Abstract
Neurodegenerative disorders with high brain iron include Parkinson disease, Alzheimer disease and several childhood genetic disorders categorized as neuroaxonal dystrophies. We mapped a locus for infantile neuroaxonal dystrophy (INAD) and neurodegeneration with brain iron accumulation (NBIA) to chromosome 22q12-q13 and identified mutations in PLA2G6, encoding a calcium-independent group VI phospholipase A2, in NBIA, INAD and the related Karak syndrome. This discovery implicates phospholipases in the pathogenesis of neurodegenerative disorders with iron dyshomeostasis.
The neuroaxonal dystrophies include INAD, NBIA and Schindler disease and share the distinctive pathologic feature of axonal degeneration with distended axons (spheroid bodies) throughout the central nervous system. INAD is characterized by progressive motor and sensory impairment, with spheroids also found in peripheral nerves1 (Supplementary Note online). NBIA comprises a clinically and genetically heterogeneous group of disorders with high basal ganglia iron and includes pantothenate kinase–associated neurodegeneration caused by mutations in PANK2 (refs. 2,3). Some individuals with INAD show high brain iron; hence, these phenotypes do overlap. To investigate the molecular bases of these two forms of neuroaxonal dystrophy, we undertook genome-wide linkage studies in 12 families with INAD and in a large consanguineous Pakistani family with NBIA (family 252). Using polymorphic microsatellite markers, we mapped a novel INAD1 locus to a 6.0-Mb region of 22q12.3–q13.2 (LOD score of 4.78 at D22S692), with evidence of locus heterogeneity (Supplementary Methods online). We also detected linkage to chromosome 22q12–q13 in family 252 with NBIA (LOD score 4.65), with a minimal region of linkage of 4.9 Mb between D22S426 and D22S276 containing the disease locus (Supplementary Fig. 1 and Supplementary Fig. 2 online).
In view of the common regions of linkage, we predicted that the NBIA variant in family 252 and INAD would be allelic and proceeded to sequence candidate genes in DNA from family 252 and three INAD probands. After sequencing 70 of ~100 positional candidate genes, we detected mutations in PLA2G6 in all four kindreds (Table 1 and Supplementary Fig. 3 online). We detected a homozygous missense mutation (leading to the amino acid change L545T) in the proband of family 252. This mutation segregated with disease status in 15 affected and unaffected family members. We did not detect this change in 192 ethnically matched control chromosomes. We also detected PLA2G6 mutations in the three INAD probands. We did not detect any of these changes in 192 control chromosomes. We also identified PLA2G6 mutations in 28 additional probands with INAD and in the Karak syndrome family with high basal ganglia iron reported previously4. We identified a total of 44 unique mutations (32 missense, five deletions leading to a frame-shift, three nonsense, two leading to amino acid deletions without a frameshift, one splice site and one large deletion) (Table 1 and Fig. 1). Of the missense mutations, 85% occurred at amino acid positions that are conserved in vertebrates.
Table 1.
Phenotypically classified individuals and their mutations
| Individual | Exon | Mutation | Result | Spheroids | Brain iron |
|---|---|---|---|---|---|
| NBIA with PLA2G6 mutations | |||||
|
| |||||
| 45 | 3 | 238G→A | A80T | u | + |
| 17 | 2370_2371delTG | Y790X | |||
| 102 | 3 | 319delC | L107fsX110 | No biopsy | + |
| 125 | 16 | 2251G→Ab | E751K | No biopsy | + |
| 252a,c | 12 | 1634A→Cb | K545T | No biopsy | + |
| 2864 | 14 | 1894C→T | R632W | No biopsy | + |
| 15 | 2070_2072delTGT | V691del | |||
| 288 | 7 | 1061T→C | L354P | No biopsy | + |
| 16 | 2215G→C | D739H | |||
|
| |||||
| Classical INAD | |||||
|
| |||||
| 73a | 17 | 2370T→Gb | Y790X | + | − |
| 81 | 3 | 404T→C | F135S | + | − |
| 11 | 1472T→G | L491R | |||
| 144a | 14:17 | PLA2G6IVS13_FLJ22582IVS2delb | Fusion protein | + | u |
| 173d | 2 | 109C→T | R37X | + | − |
| 11 | 1501G→C | E501Q | |||
| 177a | 13 | 1772G→Ab | R591Q | + | − |
| 182a | 2 | 109C→Tb | R37X | + | − |
| 184a | 7 | 1018G→Ab | G340R | + | u |
| 186 | 11 | 1538_1541delACTG | D513fsX543 | + | − |
| 17 | 2370_2371delTG | Y790X | |||
| 211-1a | 10 | 1351delCb | L451fsX470 | + | u |
| 212 | 14 | 1912G>A | G638R | + | − |
| 16 | 2221C→T | R741W | |||
| 229 | 11 | 1506G→C | K502N | + | − |
| 12 | 1612C→T | R538C | |||
| 231 | 7 | 1039G→Ab | G347R | + | − |
| 277 | 16 | IVS16+1G→Ab | Aberrant splicing | No biopsy | − |
|
| |||||
| INAD with brain iron | |||||
|
| |||||
| 16 | 13 | 1744G→T | V582L | − | + |
| 16 | 2233C→T | R745W | |||
| 171a | 4 | 469C→Tb | H157Y | + | + |
| 172-1e | 4 | 470A→G | H157R | + | + |
| 7 | 1021G→A | A341T | |||
| 172-2f | 4 | 470A→G | H157R | − | + |
| 7 | 1021G→A | A341T | |||
| 176a | 2 | 109C→Tb | R37X | + | + |
| 180 | 13 | 1770C→A | D590E | + | + |
| 16 | 2221C→T | R741W | |||
| 211-2a | 10 | 1351delCb | L451fsX470 | U | + |
| 256 | 5 | 755delA | N252fsX260 | − | + |
| 7 | 1061T→C | L354P | |||
| 273 | 12 | 1699G→A | E567K | + | + |
| 13 | 1754C→T | T585I | |||
|
| |||||
| Classical INAD by report | |||||
|
| |||||
| 178 | 6 | 848A→Gb | D283G | ||
| 179 | 7 | 929T→Ab | V310E | ||
| 188 | 4 | 439G→A | A147T | ||
| 15 | 2132C→T | P711L | |||
| 189 | 11 | 1442T→Ab | L481Q | ||
| 190 | 4 | 470A→G | H157R | ||
| 14 | 1903C→T | R635X | |||
| 191 | 14 | 1903C→Tb | R635X | ||
| 192 | 5 | 751_759del9bp | P251_G253del | ||
| 11 | 1463T→G | V488G | |||
| 193 | 11 | 1442T→Ab | L481Q | ||
| 194 | 14:17 | PLA2G6IVS13_FLJ22582IVS2delb | Fusion protein | ||
| 217 | 15 | 2070_2072delTGTb | V691del | ||
| 264 | 7 | 1058C→T | P353L | ||
| 11 | 1549G→T | G517C | |||
Survey of a large number of ESTs in the public database did not uncover any of the sequence changes described here. X indicates stop. + = present, − = absent, u = unknown. See Supplementary Table 1 online for primer sequences. Written informed consent was obtained from all study participants. This study was approved by institutional review boards of Oregon Health & Science University or the University of Birminghas
Parents are known to be consanguineous, or there is probable identity by descent based on haplotypes.
Mutation is homozygous. Individuals who are unlikely to be identical by descent and who seem to be homozygous for one mutation by sequence analysis may be hemizygous because of a deletion of one allele.
Individual from index NBIA family.
Three individuals were previously reported in ref. 1.
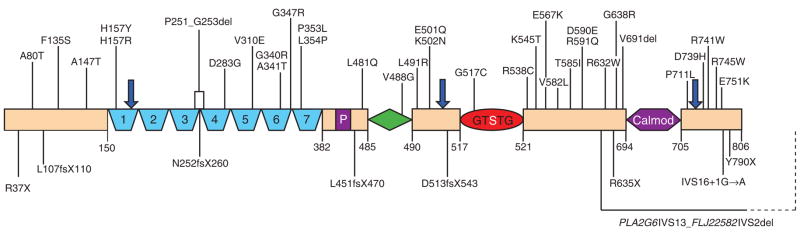
Figure 1.
Schematic showing iPLA2-VIA protein structure7 and mutation-derived changes. Variant 1 is shown with seven ankyrin repeats (blue), a proline-rich motif (purple box), a glycine-rich nucleotide binding motif (green diamond), a lipase motif (red with the active site, Ser519, highlighted), a proposed C-terminal Ca2+-dependent calmodulin binding domain (purple) and three putative caspase cleavage sites (blue arrows). Numbers indicate amino acids. Substitutions or deletions of amino acids not altering the reading frame are shown above the diagram; nonsense mutations, splice mutations and deletions causing frameshifts are shown below. A deletion encompassing exons 14 through 17, the 3′of PLA2G6 and exons 1 and 2 of a downstream putative transcript, FLJ22582, is shown at the C terminus.
We reviewed the clinical and brain magnetic resonance imaging (MRI) data from individuals with PLA2G6 mutations (Supplementary Fig. 4 online). Although brain MRI changes indicating high levels of iron have been reported only rarely in INAD5,6, 8 of 20 kindreds (40%) with mutation-positive INAD showed high iron in the globus pallidus. This high frequency may be skewed by a bias in our collection toward those with high brain iron; nevertheless, INAD should clearly be included in the differential diagnosis of neurodegeneration with high brain iron. In contrast, we identified PLA2G6 mutations in only 4 of 24 additional individuals diagnosed with NBIA. PLA2G6 genotype correlates with phenotype only insofar as we found homozygous null mutations only in INAD and not in any individuals with a clinical diagnosis of NBIA.
Historically, diagnosis of INAD has required histopathological evidence of dystrophic axons. However, we found that the presence of spheroids on biopsy did not absolutely correlate with mutations in PLA2G6. Five individuals with clinical and pathological features of INAD were negative for PLA2G6 mutations. Incomplete detection of mutations could explain this; however, we were able to identify both mutated alleles in all but one individual with mutations. An alternative explanation is that INAD is genetically heterogeneous. Indeed, although PLA2G6 is the major disease locus, our linkage data support the existence of at least one additional INAD locus (data not shown).
Although spheroids are characteristic of INAD, three individuals with a clinical diagnosis of INAD but without spheroids were found to have mutations in PLA2G6. Possible reasons for normal pathologic studies include biopsy at an age before pathological changes have developed or from a site where changes are not evident. Although individual 256 had only a single negative biopsy at age 2 years, individuals 16 and 172-2 (ref. 1) had negative biopsies at 12 years of age. The latter’s sibling (172-1) was positive both for mutations in PLA2G6 and for spheroids (at age 11 years)1. These observations indicate a role for PLA2G6 mutation detection in the diagnostic evaluation of all individuals suspected to have INAD, regardless of biopsy results. Indeed, we would argue that molecular testing should replace this more invasive procedure and that PLA2G6 mutation studies are indicated for individuals with a phenotype in the spectrum of NBIA as well.
PLA2G6 encodes iPLA2-VI, a calcium-independent phospholipase. Phospholipase A2 enzymes catalyze the hydrolysis of glycerophospholipids at the sn-2 position, generating a free fatty acid, usually arachidonic acid, and a lysophospholipid. The iPLA2-VI protein is active as a tetramer, with proposed roles in phospholipid remodeling, arachidonic acid release, leukotriene and prostaglandin synthesis and apoptosis7. PLA2G6 transcript variants encode multiple isoforms8, and the two enzymatically active forms are predicted to be affected by all of the mutations reported here. A subset of mutations would also alter the shorter enzymatically inactive isoforms, which seem to act as dominant-negative inhibitors when incorporated into the tetramer7,8. Because all of the mutations identified are likely to be inactivating, no additional effect is predicted for this subset, and indeed, no distinctive phenotype is recognized.
The calcium-independent PLA2 (iPLA2) enzymes are critical in cell membrane homeostasis. Levels of phosphatidylcholine, abundant in mammalian cell membranes and key in maintaining membrane integrity, are regulated by the opposing actions of CTP:cytidy-lylphosphocholine transferase and iPLA2-VIA9. Defects in iPLA2-VIA could lead to a relative abundance of membrane phosphati-dylcholine and secondary structural abnormalities, which may underlie the axonal pathology observed in INAD and NBIA.
Although defective phospholipid metabolism has been suspected in neurodegeneration9,10, our discovery clearly implicates phospholipase A2 dysfunction in neurodegeneration and more specifically in disease associated with brain iron dyshomeostasis. Impaired phospholipid metabolism has also been implicated in the pathogenesis of another form of NBIA, pantothenate kinase–associated neurodegeneration3. Iron accumulates with age in brain regions that are targeted in Alzheimer disease and Parkinson disease, and iron is implicated in their pathogenesis11. Hence, this new link between phospholipid defects and brain iron metabolism may generate insights into the pathogenesis of common neurodegenerative disorders and may spur ideas for new neuroprotective strategies.
Supplementary Material
Acknowledgments
We are grateful to the many contributing individuals, families and physicians. This work was funded by the Wellcome Trust and WellChild (E.M.), by the US National Eye Institute, L’Association Internationale De Dystrophie Neuro Axonale Infantile, the US National Institute of Child Health and Human Development, and the R.J. Murdock Foundation (S.J.H.) and by the NBIA Disorders Association and NORD (E.M., S.J.H.). Additional support was provided by the Paolo Zorzi Foundation, the Italian National Ministry of Health and the Oregon Alzheimer Disease Center Grant (P30 AG008017). Genotyping services were provided by the CIDR through a federal contract from the US NIH to The Johns Hopkins University, contract number N01-HG-65403 and by the OHSU GCRC (grant M01 RR000334). We thank H. Consencgo for technical assistance. J.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing interests.
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.Nardocci N, et al. Neurology. 1999;52:1472–1478. doi: 10.1212/wnl.52.7.1472. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick SJ, et al. N Engl J Med. 2003;348:33–40. doi: 10.1056/NEJMoa020817. [DOI] [PubMed] [Google Scholar]
- 3.Zhou B, et al. Nat Genet. 2001;28:345–349. doi: 10.1038/ng572. [DOI] [PubMed] [Google Scholar]
- 4.Mubaidin A, et al. J Med Genet. 2003;40:543–546. doi: 10.1136/jmg.40.7.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farina L, et al. Neuroradiology. 1999;41:376–380. doi: 10.1007/s002340050768. [DOI] [PubMed] [Google Scholar]
- 6.Simonati A, Trevisan C, Salviati A, Rizzuto N. Neuropediatrics. 1999;30:151–154. doi: 10.1055/s-2007-973482. [DOI] [PubMed] [Google Scholar]
- 7.Balsinde J, Balboa MA. Cell Signal. 2005;17:1052–1062. doi: 10.1016/j.cellsig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Larsson PK, Claesson HE, Kennedy BP. J Biol Chem. 1998;273:207–214. doi: 10.1074/jbc.273.1.207. [DOI] [PubMed] [Google Scholar]
- 9.Baburina I, Jackowski S. J Biol Chem. 1999;274:9400–9408. doi: 10.1074/jbc.274.14.9400. [DOI] [PubMed] [Google Scholar]
- 10.Farooqui AA, Ong WY, Horrocks LA. Neurochem Res. 2004;29:1961–1977. doi: 10.1007/s11064-004-6871-3. [DOI] [PubMed] [Google Scholar]
- 11.Zecca L, Youdim MB, Riederer P, Connor JR, Crichton RR. Nat Rev Neurosci. 2004;5:863–873. doi: 10.1038/nrn1537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.