Abstract
A study was initiated to construct a micro-reactor for protein digestion based on trypsin-coated fused-silica capillaries. Initially, surface plasmon resonance was used both for optimization of the surface chemistry applied in the preparation and for monitoring the amount of enzyme that was immobilized. The highest amount of trypsin was immobilized on dextran-coated SPR surfaces which allowed the covalent coupling of 11 ng mm−2 trypsin. Fused-silica capillaries were modified in a similar manner and the resulting open-tubular trypsin-reactors having a pH optimum of pH 8.5, display a high activity when operated at 37 °C and are stable for at least two weeks when used continuously. Trypsin auto-digestion fragments, sample carry-over, and loss of signal due to adsorption of the protein were not observed. On-line digestion without prior protein denaturation, followed by micro-LC separation and photodiode array detection, was tested with horse-heart cytochrome C and horse skeletal-muscle myoglobin. The complete digestion of 20 pmol μL−1 horse cytochrome C was observed when the average residence time of the protein sample in a 140 cm ×50 μm capillary immobilized enzyme reactor (IMER) was 165 s. Mass spectrometric identification of the injected protein on the basis of the tryptic peptides proved possible. Protein digestion was favorable with respect to reaction time and fragments formed when compared with other on-line and off-line procedures. These results and the easy preparation of this micro-reactor provide possibilities for miniaturized enzyme-reactors for on-line peptide mapping and inhibitor screening.
Keywords: Trypsin reactor, Dextran hydrogel, Surface plasmon resonance, Liquid chromatography, On-line digestion
Introduction
A demand for smaller enzyme reactors has emerged in recent years, as a consequence of ongoing miniaturization in the biochemical and analytical sciences. These micro-reactors have been used in biocatalysis and biosensing. In the field of proteomics the reactors are a tool in peptide mapping, in which proteins are identified via peptide fragment identification after proteolysis. Currently, in spite of its limitations, most of these analyses are conducted by means of 2D gel electrophoresis followed by digestion of the proteins, liquid chromatographic (LC) separation, and mass spectrometric (MS) identification of the peptides [1–3]. The most time-consuming step in this procedure is digestion of the protein using a protease. In general, every protein to be investigated is individually incubated with the protease at a concentration of approximately 1–2% protein weight for 2 to 18 h at an elevated temperature (typically 37 °C). In addition to the long incubation time needed, a certain level of auto-digestion of the protease can be expected. To reduce sample handling, digestion time, and the risk of sample contamination, methods for the on-line digestion of proteins have been developed that use proteases immobilized on a solid support.
Immobilized enzyme reactors have been developed and used over the years for several industrial and analytical purposes [4–6]. An obvious benefit for immobilizing biocatalysts is the fact that the enzyme can be used in several catalytic cycles and that both catalyst and reaction mixture can easily be separated. Moreover, immobilized enzymes generally show an improved stability even at more extreme reaction conditions. Several procedures have been developed for immobilization of enzymes, e.g. adsorption or encapsulation in a matrix or membrane. Alternatively, and more often used, is the covalent attachment of biocatalysts to carrier materials, which allows the immobilization of a large amount of enzyme for a high activity per surface area. Generally, particulate large-pore carrier materials are used, such as controlled-pore glass [7, 8], silica [9], or polymers like the commercially available poroszyme [10–12]. Current research in the production of immobilized enzymes is focused on the use of monolithic materials, as they enable efficient fragmentation of proteins [13–17]. Although both commercially available and self-prepared reversed-phase capillary monolithic columns have successfully passed reproducibility assessment [18, 19], synthesis of monoliths suitable for small-scale enzyme reactors can still be troublesome. Materials suitable for the fabrication of larger-scale enzyme reactors are commercially available from BIA Separations (Ljubljana, Slovenia).
Although it is possible to apply an immobilized enzyme reactor (IMER) positioned after the separation column [20], most papers dealing with on-line digestion of protein samples position the IMER upstream of the separation column. In these cases the sample is first digested and the resulting peptide fragments are separated and identified by LC–MS. This approach is often employed in multi-dimensional LC methods [13, 21, 22], and has also found application in peptide mapping using capillary electrophoresis [23, 24]. Alternatively, as recently shown by Zhao et al. [25] and Krenkova et al. [26], who covalently coupled trypsin to the wall of fused-silica nanoelectrospray emitters, a protein sample can be analyzed by direct infusion into a mass spectrometer.
This paper describes the development of trypsin-modified open-tubular micro-reactors. The chemistry was controlled and optimized using surface plasmon resonance (SPR), a technique allowing sensitive and real-time monitoring of surface reactions such as protein binding [27]. The surface modification resulting in the highest enzyme immobilization yield, was used to covalently immobilize the trypsin on the inside wall of a fused-silica capillary. The constructed trypsin micro-reactor, which is compatible with micro- and nano-LC, was further characterized. The influence of reaction time, pH, temperature, and reactor stability were investigated with the model substrate insulin B-chain. The reactor was also applied to digestion of the proteins cytochrome C and myoglobin. The produced peptides were analyzed by liquid chromatography–mass spectrometry.
Experimental
Materials
The SPR equipment used was from IBIS Technologies (currently available from Eco Chemie, Utrecht, The Netherlands) equipped with a 200-μL polycarbonate cuvet. The gold-sensor disks, purchased from SSENS (Hengelo, The Netherlands), were positioned on the IBIS-prism using index-matching oil from R.P. Cargille Laboratories (Cedar Grove, USA). PEEK nuts, unions, tubing, and loops were from Upchurch (Santa Monica, USA). Manual injections during the preparation of the reactors were performed using a Rheodyne 7010 injector (Inacom, Veenendaal, The Netherlands) equipped with a 1-mL PEEK loop. Model 10ADvp HPLC pumps from Shimadzu (Kyoto, Japan) were used for reactor preparation and activity determinations. The water used for washing and to prepare buffers was produced by a Sartorius Arium 611 ultrapure water system (Nieuwegein, The Netherlands; conductivity >18.2 MΩ cm). The model ABS759A UV absorbance detector was equipped with a capillary flow-cell (75-μm i.d.) and was obtained from Applied Biosystems (Nieuwerkerk a/d IJssel, The Netherlands).
On-line digestion experiments with micro-HPLC separations were conducted using LC-Packings instruments and columns (Amsterdam, The Netherlands). The equipment consisted of an injector (Famos), nanovalve column switcher (Switchos), nanopump (Ultimate), and a photodiode-array detector (PDA) equipped with a micro flow cell (45 nL). The reversed-phase pre-columns were 5 × 0.3 mm with 5-μm 100 Å C18 particles. The 150 × 0.3 mm reversed-phase micro-column contained 3-μm 100 Å C18 PepMap particles. The mass spectrometer was an Agilent LC/MSD XCT ion trap (Amstelveen, Netherlands).
Acetic acid, boric acid, calcium chloride (CaCl2), ethanol, ethanolamine (EA), hydrochloric acid (HCl), hydrogen peroxide, sodium dihydrogen phosphate, sodium chloride, sodium hydroxide (NaOH), and sulfuric acid were purchased from Merck (Darmstadt, Germany). HPLC grade acetonitrile and ethanol were from Biosolve (Valkenswaard, The Netherlands). 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), tris-hydroxymethylaminoethane (TRIS), urea, and acetone were purchased from Acros Organics (Geel, Belgium). Benzamidin, N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC), benzoyl-arginine ethyl ester (BAEE), iodoacetamide, N-hydroxysuccinimide (NHS), and carboxyl-modified dextran (CMD) were purchased from Fluka (Buchs, Switzerland). Horse-heart cytochrome C, insulin B (oxidized), polyoxyethylenesorbitan monolaurate (Tween 20), and porcine pancreas trypsin were purchased from Sigma (St Louis, USA). The specific activity of trypsin was determined according to the method of Schwert and Takenaka [28] using BAEE as substrate and turned out to be 14700 U mg−1. Aminopropyltriethoxysilane (APTES), carbonyl diimidazole (CDI), glycidoxypropyltrimethoxysilane (GOPS), and mercaptoethanol (ME) were from Aldrich Chemical Company (Milwaukee, USA). Amino-modified dextran (AMD) was from Unavera ChemLab (Mittenwald, Germany). Fused-silica capillaries were purchased from Bester (Mijdrecht, The Netherlands).
Piranha solution was prepared by mixing 1 part 30% hydrogen peroxide and 6 parts concentrated sulfuric acid (caution, aggressive solution). TRIS digestion buffer consisted of 50 mmol L−1 TRIS and 1 mmol L−1 CaCl2 adjusted to pH 8.2 by use of 1 mol L−1 HCl solution.
Methods
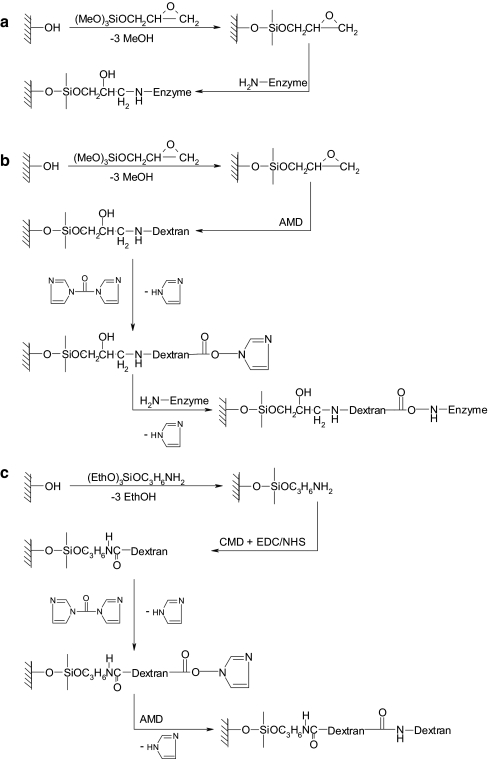
Preparation of dextran-coated SPR sensor disks (Fig. 1)
Fig. 1.
Surface modifications used to immobilize the enzyme in fused-silica capillaries. a, GOPS-modified; b, GOPS/AMD-modified; c, APTES/CMD/AMD-modified surface. In the last case the CDI activation and enzyme (H2N–enzyme) coupling are as in b
The SPR sensor disks were extensively cleaned in freshly prepared piranha solution. After 1 h the disks were thoroughly rinsed with water, dried in a stream of nitrogen gas, and immediately incubated for 6 h in a 10 mmol L−1 solution of ME in ethanol in order to produce a self-assembled monolayer (SAM) containing hydroxyl functionalities. After SAM formation, the disks were washed with ethanol and water, and dried with nitrogen gas. The resulting ME disks were incubated for 1 h with a 10% GOPS solution in 98% ethanol after which the disks were washed with ethanol, dried with nitrogen gas and stored overnight at 50 °C.
Dextran-modified sensors were prepared by incubating GOPS-modified sensor disks for 20 h at room temperature with a 10% AMD solution in a 50 mmol L−1 borate buffer, pH 9.5. After removing the solution the disks were washed with water, ethanol, and water, dried in a gentle flow of nitrogen gas, and stored at room temperature in a closed box until use.
Alternatively, ME disks were incubated for 1 h with a 10% APTES solution in acetone. After washing, drying, and storing overnight in the oven, similar to the GOPS disks, the surfaces were incubated for 1 h with a solution containing 5% CMD in water containing 200 mmol L−1 EDC and 50 mmol L−1 NHS. After removing the solution, the disks were washed with water and ethanol, and incubated for 15 min in a solution containing 100 mmol L−1 CDI in acetone. These activated surfaces were washed with ethanol, blown to dryness with nitrogen, and incubated overnight with a solution of 10% AMD in water. After removing the solution the disks were washed with water, ethanol, and water, dried in a gentle flow of nitrogen gas, and stored at room temperature in a closed box until use.
SPR experiments
The GOPS-modified SPR sensor disks were incubated overnight with 200 μL of a solution containing 2.5 mg mL−1 trypsin and 50 μg mL−1 benzamidin in 50 mmol L−1 borate buffer, pH 9.5 (Fig. 1a). Both types of dextran-modified sensor disk were activated for 60 min with 200 μL 100 mmol L−1 CDI in dry acetone. After washing the surface with water to remove the last traces of acetone, the sensors were incubated overnight with 200 μL 2.5 mg mL−1 trypsin and 50 μg mL−1 benzamidin in 50 mmol L−1 borate buffer, pH 8.5 (Fig. 1b and c, respectively). The remaining esters were inactivated by incubating the disks for 10 min with 1 mol L−1 EA in 50 mmol L−1 borate buffer, pH 8.5. The successive steps in the immobilization were monitored with SPR and the amount of covalently coupled enzyme was calculated from the recorded angle shift. During the experiments the SPR system was thermostatted at 25.0 °C.
Preparation of dextran-coated fused-silica capillaries
In order to generate a proper surface for silanization, fused-silica capillaries were cleaned for 30 min with 2 mol L−1 NaOH solution at a flow-rate of 5 μL min−1. The capillary was then washed for 30 min with 0.1 mol L−1 HCl, for 5 min with water, and finally for 5 min with ethanol. In order to prepare dextran-coated capillaries, the capillaries were flushed for 60 min with a 10% GOPS solution in ethanol. After this step, the capillaries were closed with silicon plugs and dried overnight at 50 °C. After silanization, the capillaries were flushed with methanol at a flow-rate of 10 μL min−1 after which the capillaries were chemically modified in flow, using injections with a Rheodyne 7010 manual injector equipped with a 1-mL PEEK loop, in a way similar to that described above in the SPR section. Washing steps were conducted at flow rates of 10 μL min−1, overnight incubations at a flow rate of 1 μL min−1. All chemistries are outlined in Fig. 1.
Enzyme activity determination
Insulin B chain was used as a substrate to determine the activity of trypsin. Injections of 1 μL of a concentration of 20 μmol L−1 in digestion buffer were introduced into the reactor that was kept at the indicated temperature using a water bath. To determine the enzyme activity in solution, trypsin was incubated in these solutions also (final trypsin concentration 0.4 μg mL−1). At time intervals from 0.5 to 30 min, 100-μL samples were taken and the activity was stopped by the addition of 5 μL 20% TFA in water. Buffers that were used to determine the effect of pH on activity were MES (pH 5.5 to 6.5), MOPS (pH 6.5 to 7.8), TRIS (pH 7.5 to 9), and CHES (pH 8.6 to 10.1). The buffers were adjusted to pH by use of 1 mol L−1 NaOH and were prepared at a 50 mmol L−1 concentration, also containing 5 mmol L−1 CaCl2. To determine the effect of temperature, the temperature during incubation was varied between 10 °C and 60 °C. Both the on-line and off-line samples were analyzed using micro-HPLC with PDA detection, as outlined below. The insulin B conversion of both immobilized trypsin and the enzyme in solution was calculated from the peak areas of substrate and products.
On-line protein digestion in micro-HPLC
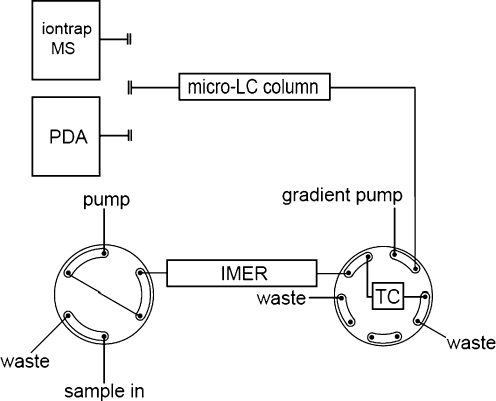
For the on-line peptide and protein digestion experiments the trypsin-modified capillaries were positioned between the LC injector and a 10-port valve, as shown in Fig. 2, and were immersed in a thermostatted water bath set at 37 °C unless mentioned otherwise. Protein samples were prepared in digestion buffer and were transported through the capillary towards a 10-port valve using a 5% acetonitrile solution containing 0.05% TFA. The peptide fragments formed during digestion were concentrated on an RP trapping column (TC) and salts and other buffer components present in the sample were removed. By switching the valve the trapping column is in series with the RP micro column and an acetonitrile gradient started. The gradient was composed of two solutions: (A) 5% acetonitrile in water containing 0.05% TFA and (B) 80% acetonitrile in water containing 0.04% TFA. In 30 min the gradient changed linearly from 0 to 50% B, followed by 10 min at 90% B and 20 min at 0% B. The eluent was monitored with the PDA detector in the range from 200 to 595 nm.
Fig. 2.
Set-up used for on-line protein digestion using a trypsin-modified fused-silica capillary. For detection a PDA or an ion-trap MS was used
ESI-MS was conducted in the positive-ion mode with the capillary voltage set at 3500 V. The flow rate and temperature of the nitrogen drying gas were 5 L min−1 and 325 °C, respectively. The sequence of the peptide fragments was determined by using the mass spectrometer in auto-MS–MS mode fragmenting the two peptides that were most abundantly present when the signal reached threshold. The MS–MS result was analyzed by a Mascot database search (http://www.matrixscience.com).
Results and discussion
Surface chemistry and enzyme immobilization
As shown before [29], modification of an SPR sensor surface with a dextran hydrogel leads to less non-specific adsorption of proteins compared with unmodified surfaces. The presence of such a layer also enhances the immobilization capacity of biomolecules compared with monolayer-based coatings. Due to the flexible nature of the dextran chains, the accessibility also is often improved compared with molecules immobilized on a flat surface. As the amount of protein present in a capillary after immobilization cannot be determined easily, SPR sensors were used as a model to investigate the effect of the different surface modifications on the amount of trypsin that could be attached covalently. Hydroxyl functionalities necessary to enable silanization are introduced using mercaptoethanol (ME), but all other surface modifications are carried out in exactly the same way, both on the SPR sensor surfaces and in the fused-silica capillaries.
Trypsin immobilization after silanization with GOPS by reaction of the trypsin primary amines and the glycidyl function of GOPS results in an SPR angle shift of 350 ± 20 m° (n = 3), which equals an amount of trypsin of 2.9 ± 0.2 ng mm–2 covalently attached to the surface, which is close to monolayer coverage of trypsin. When the GOPS-silanized surface is modified with AMD, resulting in a dextran hydrogel, the amount of trypsin that can be immobilized after CDI activation increases to 9.5 ± 0.3 ng mm−2 (n = 3). To further increase the amount of trypsin, an intermediate dextran layer was added. Therefore, fused-silica capillaries were APTES silanized and modified with CMD and AMD, subsequently. The amount of trypsin that could covalently be attached to these layers was determined with SPR and proved to be 11.1 ± 0.5 ng mm−2 (n = 2). As this amount is significantly more than obtained with the GOPS/AMD surface, all further experiments have been conducted with fused-silica capillaries the surface of which is modified with dextran in this way.
Assuming that the investigated surface modification allows a similar amount of trypsin to be immobilized per surface area in fused-silica capillaries, the quantity of enzyme immobilized on the dextran-modified capillaries is more than five times larger per surface area compared with other open-tubular, trypsin micro-reactors described in the literature [23, 30–32]. These reactors, first described by Amankwa and Kuhr [30], are based on immobilized avidin and, at saturation, 6.5 pmol biotinylated trypsin was immobilized in a 50-cm long capillary of 50-μm i.d.. This amount equals 2 ng trypsin mm−2 capillary surface, which is less than monolayer coverage.
Characterization of the enzyme reactor
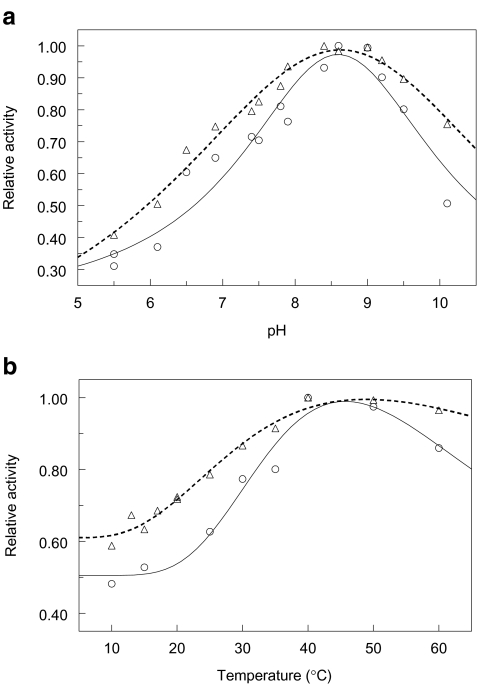
Fused-silica capillaries were modified with dextran as described above and the activity and characteristics of the trypsin micro-reactor were assessed with the oligopeptide insulin B chain, a 30 amino acids long insulin fragment. When insulin B is exposed to the enzyme present in the reactor, hydrolysis at the arginine present at position 22 and, to a lesser extent, the lysine at position 29 is observed. Using the aforementioned substrate insulin B, the pH optimum was determined for the enzyme in solution and the immobilized enzyme. As can be observed in Fig. 3a, the optimum pH value for both free and immobilized trypsin is pH 8.5. Therefore, all further experiments are conducted at this pH value.
Fig. 3.
Effect of (a) pH and (b) temperature on the relative activities of trypsin in solution (circles) and trypsin immobilized on an APTES/CMD/AMD-modified fused-silica surface (triangles). In b the maximum insulin B conversion rate determined under experimental conditions is 4.41 ± 0.02 pmol min−1 μg−1 for the enzyme in solution and 12.32 ± 0.35 pmol min−1 μg−1 for a 3-μL, 50-μm i.d. microreactor operated at a flow rate of 1 μL min−1
Also the effect of temperature on the activity of both the immobilized trypsin and the enzyme in solution was determined in the range 10 °C to 60 °C. As can be observed in Fig. 3b, the activity of trypsin increases with temperature to reach a maximum around 40 °C and decreases at higher temperatures. The immobilized enzyme shows a higher activity compared with the enzyme in solution, which may be due to an often observed higher stability of immobilized enzymes, also under more extreme conditions. From the data the activation energy can be determined. For the immobilized enzyme a value of 12.2 ± 0.3 kJ mol−1 can be calculated and for the enzyme in solution a value of 17.2 ± 0.5 kJ mol−1. This means that for the immobilized enzyme the temperature has less effect on the activity than for the enzyme in solution. As the highest activity is observed at 37 °C, all further experiments are conducted at that temperature.
For the enzyme in solution a specific activity of 4.41 ± 0.02 pmol min−1 μg−1 is determined at a concentration of 20 pmol μL−1 insulin B and pH 8.5 and 37 °C. Under similar conditions, in a microreactor of 3 μL with a 50 μm i.d. and operated at a flow rate of 1 μL min−1, an amount of 12.32 ± 0.35 pmol insulin B is converted. In such a reactor a total of 2.67 μg trypsin might be present taking the earlier SPR observations into account, which means that the immobilized enzyme is both accessible and still active after immobilization. The specific activity after immobilization is higher than observed in recent papers for the protease pepsin immobilized on beaded chitosan [33, 34] and trypsin photoimmobilized in a fused-silica capillary [34].
The stability during operation of the open-tubular reactor was tested at 37 °C and pH 8.5. Activity tests using the model substrate show that the activity of a reactor that is continuously in operation, is constant for at least two weeks. The enzyme trypsin dissolved in digestion buffer and incubated for 24 h at 37 °C loses 60% of its activity, and after three days no activity is measured.
On-line digestion of proteins
The on-line digestion of horse cytochrome C is accomplished as described in the Methods section. Samples are submitted to on-line digestion in a 22-cm long trypsin-modified APTES/CMD/AMD-coated capillary with an i.d. of 75 μm (total volume 1 μL) or with an i.d. of 50 μm and a length of 51 cm (1 μL) or 140 cm (2.75 μL). When the protein is not reduced, the heme-moiety will remain covalently attached to the peptides containing the protein sulfhydryls. By monitoring the heme-containing peptides at 395 nm, the progress of the digestion and the amount of undigested protein can be determined. The results are summarized in Table 1.
Table 1.
Summary of the on-line digestion experiments for cytochrome C. The experiments were conducted at pH 8.5 and 37°C (n = 3)
| capillary ID (μm) | reactor volume (μL) | amount injected | digestion time (s) | % undigested protein (SD) | |
|---|---|---|---|---|---|
| (μg mL−1) | (pmol) | ||||
| 75 | 1 | 248 | 20 | 60 | 65.9 (4.8) |
| 75 | 1 | 124 | 10 | 60 | 35.8 (3.4 |
| 75 | 1 | 12.4 | 10 | 60 | 23.6 (1.3) |
| 50 | 1 | 248 | 20 | 60 | 14.0 (1.9) |
| 50 | 1 | 124 | 10 | 60 | 2.3 (2.0) |
| 50 | 2.75 | 248 | 20 | 165 | 0 |
| 50 | 2.75 | 124 | 10 | 165 | 0 |
| 50 | 2.75 | 12.4 | 10 | 165 | 0 |
The experiments were conducted at pH 8.5 and 37 °C (n = 3)
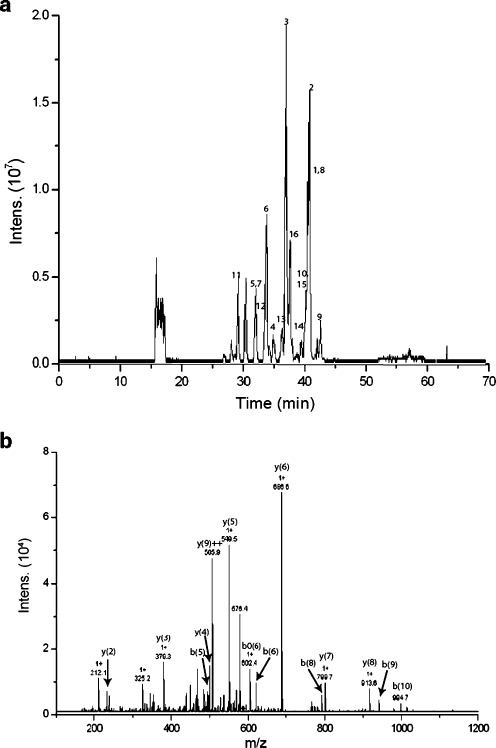
As expected when a limited amount of enzyme activity is present in a reactor (22 cm × 75 μm), for increasing concentrations cytochrome C a larger amount of protein is undigested. Nevertheless, many tryptic peptides are still generated. As can be expected, an increase in exposure time of the substrate with the immobilized enzyme will result in an improved digestion yield. A longer contact time is achieved by increasing the reactor volume by using a longer enzyme-modified capillary. Additionally, the enzyme-to-substrate ratio is increased, which is accomplished by changing the surface-to-volume ratio by using a capillary with a smaller internal diameter. As can be observed in Table 1, decreasing the i.d. of the capillary leads to improved digestion for a reactor of equal volume due to a higher surface-to-volume ratio and hence a higher amount of enzyme. The use of a 140 cm ×50 μm capillary allows the complete digestion of up to 20 pmol (248 μg mL−1) of cytochrome C in less than 5 min including the sample concentration and removal of salts by the trapping column.
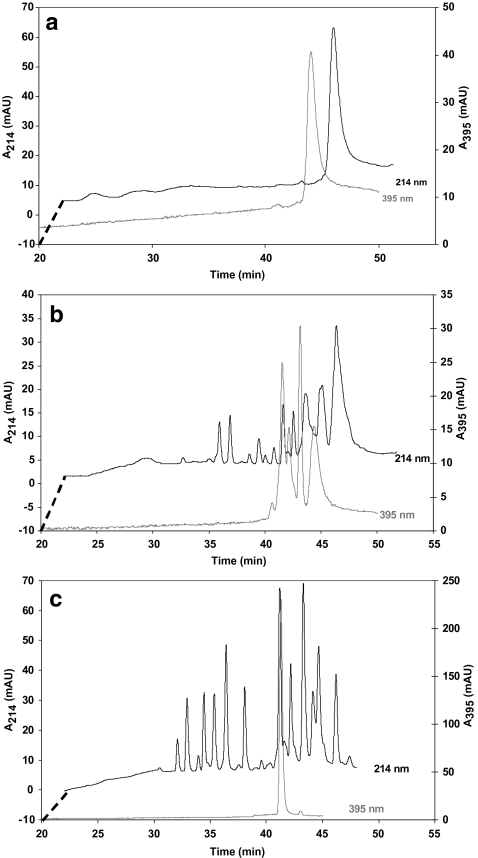
Chromatograms for the on-line digestions obtained with capillaries with an i.d. of 50 μm are shown in Fig. 4. Figure 4a shows a blank run in a capillary containing no enzyme and Fig. 4b and c present on-line digestions for a micro-reactor of 2.75 μL operated at 5 μL min−1 and 1 μL min−1, respectively. As discussed above, incomplete digestion of the injected cytochrome C will lead to the presence of multiple peptide fragments containing the heme group, as is visible in Fig. 4b. The digestion is complete when the sample exposure time is 165 s (flow rate 1 μL min−1) as both intermediate products and the undigested protein (retention time 44 min in Fig. 4a), which are visible at a wavelength of 395 nm as outlined above, are no longer observed. An injection of off-line-digested cytochrome C showed a similar chromatogram as is shown in Fig. 4c.
Fig. 4.
Chromatograms obtained from injection of 10 pmol horse cytochrome C in capillary digestion systems monitored at 214 nm and 395 nm. The experiments were conducted with an APTES-CMD-AMD derivatised fused-silica capillary of (a) 510 × 0.050 mm, not containing trypsin, operated at 1 μL min−1 (blank); (b) 1400 × 0.050 mm, trypsin-modified, operated at 5 μL min−1 (average sample residence time 33 s); (c) as (b) but operated at 1 μL min−1 (average sample residence time 165 s). For clarity the beginning of the chromatogram displaying the 214 nm signal is offset as indicated
The on-line digestion of horse cytochrome C is also monitored with mass spectrometry. The effect of flow rate and hence incubation time on the digestion of the protein and the number of peptides identified with a Mascot database search is determined and summarized in Table 2. In this table the undigested amounts of protein which have been determined using the PDA detector are also shown. At a flow rate of 1 μL min−1 protein digestion is complete and many peptides are matched, resulting in high sequence coverage and Mascot score. With higher flow, and hence decreasing incubation time, the amount of protein that remains undigested increases and consequently fewer peptides are produced and observed. However, even at relatively high flow rates an adequate amount of peptides is still formed and the protein can be identified on the basis of the fragments present. Nevertheless, the ion intensity is low and some peptides are not retrieved as they are below the threshold for auto-MS–MS. Table 3 summarizes the peptides observed and matched using the MS–MS data and a Mascot.database search of proteins digested at a flow rate of 1 μL min−1. A base-peak chromatogram (BPC) of the digestion of cytochrome C under these conditions is shown in Fig. 5a. The MS–MS fragmentation of one of the peptides is presented in Fig. 5b.
Table 2.
General overview of the on-line digestion experiments for cytochrome C (n = 3)
| flow rate (μL min-1) | digestion time (s) | sequence coverage (%) | total no peptides matched | Mascot score | undigested protein (%) |
|---|---|---|---|---|---|
| 10 | 17 | 66 | 11 | 347 | 60.3 |
| 5 | 33 | 66 | 21 | 501 | 31.8 |
| 2 | 83 | 66 | 23 | 649 | 18.5 |
| 1 | 165 | 87 | 27 | 833 | 0 |
Table 3.
Peptide fragments observed by MS in the on-line digestion of cytochrome C and myoglobin The experiments were conducted at pH 8.5 and 37 °C at a flow rate of 1 μL min−1 using a 2.75-μL trypsin reactor
| Peptide sequence | Theoretical m/z | Experimental m/z | Position | Missed cleavage |
|---|---|---|---|---|
| Cytochrome C (sequence coverage 87%) | ||||
| 1....IFVQKCAQCHTVEK | 1632.8 | 816.9 (2+) | 9–22 | 1 |
| 2....HKTGPNLHGLFGR | 1432.8 | 717.1 (2+) | 26–38 | 1 |
| 3....TGPNLHGLFGR | 1167.6 | 584.6 (2+) | 28–38 | 0 |
| 4....TGPNLHGLFGRK | 1295.7 | 648.6 (2+) | 28–39 | 1 |
| 5....KTGQAPGFTYTDANK | 1597.8 | 533.4 (3+) | 39–53 | 1 |
| 6....TGQAPGFTYTDANK | 1469.7 | 735.6 (2+) | 40–53 | 0 |
| 7....TGQAPGFTYTDANKNK | 1711.8 | 571.4 (3+) | 40–55 | 1 |
| 8....GITWKEETLMEYLENPKK | 2208.1 | 736.8 (3+) | 56–73 | 2 |
| 9....EETLMEYLENPK | 1494.7 | 748.0 (2+) | 61–72 | 0 |
| 10 EETLMEYLENPKK | 1622.8 | 812.1 (2+) | 61–73 | 1 |
| 11...YIPGTK | 678.4 | 678.1 (1+) | 74–79 | 0 |
| 12...MIFAGIKK | 906.5 | 454.0 (2+) | 80–87 | 1 |
| 13...KTEREDLIAYLKK | 1477.8 | 739.6 (2+) | 88–99 | 2 |
| 14...TEREDLIAYLKK | 1349.7 | 675.5 (2+) | 89–99 | 1 |
| 15...EDLIAYLK | 963.5 | 482.6 (2+) | 92–99 | 0 |
| 16...EDLIAYLKK | 1091.6 | 546.6 (2+) | 92–100 | 1 |
| Myoglobin (sequence coverage 88%) | ||||
| 1....GLSDGEWQQVLNVWGK | 1814.9 | 908.7 (2+) | 1–16 | 0 |
| 2....VEADIAGHGQEVLIR | 1605.8 | 803.6 (2+) | 17–31 | 0 |
| 3....VEADIAGHGQEVLIR | 1605.8 | 536.1 (3+) | 17–31 | 0 |
| 4....LFTGHPETLEK | 1270.7 | 636.0 (2+) | 32–42 | 0 |
| 5....HLKTEAEMK | 1085.6 | 543.5 (2+) | 48–56 | 1 |
| 6....HGTVVLTALGGILK | 1377.8 | 689.7 (2+) | 64–77 | 0 |
| 7....HGTVVLTALGGILKK | 1505.9 | 502.7 (3+) | 64–78 | 1 |
| 8....KKGHHEAELKPLAQSHATK | 2109.1 | 703.7 (3+) | 78–96 | 2 |
| 9....KGHHEAELKPLAQSHATK | 1981.0 | 661.1 (3+) | 79–96 | 1 |
| 10 GHHEAELKPLAQSHATK | 1853.0 | 618.8 (3+) | 80–96 | 0 |
| 11...YLEFISDAIIHVLHSK | 1884.0 | 628.8 (3+) | 103–118 | 0 |
| 12...HPGNFGADAQGAMTK | 1500.7 | 751.5 (2+) | 119–133 | 0 |
| 13...ALELFRNDIAAK | 1359.8 | 680.6 (2+) | 134–145 | 1 |
| 14...YKELGFQG | 940.5 | 471.0 (2+) | 146–153 | 1 |
Fig. 5.
On-line digestion of 10 pmol cytochrome C at a flow rate of 1 μL min−1: (a) base peak chromatogram, the numbers correspond to the matched peptides in Table 3, and (b) MS–MS fragmentation of the peptide TGPNLHGLFGR with m/z 584.9 showing several of the matched fragment ions
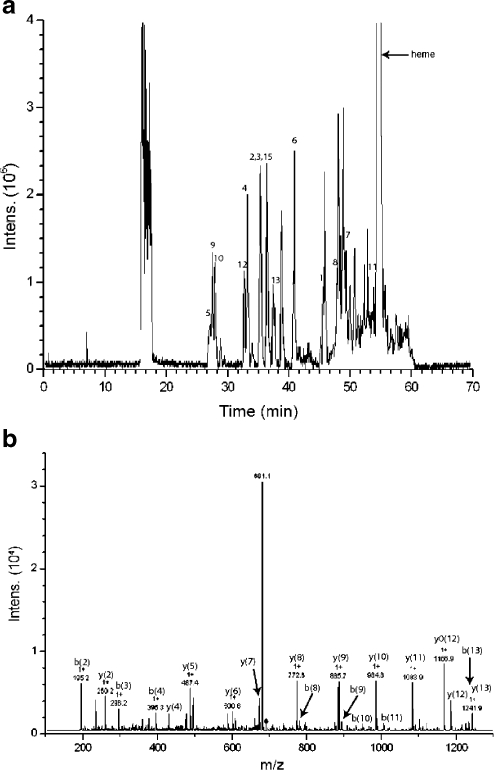
In further experiments horse myoglobin was digested on-line. This protein is generally regarded as difficult to digest [15, 35]. When 1 μL of a 10 μmol L−1 solution in buffer is injected at a flow rate of 1 μL min−1 (165 s exposure time), the injected protein is completely digested, as was observed with UV detection (data not shown). Using mass spectrometric analysis, 13 different peptides are observed and matched using the MS–MS data and a Mascot database search, resulting in a sequence coverage of 88%. The matched peptides are summarized in Table 3 and a BPC of the on-line digestion of myoglobin is shown in Fig. 6a, with an MS–MS spectrum of one of the tryptic peptides. Both the degree of digestion and the sequence coverage are adequate compared with other systems that often use a high percentage of modifier to enhance digestion, as the absence of denaturing agents during digestion leads to little or no digestion of myoglobin [35]. Therefore these reactors are generally used for direct infusion into MS or off-line protein digestion as the presence of high concentrations of methanol or acetonitrile in the digestion buffer will seriously impede on-line protein digestion in combination with RP-LC.
Fig. 6.
Results from on-line digestion of 10 pmol myoglobin at a flow rate of 1 μL min−1: (a) base peak chromatogram, the numbers correspond with the matched peptides in Table 3, and (b) the MS–MS spectrum of the peptide HGTVVLTALGGILK with m/z 689.7
Amankwa and Kuhr [30] reported that proteins and peptides adsorb to the capillary surface of their trypsin reactor. Reactor fouling, and as a result sample carry-over, is also observed when trypsin-modified particulate polymers or monoliths are used [36]. Adsorption of proteins and peptides to these relatively hydrophobic columns is prevented by the addition of up to 20% methanol to the digestion buffer. For dextran-coated surfaces, it is known that the non-specific adsorption of protein is minimal and depends on the character of the oligosaccharide layer [29, 37, 38]. During protein-digestion experiments on SPR sensor surfaces modified identically with the capillary reactors no adsorption during incubation is observed (data not shown). Similarly, during on-line digestion using the dextran coated capillaries, sample adsorption and carry-over are not observed, as repeated sample injections result in identical chromatograms and blank buffer injections do not show peptide fragments. On basis of the latter result and the long lifetime of the reactor, trypsin auto-digestion is probably absent.
The developed IMER has been applied for the digestion of horse cytochrome C and myoglobin in buffer without prior pretreatment followed by separation and MS–MS sequencing. The time needed to obtain complete protein digestion and sample trapping is 5 min with an average sample residence time of 165 s. The total analysis time, digestion and LC separation, is 65 min. The developed micro-reactor is competitive when compared with other systems used for on-line protein digestion described in literature [13, 14, 39–41]. In the capillary IMERs described by Amankwa and Kuhr [30] and Bossi et al. [34] the complete digestion of native proteins was only obtained when the protein resided in the trypsin-coated capillary for 15–25 min. The peptide separation was carried out with CE in these latter cases.
Many reactors use a protein denaturation step to make the protein more susceptible to proteolytic action. The digestion efficiency can be enhanced by partial denaturation by addition of 35–45% acetonitrile to the buffer [11, 15, 35, 36] or by sonication [42, 43]. However, the latter has not yet been used in an automated set-up and the presence of high amounts of modifier will make reversed-phase chromatography in a subsequent step for the separation of the fragments produced during protein digestion very difficult. The described reactors were efficient in terms of time needed for digestion and the digestion result, but were, as a rule, used for direct infusion into the MS or used for off-line digestion.
Alternatively, protein denaturation is accomplished using chemicals such as SDS, guanidine HCl or urea. A recent example is the miniaturized on-line proteolysis–capillary LC system as described by Samskog et al. [12], who employed a 10-μL column packed with poroszyme. The sample contained guanidine.HCl for protein denaturation, and was diluted with buffer, also containing methanol, prior to injection. The total time needed for digestion, trapping, and removal of the high concentration of salt was 15 min. The system needed periodic regeneration to counteract the effects of residual salts in the analyte.
Conclusions
To study the immobilization of the protein trypsin in a fused-silica capillary, a number of surface modifications was tested. In order to investigate the amount of protein that can be covalently attached to these surfaces, SPR sensor disks are modified with these coatings to mimic the capillary surface. The SPR measurements show that the best results were obtained using a dextran coating with an intermediate layer. The resulting open-tubular trypsin reactors having a pH optimum of pH 8.5 display a high activity when operated at 37 °C and are stable for at least two weeks when used continuously.
The capillary reactors show flow-dependent catalysis. For a capillary IMER, the conversion of the insulin B chain increases with decreasing flow and hence a longer residence time. The same is observed for the digestion of horse cytochrome C. The complete digestion of 20 pmol μL−1 horse cytochrome C is observed without the need of protein denaturation, reduction, or alkylation when the average residence time of the protein sample in a 140 cm × 50 μm capillary IMER is 165 s. For the proteins used in this study trypsin-reactors were described that were capable of faster digestion using denaturing agents like acetonitrile to enhance the digestion process. However, the presence of such agents would seriously hamper direct analysis of the peptides formed using RPLC and MS. Identification of the proteins cytochrome C and myoglobin is possible by the tryptic peptides that are produced on-line, separated by micro-RPLC, and analyzed using mass spectrometry with auto MS–MS.
The open-tubular reactor can be produced easily, reproducibly, and inexpensively, and can be used for other applications such as enzyme-inhibitor studies. Future research will focus on the development of miniaturized multi-dimensional analysis systems employing on-line digestion using these IMERs. The capillary enzyme reactors show no backpressure and seem promising for coupling to other analytical techniques such as capillary electrophoresis and surface plasmon resonance.
References
- 1.Courchesne PL, Patterson SD (1999) Identification of proteins by matrix-assisted laser desorption/ionization mass spectrometry using peptide and fragment ion masses. In: Link AJ (ed) Methods in molecular biology: 2-D proteome analysis protocols. Humana Press, Totowa, pp 487–511 [DOI] [PubMed]
- 2.Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C (1993) Proc Natl Acad Sci USA 90:5011–5015 [DOI] [PMC free article] [PubMed]
- 3.Garbis S, Lubec G, Fountoulakis M (2005) J Chromatogr A 1077:1–18 [DOI] [PubMed]
- 4.Girelli AM, Mattei E (2005) J Chromatogr B 819:3–16 [DOI] [PubMed]
- 5.Urban PL, Goodall DM, Bruce NC (2006) Biotechnol Adv 24:42–57 [DOI] [PubMed]
- 6.Hermanson GT, Mallia AK, Smith PA (1992) Immobilized affinity ligand techniques. Academic Press, San Diego
- 7.Bonneil E, Waldron KC (2000) Talanta 53:687–699 [DOI] [PubMed]
- 8.Bonneil E, Mercier M, Waldron KC (2000) Anal Chim Acta 404:29–45 [DOI]
- 9.Davis MT, Lee TD, Ronk M, Hefta SA (1995) Anal Biochem 224:235–244 [DOI] [PubMed]
- 10.Hara S, Katta V, Lu HS (2000) J Chromatogr A 867:151–160 [DOI] [PubMed]
- 11.Slysz GW, Schriemer DC (2003) Rapid Commun Mass Spectrom 17:1044–1050 [DOI] [PubMed]
- 12.Samskog J, Bylund D, Jacobsson SP, Markides KE (2003) J Chromatogr A 998:83–91 [DOI] [PubMed]
- 13.Calleri E, Temporini C, Perani E, Stella C, Rudaz S, Lubda D, Mellerio G, Veuthey J-L, Caccialanza G, Massolini G (2004) J Chromatogr A 1045:99–109 [DOI] [PubMed]
- 14.Calleri E, Temporini C, Perani E, De Palma A, Lubda D, Mellerio G, Sala A, Galliano M, Caccialanza G, Massolini G (2005) J Proteome Res 4:481–490 [DOI] [PubMed]
- 15.Palm AK, Novotny M (2004) Rapid Commun Mass Spectrom 18:1374–1382 [DOI] [PubMed]
- 16.Kato M, Inuzuka K, Sakai-Kato K, Toyo’oka T (2005) Anal Chem 77:1813–1818 [DOI] [PubMed]
- 17.Peterson DS, Rohr T, Svec F, Frechet JMJ (2002) Anal Chem 74:4081–4088 [DOI] [PubMed]
- 18.Geiser L, Eeltink S, Svec F, Frechet JMJ (2007) J Chromatogr A 1140:140–146 [DOI] [PMC free article] [PubMed]
- 19.Kele M, Guiochon G (2002) J Chromatogr A 960:19–49 [DOI] [PubMed]
- 20.Slysz GW, Schriemer DC (2005) Anal Chem 77:1572–1579 [DOI] [PubMed]
- 21.Nadler T, Blackburn C, Mark J, Gordon N, Regnier FE, Vella G (1996) J Chromatogr A 743:91–98 [DOI]
- 22.Riggs L, Sioma C, Regnier FE (2001) J Chromatogr A 924:359–368 [DOI] [PubMed]
- 23.Licklider L, Kuhr WG, Lacey MP, Keough T, Purdon MP, Takigiku R (1995) Anal Chem 67:4170–4177 [DOI]
- 24.Ye M, Hu S, Schoenherr RM, Dovichi NJ (2004) Electrophoresis 25:1319–1326 [DOI] [PubMed]
- 25.Zhao C, Jiang H, Smith DR, Bruckenstein S, Wood TD (2006) Anal Biochem 359:167–175 [DOI] [PubMed]
- 26.Krenkova J, Kleparnik K, Foret F (2007) J Chromatogr A 1159:110–118 [DOI] [PubMed]
- 27.Stenberg E, Persson B, Roos B, Urbanicky C (1991) J Colloid Interface Sci 143:513–526 [DOI]
- 28.Schwert GW, Takenaka Y (1955) Biochim Biophys Acta 16:570 [DOI] [PubMed]
- 29.Stigter ECA, De Jong GJ, Van Bennekom WP (2005) Biosens Bioelectron 21:474–482 [DOI] [PubMed]
- 30.Amankwa LN, Kuhr WG (1992) Anal Chem 64:1610–1613 [DOI]
- 31.Amankwa LN, Kuhr WG (1993) Anal Chem 65:2693–2697 [DOI]
- 32.Guo Z, Xu S, Lei Z, Zou H, Guo B (2003) Electrophoresis 24:3633–3639 [DOI] [PubMed]
- 33.Durgun G, Senay A, Cetinus A (2007) Food Chem 100:964–971 [DOI]
- 34.Bossi A, Guizzardi L, D’Acunto MR, Righetti PG (2004) Anal Bioanal Chem 378:1722–1728 [DOI] [PubMed]
- 35.Freije JR, Mulder PPMFA, Werkman W, Rieux L, Niederlander HAG, Verpoorte E, Bisschoff R (2005) J Proteome Res 4:1805–1813 [DOI] [PubMed]
- 36.Krenková J, Bilkova Z, Foret F (2005) J Sep Sci 28:1675–1684 [DOI] [PubMed]
- 37.Frazier RA, Matthijs G, Davies MC, Roberts CJ, Schacht E, Tendler SJB (2000) Biomaterials 21:957–966 [DOI] [PubMed]
- 38.McArthur SL, Mclean KM, Kingshott P, St John HAW, Chatelier RC, Griesser HJ (2000) Colloids Surf B 17:37–48 [DOI]
- 39.Jiang Y, Lee CS (2001) J Chromatogr A 924:315–322 [DOI] [PubMed]
- 40.Craft D, Li L (2005) Anal Chem 77:2649–2655 [DOI] [PubMed]
- 41.Feng S, Ye M, Jiang X, Jin W, Zou H (2006) J Proteome Res 5:422–428 [DOI] [PubMed]
- 42.Licklider L, Kuhr WG (1998) Anal Chem 70:1902–1908 [DOI] [PubMed]
- 43.López-Ferrer D, Capelo JL, Vázquez J (2005) J Proteome Res 4:1569–1574 [DOI] [PubMed]