were positively selected with magnetic beads coated with anti-human CD4 antibodies (Dynabeads M-450CD4, Dynal). The purified cells were cultured in the presence of IL-2 alone or with the addition of recombinant human IL-4 (1.5 units/ml, Boehringer Mannheim) every 2 days. Monocyte-derived macrophage cultures were established as described (29).
The tat(−) molecular clone was derived from HXB-2 and contains 10 point mutations introducing 7 stop codons throughout the first exon of the tat gene. The Rev(−)RRE(−) infectious molecular clone contains the previously described heterologous posttranscriptional control element CTE in the nef region (30). The nef(−) molecular clone was derived from pNL4–3 (31). pNL43GFP11 expresses the green fluorescent protein (GFP) mutant GFPsg11 (32, 33) as a fusion protein with the 24 N-terminal amino acids of Nef.
For viral infections, 107 phytohemagglutinin-stimulated PBMC were incubated at 37°C with 1 ml of cell-free supernatants from HIV-1-infected PBMC or transfected 293 cells. After a 2-hr incubation, the cells were washed twice with PBS and resuspended in RPMI 1640 medium B supplemented with IL-2 (5 units/ml, Boehringer Mannheim) at a density of 106 cells/ml. Each infected sample was split into two independent cultures, one of which was treated with human recombinant IL-4 (1.5 units/ml, Boehringer Mannheim). For blocking experiments, an anti-IL-4 neutralizing mAb (PharMingen) and the soluble form of the IL-4 receptor (IL-4R) were used at a concentration of 200 ng/ml and 100 ng/ml, respectively. Viral replication was monitored by measuring p24 gag in culture supernatants by a commercial ELISA kit (Cellular Products) at the indicated time points. The following clinical isolates were obtained through the AIDS Research and Reference Reagent Program: 92UG029 and 92UG031 (subtype A), 92BR004 and 92BR014 (subtype B), 93IN101 and 93IN904 (subtype C), 92UG035 and 92UG038 (subtype D), CMU08 and CMU10 (subtype E), and 93BR020 and 93BR029 (subtype F). The biological phenotype (SI/NSI) of all the clinical isolates was confirmed in our laboratory. Isolate CMU10 was catalogued as NSI but displayed SI phenotype (replication and syncytia formation in human T cell lines) in our assay. SI/NSI determination was performed by infection of Jurkat T cells and inspection of cultures up to 3 weeks after infection.
Immunostaining and Fluorescence-Activated Cell Sorter (FACS) Analysis.
For indirect immunofluorescence, 106 cells were washed once with PBS containing 1% human serum and 0.1% sodium azide and incubated on ice for 30 min with the mouse anti-CCR5 mAb 2D7 or the anti-CXCR4 mAb 12G5. After incubation, the cells were washed twice and resuspended in 100 μl of buffer containing fluorescein isothiocyanate-conjugated F(ab′)2 goat anti-mouse IgG (Boehringer Mannheim). After a 30-min incubation on ice, the cells were washed twice and analyzed by FACS (Becton Dickinson). Dead cells were excluded by propidium iodide staining. Phycoerythrin-conjugated mAbs anti-CD4, CD45RO, and CD28 (PharMingen) were used for double staining experiments. FACS analysis of NL43GFP11-infected samples was performed in cells fixed with 2% paraformaldehyde.
RNase Protection Assay.
Total RNA from CD4+ T lymphocytes or primary macrophages was extracted by the RNazol procedure. For CCR5 mRNA analysis, 5 μg of total RNA were hybridized to radiolabeled probes produced by an in vitro transcription kit (PharMingen), and digested with ribonuclease. Samples were electrophoresed on 5% acrylamide 7 M urea gels. Cellular transcripts glyceraldehyde-3-phosphate dehydrogenase and L32 were used as internal controls. For quantification of CCR5 mRNA expression, the amount of RNA per sample was normalized to the control glyceraldehyde-3-phosphate dehydrogenase or L32 by using the imagequant program.
RESULTS
IL-4 Stimulates SI and Inhibits NSI HIV-1 Variants in Primary Lymphocytes.
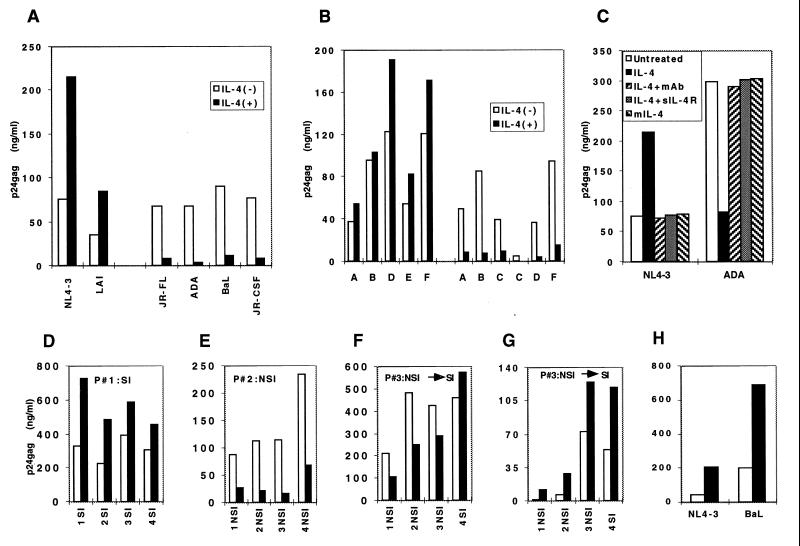
To study the effects of TH2 cytokines on HIV-1 propagation, we infected PBMC with a panel of HIV-1 strains including both SI and NSI viruses. IL-4 treatment of cells infected with either the SI strain NL4–3 or the NSI strain JR-FL demonstrated increased replication of NL4–3 and decreased replication of JR-FL (Fig. 1A). Increased viral production in the presence of IL-4 was also observed with the SI HIV-1 strain LAI (2- to 5-fold in different experiments), whereas expression of the NSI HIV-1 strains JR-CSF, BaL, and ADA was decreased by 5- to 10-fold. Therefore, these experiments demonstrated a dual effect of IL-4 on HIV-1 expression, depending on the biological phenotype of the virus. Similar results were obtained with several SI and NSI HIV-1 clinical isolates from clades A–F (Fig. 1B). In identical experiments, cells treated with either IL-5, IL-10, or IL-13 did not show any difference in the level of HIV-1 expression as compared with untreated cells (data not shown), indicating that the effect of IL-4 is not a common property of TH2 cytokines. The effects of IL-4 on HIV-1 replication were also similar in CD8(+)-depleted PBMC and in purified CD4+ T lymphocytes, indicating direct action of IL-4 on CD4+ cells in the absence of interactions with other cells or with soluble factors released by other cell types (data not shown). The use of an anti-IL-4 neutralizing antibody or of the soluble extracellular domain of the IL-4R abolished both the positive and the negative effects of IL-4 on HIV-1 expression (Fig. 1C). Furthermore, the use of an IL-4 mutant able to bind to the IL-4R but lacking biological activity (34) demonstrated that receptor signaling is required for the IL-4 effect on viral expression, excluding the possibility that IL-4R occupancy interferes with entry of the down-regulated HIV-1 strains (Fig. 1C).
Figure 1.
Dual effect of IL-4 in HIV-1 infection.(A) Human PBMC were infected with the indicated clones and cultured in the absence (□) or presence (■) of IL-4. Similar results were obtained in more than 10 independent experiments with well-characterized HIV-1 SI and NSI strains such as NL4–3, LAI, JR-FL, JR-CSF, BaL, ADA, or YU-2. (B) Infection with different clinical isolates from clades A–F. Bars represent p24 gag values in culture supernatants in the absence (□) or presence (■) of IL-4 at 7 days postinfection. (C) Neutralization of the IL-4 effects on the expression of the HIV-1 molecular clones NL4–3 (SI) and ADA (NSI) by anti-IL-4 neutralizing mAb and the soluble form of IL-4 receptor. A mutant IL-4 (mIL-4) able to bind to the IL-4 receptor but lacking biological activity did not affect HIV-1 expression. (D–F) Infection with sequential HIV-1 isolates from three patients with different clinical course. Bars represent p24 gag values in culture supernatants at 7 days postinfection. (G) Macrophage infection with clinical isolates from patient no. 3 in the presence or absence of IL-4. (H) Macrophage infection with either SI (NL4–3) or NSI (BaL) results in up-regulation of expression in the presence of IL-4.
To investigate whether primary HIV-1 isolates were regulated by IL-4 in a similar way, we examined sequential isolates obtained over a period of several years from HIV-1 infected individuals with different clinical course. Infection studies with HIV-1 isolates from three representative patients, a rapid progressor with SI virus, a nonprogressor with NSI and a patient that switched from NSI to SI are shown in Fig. 1 D–F, respectively. Sequential isolates from nonswitching individuals with either SI or NSI showed consistent up- or down-regulation by IL-4, respectively. In the third patient, switching correlated with a change in the pattern of regulation mediated by IL-4 (Fig. 1F). Similar regulation of HIV by IL-4 was found using 17 sequential HIV-1 isolates from three additional patients (data not shown).
IL-4 Stimulates All HIV-1 Variants in Primary Macrophages.
In addition to CD4(+) T lymphocytes, cells of the mononuclear phagocyte lineage are targets for HIV-1 infection and are believed to be important reservoirs for the maintenance of this persistent viral infection. To investigate whether IL-4 was able to regulate HIV-1 expression in human macrophages, we performed infection experiments with four sequential clinical isolates obtained from patient no. 3 described in Fig. 1F, that switched from NSI to SI. In contrast to the results in T lymphocytes, all clinical isolates were up-regulated by IL-4 in blood derived macrophages irrespective of their biological phenotype (Fig. 1G). Because SI HIV-1 clinical isolates are heterogeneous mixtures of viral variants able to use either CCR5 or CXCR4 as coreceptors, we extended our studies to HIV-1 strains restricted to CCR5 (BaL) or CXCR4 (NL4–3) coreceptor usage. These experiments established that the effect of IL-4 on HIV-infected primary macrophages is stimulatory in all cases (Fig. 1H).
IL-4 Down-Regulates CCR5 in Primary CD4+ T Lymphocytes.
Because the SI and NSI HIV-1 isolates use primarily the chemokine receptors CXCR4 and CCR5 for entry, respectively (4–9), we studied the effect of IL-4 on the expression of these receptors by FACS analysis of both PBMC and purified CD4+ T cells. In all cases, we measured a 2- to 5-fold decrease in the number of CCR5-expressing cells in the presence of IL-4 (Fig. 2 and Table 1). FACS analysis after double staining with anti-CCR5 antibodies and phosphatidylethanolamine-conjugated anti-CD45RO demonstrated, in agreement with a recent report (35), that CCR5 expression is mainly restricted to cells expressing CD45RO antigen (Fig. 2B). The CCR5 down-regulation induced by IL-4 was more prominent in CD4(+) than in CD4(−) cells (5- vs. 2-fold decrease, respectively, Fig. 2). No changes in the levels of CD3, CD4, CD45RO, CD45RA, or CD28 were detected in IL-4-treated cultures.
Figure 2.
CCR5 surface expression. FACS analysis of human PBMC doubly-stained with anti-CCR5 and either anti-CD4 (Left) or anti-CD45RO. Quadrants were set according to the staining of control cells. Numbers indicate the percentage of positive cells in each quadrant.
Table 1.
FACS analysis of CCR5 and CXCR4 expression in human PBMC of different healthy donors
| Donor | CCR5
|
CXCR4
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| IL-4 (−)
|
IL-4 (+)
|
IL-4 (−)
|
IL-4 (+)
|
||||||
| % Positive cells | MFI | % Positive cells | MFI | Donor | % Positive cells | MFI | % Positive cells | MFI | |
| 1 | 22 | 135 | 10 | 164 | 1 | 89 | 183 | 84 | 256 |
| 2 | 29 | 146 | 17 | 202 | 2 | 94 | 219 | 92 | 292 |
| 3 | 27 | 63 | 10 | 62 | 3 | 88 | 159 | 96 | 226 |
| 4 | 46 | 71 | 21 | 128 | 4 | 82 | 58 | 85 | 91 |
| 5 | 35 | 42 | 15 | 50 | |||||
| 6 | 55 | 49 | 26 | 57 | |||||
The assays were performed as in Fig. 2A. The mouse mAb 12G5(45) was used for CXCR4 detection. MFI, mean fluorescence intensity. The values shown were obtained from the complete phytohemagglutinen-stimulated PBMC samples.
To study the mechanism of CCR5 down-regulation, we analyzed the levels of expression of CCR5 mRNA in primary lymphocytes by an RNase protection assay (Fig. 3 A and B). These experiments showed a 2- to 4-fold down-regulation of CCR5 RNA after 3 to 7 days of IL-4 treatment. Similar results were obtained in quantitative reverse transcription–PCR experiments (data not shown). Therefore, down-regulation of CCR5 occurs at the level of gene expression. No down-regulation of CCR5 mRNA was observed in IL-4 treated macrophages, demonstrating that the IL-4 effect on CCR5 is cell type specific (Fig. 3 C and D).
Figure 3.
(A) RNase protection assay for CCR5 mRNA in CD4+ lymphocytes of four healthy donors (1–4) in the absence and presence of IL-4. Cellular transcripts glyceraldehyde-3-phosphate dehydrogenase and L32 were used as internal controls. Each RNA sample was analyzed in two independent experiments with the same results. (B) Quantification of CCR5 mRNA expression in A. The amount of RNA per sample was normalized to the control glyceraldehyde-3-phosphate dehydrogenase by using the imagequant program. Similar results were obtained with L32 as internal standard. (C) RNase protection assay for CCR5 mRNA in 14-day-old attached macrophages from a healthy donor in the absence of any cytokines (−) and in the presence of 3 ng/ml of either IL-4, IL-5, or IL-10, as indicated. (D) Quantification of CCR5 mRNA expression in C.
In contrast to our findings with CCR5, the number of CXCR4-positive cells did not change significantly in the IL-4-treated PBMC, whereas the overall expression of the CXCR4 receptor increased 35–40% at the single-cell level as demonstrated by the increased mean fluorescence intensity in the positive cell population (Table 1). Because the expression of CXCR4 in the absence of IL-4 treatment was already high, the impact of CXCR4 up-regulation on HIV-1 infection is unclear. Other tested cytokines (IL-5, IL-10, and IL-13) did not affect the expression of either CCR5 or CXCR4 under the same experimental conditions (data not shown).
IL-4 Activates HIV-1 Transcription.
The selective inhibition by IL-4 of HIV-1 strains with NSI phenotype was reminiscent of that described for the CC-chemokines RANTES, MIP-1α, and MIP-1β (36). To study any potential chemokine involvement, we measured the levels of CC-chemokines in culture supernatants of uninfected or JR-CSF infected human PBMC in the presence of IL-4. No significant changes were detected, excluding the possibility that NSI inhibition by IL-4 is caused by increased concentrations of the natural ligands for CCR5 (data not shown).
To investigate whether the IL-4 effects on HIV-1 are mediated exclusively through the regulation of the HIV-1 coreceptors, we infected PBMC with different HIV strains, and after 2 days treated the cells with 1 μM 3′-azido-3′-dioxythymidine. These culture conditions block viral spread and allow the study of IL-4 effects on postintegration steps of the viral life cycle. The results demonstrated that, under these conditions, IL-4 increased the expression of both SI and NSI isolates of HIV-1 (Table 2). We also transfected CD4(−) human 293 cells with DNA molecular clones of HIV-1, thus bypassing entry and early events in the viral life cycle. In agreement with the results of the 3′-azido-3′-dioxythymidine-treated PBMC, IL-4 treatment of cells transfected with either the SI NL4–3 or the NSI JR-CSF resulted in increased expression of both molecular clones (Fig. 4). These experiments strongly suggested an additional mechanism by which IL-4 was able to increase the expression of all HIV-1 isolates.
Table 2.
IL-4 effect on HIV-1 in the absence of viral propagation
| Virus | AZT | p24 (ng/ml)
|
Induction | |
|---|---|---|---|---|
| IL-4 (−) | IL-4 (+) | |||
| NL4-3 | (−) | 606 | 979 | 1.6× |
| (+) | 54 | 118 | 2.2× | |
| JR-FL | (−) | 473 | 243 | 0.5× |
| (+) | 3.6 | 5.2 | 1.4× | |
PHA-stimulated PBMC were infected with NL4-3 or JR-FL as described. At 2 days postinfection, the cells infected with each of the viruses were washed with PBS and split into four independent cultures. These cultures were kept untreated or were treated with IL-4 (1.5 units/ml). 3′-azido-3+-dioxythymidine (1 μM), or IL-4 + AZT 3′-azido-3′-dioxythymidine, respectively. p24gag concentrations in cell-free supernatants were measured at 7 days posttreatment. Similar results were obtained in two independent experiments.
Figure 4.
Activation of HIV-1 expression in 293 cells transfected with 5 μg of the indicated molecular clones. After overnight incubation, the cells were washed and cultured in DMEM medium in the presence (5 units/ml) or absence of recombinant human IL-4. Presented results were obtained in supernatants harvested 48 hours after washing the DNA. Similar results were obtained in three independent experiments.
To further analyze the mechanism of IL-4 up-regulation, we used HIV-1 molecular clones with mutations in the regulatory genes tat, rev. or nef. An IL-4-mediated increase in viral replication was observed in PBMC-infected with either the rev-independent or the nef(−) HIV-1 clones (Table 3). To study the effect of Tat, we transfected a tat(−) HIV-1 molecular clone into human 293 cells, which allow efficient Tat-independent HIV-1 expression. Addition of IL-4 failed to increase viral expression (Fig. 4). The supernatant from the transfected cells was used to infect PBMC. This resulted in low levels of viral expression, which did not increase upon IL-4 treatment (Table 3). These results indicated that the presence of Tat is required for the stimulatory effect of IL-4 on HIV-1 expression.
Table 3.
IL-4 effect on HIV-1 mutant molecular clones
| Virus | p24 (ng/ml)
|
Induction | |
|---|---|---|---|
| IL-4 (−) | IL-4 (+) | ||
| NL4-3 | 31.3 | 72.3 | 2.3× |
| tat(−) | 1.5 | 1.5 | 1× |
| Rev(−)RRE(−) | 5.2 | 10.7 | 2× |
| nef(−) | 27.1 | 59.6 | 2.2× |
Phytohemagglutinen-stimulated PBMC were infected with the different HIV-1 mutant molecular clones. Supernatants for cell-free infection were obtained from transfected 293 cells. Values represent p24gag concentrations in culture supernatants at 5 days postinfection. Similar results were obtained in three independent experiments using cells from different donors.
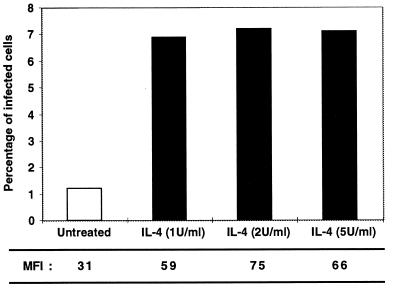
We obtained additional evidence of increased HIV-1 transcriptional activity in the presence of IL-4 by using NL43GFP11, an infectious NL4–3-derived molecular clone tagged with a GFP mutant in the nef region. FACS analysis of PBMC infected with NL43GFP11 demonstrated that IL-4 treatment increased both the proportion of infected cells and the intracellular concentration of GFP per cell (Fig. 5). These results suggest that IL-4 increases HIV-1 expression via a Rev-independent but Tat-dependent mechanism. The observed activation of 2-fold per infection cycle can result in high induction over several cycles of virus propagation.
Figure 5.
Stimulation of HIV-1 expression by IL-4. IL-4 increases GFP expression in PBMC infected with the molecular clone NL43GFP11. The mean fluorescence intensity per cell (MFI) for each sample is shown at the bottom. Dose response analysis of IL-4 showed that 0.1 and 0.2 units/ml did not show activation, and 0.5 units/ml showed partial activation of HIV-1 expression. The cells were analyzed 7 days after infection.
DISCUSSION
The ability of cytokines to influence HIV-1 propagation has been studied extensively (37). Despite this, the effects of IL-4 and the other TH2 cytokines on HIV-1 of different biological phenotypes in primary cells have not been fully elucidated. The present studies show that IL-4 affects HIV-1 expression in primary cells via multiple mechanisms. IL-4 activates directly viral expression of all HIV-1 isolates through a transcriptional mechanism. In addition, IL-4 down-regulates CCR5 expression in T cells, thus inhibiting infection by NSI HIV-1 isolates. In macrophages, where IL-4 treatment failed to down-regulate CCR5 expression, a stimulatory effect for both NSI and SI viruses was observed after treatment with this cytokine.
The importance of CCR5 in HIV-1 transmission has been demonstrated by epidemiological studies showing resistance to HIV-1 infection by individuals homozygous for a deletion in this gene (38–40). Therefore, it is anticipated that the down-modulation of CCR5 will have a strong impact on viral propagation. A correlation between low expression of CCR5 and reduced infectibility of T cells by HIV-1 isolates of NSI phenotype has been reported (41). The combination of the IL-4 effects on viral and cellular genes results in up-regulation of SI and down-regulation of NSI HIV-1 isolates. These results suggest that IL-4 may be involved in the switch from NSI to SI and is an important factor for viral evolution in vivo and AIDS pathogenesis. The increased susceptibility of IL-4-treated macrophages to infection with SI variants is an additional indication that IL-4 is an important factor in the emergence and maintenance of CXCR4-using HIV strains.
The appearance of SI variants that takes place in more than half of HIV-1-infected individuals is a sign of poor prognosis and correlates with faster CD4+ cell depletion and rapid disease progression (10–13). Increased IL-4 production in lymphatic tissues may transcriptionally activate the expression of all HIV-1 quasispecies. Because of CCR5 down-regulation, NSI viruses could be eventually counterselected. Combination of the two mechanisms should result in a viral population enriched in HIV-1 variants that are able to infect through coreceptors other than CCR5. Therapeutic interventions against virus entry by targeting the HIV-CCR5 interaction may soon be possible. In such interventions the possibility of an accelerated selection for SI variants, leading to a more aggressive disease course should be considered.
Because IL-4 plays a central role in determining the phenotype of naive T cells, and it is the principal inducer of TH2 state (42), our results also suggest that the induction of TH2 phenotype may correlate with CCR5 down-regulation. The absence of CCR5 expression among activated TH2 lymphocytes may result in protection of these cells against infection with NSI HIV-1 isolates and may be one reason for the increased frequency of T cell clones with TH2 phenotype derived from HIV-infected individuals. Indeed, recent data indicate that TH1 cell clones propagated in vitro do express CCR5 (43, 44), while TH2 cell clones express CCR5 less frequently. Such a pattern may protect TH2 cells from infection by NSI isolates. Our results suggest that IL-4 is directly involved in this differential receptor expression via intracellular mechanisms requiring signaling through the IL-4R.
Acknowledgments
We thank K. Noer for FACS analysis, B. K. Felber, W. Paul, C. Schalk-Hihi, I. Chen, D. Ho, S. Gartner, M. Popovic, R. Gallo, J. Hoxie, B. Levine, and C. June for materials and discussions, and A. Arthur for editing. Research was sponsored by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviatons: SI, syncytia-inducing; NSI, non-SI; IL, interleukin; IL-4R, IL-4 receptor; PBMC, peripheral blood mononuclear cells; GFP, green fluorescent protein; FACS, fluorescence-activated cell sorter.
References
- 1.Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo E M. Lancet. 1986;2:660–662. [PubMed] [Google Scholar]
- 2.Fenyo E M, Morfeldt-Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Asjo B. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tersmette M, Gruters R A, de Wolf F, de Goede R E, Lange J M, Schellekens P T, Goudsmit J, Huisman H G, Miedema F. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 5.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, et al. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 7.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 8.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 9.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 10.Richman D D, Bozzette S A. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 11.Koot M, Keet I P, Vos A H, de Goede R E, Roos M T, Coutinho R A, Miedema F, Schellekens P T, Tersmette M. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 12.Koot M, van ’t Wout A B, Kootstra N A, de Goede R E, Tersmette M, Schuitemaker H. J Infect Dis. 1996;173:349–354. doi: 10.1093/infdis/173.2.349. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson A, Parsmyr K, Sandstrom E, Fenyo E M, Albert J. J Clin Microbiol. 1994;32:364–370. doi: 10.1128/jcm.32.2.364-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pahwa S, Pahwa R, Good R A, Gallo R C, Saxinger C. Proc Natl Acad Sci USA. 1986;83:9124–9128. doi: 10.1073/pnas.83.23.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edelman A S, Zolla-Pazner S. FASEB J. 1989;3:22–30. doi: 10.1096/fasebj.3.1.2562947. [DOI] [PubMed] [Google Scholar]
- 16.Schnittman S M, Lane H C, Higgins S E, Folks T, Fauci A S. Science. 1986;233:1084–1086. doi: 10.1126/science.3016902. [DOI] [PubMed] [Google Scholar]
- 17.Clerici M, Shearer G M. Immunol Today. 1994;15:575–581. doi: 10.1016/0167-5699(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 18.Maggi E, Mazzetti M, Ravina A, Annunziato F, de Carli M, Piccinni M P, Manetti R, Carbonari M, Pesce A M, del Prete G, et al. Science. 1994;265:244–248. doi: 10.1126/science.8023142. [DOI] [PubMed] [Google Scholar]
- 19.Graziosi C, Pantaleo G, Gantt K R, Fortin J P, Demarest J F, Cohen O J, Sekaly R P, Fauci A S. Science. 1994;265:248–252. doi: 10.1126/science.8023143. [DOI] [PubMed] [Google Scholar]
- 20.Meyaard L, Hovenkamp E, Keet I P, Hooibrink B, de Jong I H, Otto S A, Miedema F. J Immunol. 1996;157:2712–2718. [PubMed] [Google Scholar]
- 21.Waldrop S L, Pitcher C J, Peterson D M, Maino V C, Picker L J. J Clin Invest. 1997;99:1739–1750. doi: 10.1172/JCI119338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein S A, Dobmeyer J M, Dobmeyer T S, Pape M, Ottmann O G, Helm E B, Hoelzer D, Rossol R. AIDS. 1997;11:1111–1118. doi: 10.1097/00002030-199709000-00005. [DOI] [PubMed] [Google Scholar]
- 23.Navikas V, Link J, Wahren B, Persson C, Link H. Clin Exp Immunol. 1994;96:59–63. doi: 10.1111/j.1365-2249.1994.tb06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maggi E, Giudizi M G, Biagiotti R, Annunziato F, Manetti R, Piccinni M P, Parronchi P, Sampognaro S, Giannarini L, Zuccati G, et al. J Exp Med. 1994;180:489–495. doi: 10.1084/jem.180.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyaard L, Otto S A, Keet I P, van Lier R A, Miedema F. Blood. 1994;84:4262–4268. [PubMed] [Google Scholar]
- 26.Paganelli R, Scala E, Ansotegui I J, Ausiello C M, Halapi E, Fanales-Belasio E, D’Offizi G, Mezzaroma I, Pandolfi F, Fiorilli M, et al. J Exp Med. 1995;181:423–428. doi: 10.1084/jem.181.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyjek E, Lischner H W, Hyslop T, Bartkowiak J, Kubin M, Trinchieri G, Kozbor D. J Immunol. 1995;155:4060–4071. [PubMed] [Google Scholar]
- 28.Sei S, Akiyoshi H, Bernard J, Venzon D J, Fox C H, Schwartzentruber D J, Anderson B D, Kopp J B, Mueller B U, Pizzo P A. J Infect Dis. 1996;173:1485–1490. doi: 10.1093/infdis/173.6.1485. [DOI] [PubMed] [Google Scholar]
- 29.Valentin A, Albert J, Fenyo E M, Asjo B. J Virol. 1994;68:6684–6689. doi: 10.1128/jvi.68.10.6684-6689.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibbs J S, Regier D A, Desrosiers R C. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 32.Stauber R H, Horie K, Carney P, Hudson E A, Tarasova N I, Gaitanaris G A, Pavlakis G N. BioTechniques. 1998;24:462–471. doi: 10.2144/98243rr01. [DOI] [PubMed] [Google Scholar]
- 33.Palm G J, Zdanov A, Gaitanaris G A, Stauber R H, Pavlakis G N, Wlodawer A. Nat StructBiol. 1997;4:361–365. doi: 10.1038/nsb0597-361. [DOI] [PubMed] [Google Scholar]
- 34.Kruse N, Tony H P, Sebald W. EMBO J. 1992;11:3237–3244. doi: 10.1002/j.1460-2075.1992.tb05401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1185. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 37.Poli G, Fauci A S. In: Role of Cytokines in the Pathogenesis of HIV Infection. Aggarwal B B, Puri R L, editors. Cambridge, MA: Blackwell Science; 1995. pp. 421–449. [Google Scholar]
- 38.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 39.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 40.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, et al. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 41.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul W E. Ciba Found Symp. 1997;204:208–16. doi: 10.1002/9780470515280.ch14. ; 216–9. [DOI] [PubMed] [Google Scholar]
- 43.Bonecchi R, Bianchi G, Bordignon P P, D’Ambrosio D, Lang R, Borsatti A, Sozzani S, Allavena P, Gray P A, Mantovani A, et al. J Exp Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loetscher P, Uguccioni M, Bordoli L, Baggiolini M, Moser B, Chizzolini C, Dayer J M. Nature (London) 1998;391:344–345. doi: 10.1038/34814. [DOI] [PubMed] [Google Scholar]
- 45.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, et al. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]