Abstract
Swine Hepatitis E virus (HEV) could be a zoonotic agent for HEV infection in humans. In Canada, approximately 60% of 6-mo-old commercial pigs are seropositive for HEV; the prevalence is higher in the provinces of Quebec and Ontario. A study was set up to evaluate the presence of swine HEV in Quebec farms and to compare the strains detected in fecal samples with human and swine HEV strains reported worldwide. Fecal samples were collected randomly from May 2003 to January 2004 from 70 swine farms in Quebec. In 24 specimens, representing 34% of the visited farms, HEV RNA was extracted and detected by nested reverse-transcription polymerase chain reaction (RT-PCR). All amplified nested RT-PCR products were purified, cloned, and sequenced. The nucleotide sequences of a 304 base pair fragment at the 5′ end of the open reading frame 2 gene were determined. Phylogenetic analysis revealed that the 24 amplified fragments clustered in genotype 3 and had 85% to 99% nucleotide-sequence similarity with HEV strains identified in Japan, the United States, and Canada. Three strains identified in the study (swSTHY1, swSTHY31, and swSTHY47) showed 95% homology with 2 Japanese (swJ1-1 and HE-JA10) and 1 American (US1) HEV human strains.
Résumé
Le virus de l’Hépatite E (HEV) chez le porc pourrait être un agent zoonotique responsable d’infections chez l’humain. Au Canada, approximativement 60 % des porcs commercialisés de 6 mois sont séropositifs pour HEV, avec une prévalence plus élevée pour les provinces de Québec et de l’Ontario. Dans cette étude nous avons évalué la présence de HEV porcin dans différentes fermes du Québec et comparé les souches détectées dans les matières fécales avec les souches de HEV d’origine porcine et humaine retrouvées à travers le monde. Des échantillons de matière fécale de porc ont été récupérés de façon aléatoire de mai 2003 à janvier 2004 dans 70 fermes du Québec. L’ARN de HEV a été extrait des matières fécales et détecté par réaction d’amplification en chaîne nichée à l’aide de la transcriptase reverse (RT-PCR niché) dans 24 échantillons, représentant 34 % des fermes visitées. Tous les produits d’amplification obtenus par RT-PCR niché ont été purifiés, clonés et séquencés. Les séquences obtenues, d’une longueur de 304 paires de base, sont situées à l’extrémité 5′ du gène ORF2. L’analyse phylogénétique démontre que les 24 produits d’amplification de HEV porcin sont regroupés dans le génotype 3 et que ceux-ci possèdent une similarité de la séquence en nucléotides qui se situe entre 85 % et 99 % avec des séquences de HEV déjà identifiées au Japon, aux États-Unis et au Canada. Trois séquences identifiées dans cette étude (swSTHY1, swSTHY31 et swSTHY47) démontrent 95 % d’homologie avec deux souches japonaise (swJ1-1 et HE-JA10) et une souche américaine (US1) de HEV d’origine humaine.
(Traduit par les auteurs)
Introduction
Hepatitis E virus (HEV) is a nonenveloped icosahedral virus, approximately 27 to 34 nm in diameter, in the genus Hepevirus of the family Hepeviridae (1–4). The genome is a single-stranded RNA of approximately 7.2 kb. The viral RNA contains a short 5′ untranslated region (27 to 35 nucleotides) followed by 3 partially overlapping open reading frames (ORFs) and a 3′ untranslated region (65 to 74 nucleotides) and is polyadenylated. The ORF1 (5073 to 5124 nucleotides) encodes nonstructural proteins, ORF2 (1977 to 1980 nucleotides) encodes the capsid protein, and ORF3 (366 to 369 nucleotides) encodes a small cytoskeleton-associated phosphoprotein (4–8).
Viruses in the genus Hepevirus are subdivided into 4 genotypes (1, 2, 3, and 4), which are further subdivided into 5, 2, 10, and 7 subtypes, respectively. These 24 subtypes (1a, 1b, 1c, 1d, 1e, 2a, 2b, 3a, 3b, 3c, 3d, 3e, 3f, 3g, 3h, 3i, 3j, 4a, 4b, 4c, 4d, 4e, 4f, and 4g) are based on nucleotide differences from 5 phylogenies: 5′ ORF1, 3′ ORF1, 5′ ORF2, 3′ ORF2, and complete genome (4). Genotypes 1 and 2 are mostly associated with epidemics and outbreaks of hepatitis E that are usually due to fecal contamination of water supplies. These 2 genotypes are more conserved and comprised until recently only human isolates (4,9). Identification of genotype 1 HEV in samples from swine in Cambodia was reported in 2006 (9). Genotypes 3 and 4 include isolates from humans, swine, wild boar, and deer and show high diversity, most likely owing to their zoonotic origin (4). Genotype 1 has been found in tropical and several subtropical countries in Asia and Africa; genotype 2 has been found in Mexico and Nigeria; genotype 3 is distributed worldwide, having been isolated in Asia, Europe, Oceania, and North and South America; and genotype 4 has been found exclusively in Asia (4,10,11). The fecal–oral route is the predominant mode of transmission of HEV infection (5,7,12,13).
Although man is the natural host for HEV, successful infections of nonhuman primates and swine with human HEV strains have been reported (14,15). In addition, a swine HEV strain was able to infect rhesus monkeys, experimental surrogates of humans (13). The ability of HEV to cross species barriers puts humans at risk for infection with swine HEV. Studies have revealed HEV in swine populations from Australia, Britain, Canada, China, India, Indonesia, Japan, Korea, Mexico, the Netherlands, New Zealand, Spain, Taiwan, Thailand, and the United States (4,6,7,12,15–19). Meng et al (20,21) reported that pig handlers and veterinarians tend to have a higher prevalence of anti-HEV antibodies in their blood than the general population. Between 2% and 21% of blood donors are seropositive for HEV in countries where HEV is not endemic, such as Canada, France, Korea, Taiwan, and the United States (1,12,18,21–24). This strongly suggests that swine HEV could be a zoonotic agent for HEV infection in humans. In Canada, Yoo et al (25) estimated that approximately 60% of 6-mo-old commercial pigs are seropositive for HEV, the prevalence being higher in Quebec (88.8%) and Ontario (80.1%) than in the western and eastern Canadian provinces.
The aim of this study was to evaluate the presence of swine HEV RNA in various swine farms in the province of Quebec by nested reverse-transcription polymerase chain reaction (RT-PCR) and to characterize the 5′ end of the ORF2 region of amplified RNA fragments obtained from fecal samples and compare the nucleotide sequences with those of human and swine HEV isolates reported worldwide.
Materials and methods
Sample collection and RNA extraction
Fecal samples were randomly collected from May 2003 to January 2004 from 70 pig-fattening farms in different Quebec regions. In these farms the pigs were between 2 and 4 mo of age. Samples were collected directly from the pen’s floor (1 g of fecal material from 5 sites at each farm) and diluted in minimum essential medium, pH 7.4 (Invitrogen Canada, Burlington, Ontario) to obtain a final 20% suspension (w/v). Viral RNA was extracted from the clarified fecal suspensions with use of the QIAamp Viral RNA Mini Kit (Qiagen, Mississauga, Ontario) according to the manufacturer’s instructions.
Detection of HEV RNA by nested RT-PCR
We performed nested RT-PCR (26) using the external primer set 3156N (forward, 5′-AATTATGCYCAGTAYCGRGTTG-3′ ; nucleotide positions 5663 to 5684) and 3157N (reverse, 5′-CCCTT RTCYTGCTGMGCATTCTC-3′ ; nucleotide positions 6371 to 6393) to obtain an amplification product of 731 base pairs (bp) and the internal primer set 3158N (forward 5′-GTWATGCTYTGCATWCAT GGCT-3′ ; nucleotide positions 5948 to 5969) and 3159N (reverse 5′-AGCCGACGAAATCAATTCTGTC-3′ ; nucleotide positions 6274 to 6295). The nucleotide positions were based on the HEV reference sequence (GenBank accession number NC_001434). The expected nested PCR product size was 348 bp. Amplifications were performed with the Qiagen OneStep RT-PCR kit, according to the manufacturer’s instructions, and an Eppendorf Mastercycler gradient system (Brinkmann Instruments Canada, Mississauga, Ontario).
Cloning and sequencing of the RT-PCR products
The amplified 348-bp fragments were excised from the gel and purified by means of the QIAquick Gel Extraction Kit (Qiagen). The purified products were cloned into pCR 2.1 TOPO vector with use of the TOPO TA Cloning kit (Invitrogen) with TOP10 electrocompe-tent cells, in accordance with the manufacturer’s recommendations. Sequencing was performed on recombinant plasmids in both directions by means of a CEQ 8000 Genetic Analysis System (Beckman Coulter, Fullerton, California, USA) and a CEQ Dye Terminator Cycle Sequencing Kit (Beckman Coulter) with M13 forward and reverse primers. Nucleotide alignment was undertaken with the CLUSTAL W program (www.ebi.ac.uk/clustalw). The sequences of the forward and reverse internal primers (3158N and 3159N) were deleted for sequence analysis, which left a 304-bp sequence. The phylogenetic tree was created by the neighbor-joining method with CLC free Workbench 2.5.2 software (www.clcbio.com). The statistical confidence of the phylogenetic relationships was determined by bootstrap analysis. All sequences were deposited in GenBank under accession numbers DQ859989 to DQ860012.
Results
Presumptive positive 348-bp DNA fragments were detected in 24 of the 70 fecal samples, representing HEV detection at 34% of the visited swine farms. In this study, HEV RNA was detected mainly in regions C (43%) and D (41%); no HEV RNA was detected in the 8 farms visited in region A (Table I).
Table I.
Detection of Hepatitis E virus (HEV) in porcine feces from swine farms in 4 regions of the province of Quebec by reverse-transcription polymerase chain reaction
| Region | No. (and %) of HEV-positive farms | Strain identification |
|---|---|---|
| A | 0/8 (0) | |
| B | 4/14 (28) | swSTHY1, swSTHY21, |
| swSTHY31, swSTHY40 | ||
| C | 9/21 (43) | swSTHY9, swSTHY13, |
| swSTHY19, swSTHY20, | ||
| swSTHY22, swSTHY26, | ||
| swSTHY42, swSTHY51, | ||
| swSTHY62 | ||
| D | 11/27 (41) | swSTHY7, swSTHY10, |
| swSTHY11, swSTHY12, | ||
| swSTHY14, swSTHY17, | ||
| swSTHY18, swSTHY46, | ||
| swSTHY47, swSTHY66, | ||
| swSTHY67 | ||
| Total | 24/70 (34) |
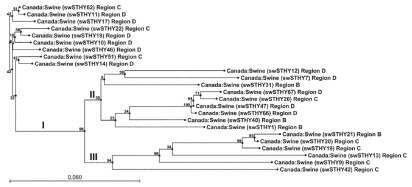
Nucleotide sequence analysis revealed that the 24 amplified fragments were all different in this genomic region and had 85% to 99% sequence similarity. The phylogenetic tree of the sequences showed 3 clusters: the sequences from region C were mainly in cluster III, those from region B were mainly in cluster II, and those from region D were distributed in clusters I and II (Figure 1). Translation into amino acid sequences revealed only 8 distinct HEV patterns, with differences of only 1 or 2 amino acids (data not shown), suggesting that most of the sequence variations occurred in the wobble position of amino acid codons.
Figure 1.
Phylogenetic tree created by the neighbor-joining method and based on a 304 base pair nucleotide sequence at the 5′ end of the open reading frame 2 gene of swine Hepatitis E virus (HEV) isolated in this study. Statistical confidence for the tree was assessed by bootstrap analysis; the bootstrap values are given on the tree. The bar represents 6% sequence divergence.
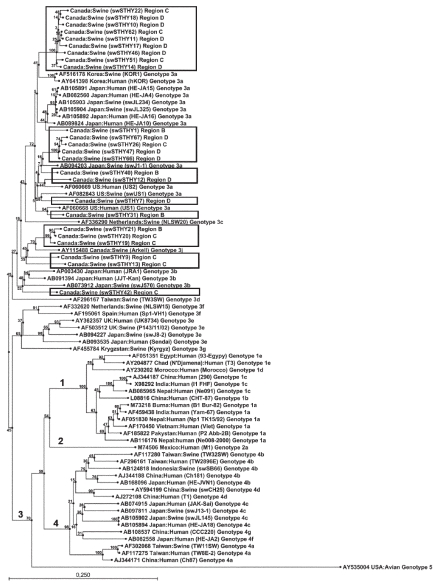
Comparison of the amplified fragments with human and swine HEV strains known worldwide obtained from GenBank confirmed that all the Quebec strains from this study belonged to genotype 3 and most (18/24) to genotype 3a, the other 6 belonging to genotypes 3j (5) and 3b (1) (Figure 2). Strains swSTHY22, swSTHY18, swSTHY10, swSTHY62, swSTHY11, swSTHY17, swSTHY46, swSTHY51, and swSTHY14 were dissimilar from other strains in genotype 3a and formed an independent cluster. Interestingly, swSTHY1 appeared in the same cluster as strains HE-JA4, HE-JA15, and HE-JA16, which had been isolated from humans after consumption of undercooked pig liver, and strains swJL234 and swJL325, which had been isolated from raw pig liver sold in grocery stores in Japan (11). However, the nucleotide homology between swSTHY1 and these Japanese strains ranged from 93% to 94%. Strains swSTHY67, swSTHY26, swSTHY47, swSTHY66, and swSTHY40 clustered with Japanese strain swJ1-1 as well as swSTHY12 and swSTHY7. Strain swSTHY31 clustered with the US swine strain swUS1 and the human strains US1 and US2. Strains swSTHY21, swSTHY20, swSTHY19, swSTHY9, and swSTHY13 all clustered with the Canadian swine HEV strain Arkell (18), forming a specific cluster. Finally, strain swSTHY42 was included in a different cluster with Japanese human and swine strains.
Figure 2.
Phylogenetic tree similarly created for human and swine HEV strains in GenBank and the swine strains isolated in this study (within the boxes). The bar represents 25% sequence divergence.
Discussion
In previous studies, the reported prevalence of HEV antibodies varied from 25% to 89% in adult swine in different provinces in Canada (25) and from 56% to 100% in adult swine in different herds in the Midwestern United States, Thailand, and Korea (15,20). Despite this high prevalence, the rate of detection of HEV RNA in serum samples was generally lower (27). The age of the animal and the sample type may have a strong influence on the detection results (26,27). Because the concentration of HEV particles excreted is often higher in feces than in blood (12), molecular detection seems more sensitive in fecal samples. Pooled swine feces collected from the pen floor can provide a good indication of the presence of HEV in the environment and avoids direct sampling from animals. Recent studies in Iowa and the Netherlands revealed that 25% and 22%, respectively, of the swine farms tested were positive for HEV with the use of pooled pig feces (28,29).
The MJ-C region of the HEV RdRp domain is sufficient in length and variability to provide a phylogenetic analysis that correlates with that of the full genome (30). The 5′ end of the ORF2 gene is one of the most conserved regions among known swine and human HEV strains but is also used for HEV molecular characterization (4,11,17,26,31). All the swine HEV detected in Quebec in this study belonged to genotype 3; however, phylogenetic analysis based on a 304-bp nucleotide sequence of the 5′ end of the ORF2 gene did not regroup all these strains in the same subtype. The Japanese strains are also distributed in different clusters within genotype 3 (4,31–33). Phylogenetic analysis in a US study revealed that all their HEV swine isolates clustered with human HEV strains US1 and US2 and swine US1 in subtype 3b (26). In Korea, isolates were also very highly associated with subtype 3a (1).
Direct evidence of HEV transmission from pigs to humans is not clearly demonstrated, but some swine and human sequences were 97% to 100% homologous (4). Moreover, 100% homology has been found between a swine HEV isolate from raw pig liver sold in a Japanese grocery store and a human isolate from a person who had severe HEV clinical infection (13). Another pair of swine and human genotype 4 HEV strain sequences with 99% homology over the entire genome was recently reported in Japan (34). In addition, a phylogenetic study conducted in Korea in 2005 demonstrated that HEV Korean isolates from humans and swine were very highly associated, showing as much as 99.6% homology (1). To our knowledge, no clinical human cases have yet been associated with food or waterborne HEV in Canada. Consequently, no association can be made between human and swine HEV in this country. However, strains swSTHY1, swSTHY31, and swSTHY47 identified in this study showed 95% homology with the human Japanese strains swJ1-1 and HE-JA10 and the US human strain US1. It will be important to continue monitoring HEV because of its zoonotic potential.
The present study indicates that HEV is present in some swine farms in different regions of Quebec, as suggested by the pigs’ serologic status (25). Sequence analysis revealed that the 24 amplified sequences were distinct at the nucleotide level in the 5′ ORF2 region and clustered in genotype 3. Most of these strains were more closely related to the Japanese strains than to the Canadian Arkell strain and the US strains.
References
- 1.Ahn JM, Kang SG, Lee DY, Shin SJ, Yoo HS. Identification of novel human hepatitis E virus (HEV) isolates and determination of the seroprevalence of HEV in Korea. J Clin Microbiol. 2005;43:3042–3048. doi: 10.1128/JCM.43.7.3042-3048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerson SU, Anderson D, Arankalle A, et al. Hepevirus. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy; Eighth Report of the International Committee on Taxonomy of Viruses. London: Elsevier/Academic Press; 2005. pp. 853–857. [Google Scholar]
- 3.Goens SD, Perdue ML. Hepatitis E viruses in humans and animals. Anim Health Res Rev. 2004;5:145–156. doi: 10.1079/ahr200495. [DOI] [PubMed] [Google Scholar]
- 4.Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- 5.Aggarwal R, Krawczynski K. Hepatitis E: an overview and recent advances in clinical and laboratory research. J Gastroenterol Hepatol. 2000;15:9–20. doi: 10.1046/j.1440-1746.2000.02006.x. [DOI] [PubMed] [Google Scholar]
- 6.Chobe LP, Lole KS, Arankalle VA. Full genome sequence and analysis of Indian swine hepatitis E virus isolate of genotype 4. Vet Microbiol. 2006;114:240–251. doi: 10.1016/j.vetmic.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Emerson SU, Purcell RH. Hepatitis E virus. Rev Med Virol. 2003;13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 8.Erker JC, Desai SM, Schlauder GG, Dawson GJ, Mushahwar IK. A hepatitis E virus variant from the United States: molecular characterization and transmission in cynomolgus macaques. J Gen Virol. 1999;80:681–690. doi: 10.1099/0022-1317-80-3-681. [DOI] [PubMed] [Google Scholar]
- 9.Caron M, Enouf V, Than SC, Dellamonica L, Buisson Y, Nicand E. Identification of genotype 1 hepatitis E virus in samples from swine in Cambodia. J Clin Microbiol. 2006;44:3440–3442. doi: 10.1128/JCM.00939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buisson Y, Grandadam M, Nicand E, et al. Identification of a novel hepatitis E virus in Nigeria. J Gen Virol. 2000;81:903–909. doi: 10.1099/0022-1317-81-4-903. [DOI] [PubMed] [Google Scholar]
- 11.Yazaki Y, Mizuo H, Takahashi M, et al. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J Gen Virol. 2003;84:2351–2357. doi: 10.1099/vir.0.19242-0. [DOI] [PubMed] [Google Scholar]
- 12.Choi IS, Kwon HJ, Shin NR, Yoo HS. Identification of swine hepatitis E virus (HEV) and prevalence of anti-HEV antibodies in swine and human populations in Korea. J Clin Microbiol. 2003;41:3602–3608. doi: 10.1128/JCM.41.8.3602-3608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng XJ, Halbur PG, Shapiro MS, et al. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol. 1998;72:9714–9721. doi: 10.1128/jvi.72.12.9714-9721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbur PG, Kasorndorkbua C, Gilbert C, et al. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J Clin Microbiol. 2001;39:918–923. doi: 10.1128/JCM.39.3.918-923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng XJ, Purcell RH, Halbur PG, et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banks M, Heath GS, Grierson SS, et al. Evidence for the presence of hepatitis E virus in pigs in the United Kingdom. Vet Rec. 2004;154:223–227. doi: 10.1136/vr.154.8.223. [DOI] [PubMed] [Google Scholar]
- 17.Cooper K, Huang FF, Batista L, et al. Identification of genotype 3 hepatitis E virus (HEV) in serum and fecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. J Clin Microbiol. 2005;43:1684–1688. doi: 10.1128/JCM.43.4.1684-1688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei Y, Yoo D. Genetic characterization and sequence heterogeneity of a Canadian isolate of swine hepatitis E virus. J Clin Microbiol. 2002;40:4021–4029. doi: 10.1128/JCM.40.11.4021-4029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi M, Nishizawa T, Okamoto H. Identification of a genotype III swine hepatitis E virus that was isolated from a Japanese pig born in 1990 and that is most closely related to Japanese isolates of human hepatitis E virus. J Clin Microbiol. 2003;41:1342–1343. doi: 10.1128/JCM.41.3.1342-1343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng XJ, Dea S, Engle RE, et al. Prevalence of antibodies to the hepatitis E virus in pigs from countries where hepatitis E is common or is rare in the human population. J Med Virol. 1999;59:297–302. [PubMed] [Google Scholar]
- 21.Meng XJ, Wiseman B, Elvinger F, et al. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J Clin Microbiol. 2002;40:117–122. doi: 10.1128/JCM.40.1.117-122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin CC, Wu JC, Chang TT, et al. Diagnostic value of immunoglobulin G (IgG) and IgM anti-hepatitis E virus (HEV) tests based on HEV RNA in an area where hepatitis E is not endemic. J Clin Microbiol. 2000;38:3915–3918. doi: 10.1128/jcm.38.11.3915-3918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas DL, Yarbough PO, Vlahov D, et al. Seroreactivity to hepatitis E virus in areas where the disease is not endemic. J Clin Microbiol. 1997;35:1244–1247. doi: 10.1128/jcm.35.5.1244-1247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansuy JM, Peron JM, Abravanel F, et al. Hepatitis E in the south west of France in individuals who have never visited an endemic area. J Med Virol. 2004;74:419–424. doi: 10.1002/jmv.20206. [DOI] [PubMed] [Google Scholar]
- 25.Yoo D, Wilson P, Pei Y, et al. Prevalence of hepatitis E virus antibodies in Canadian swine herds and identification of a novel variant of swine hepatitis E virus. Clin Diagn Lab Immunol. 2001;8:1213–1219. doi: 10.1128/CDLI.8.6.1213-1219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang FF, Haqshenas G, Guenette DK, et al. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol. 2002;40:1326–1332. doi: 10.1128/JCM.40.4.1326-1332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi M, Nishizawa T, Tanaka T, Tsatsralt-Od B, Inoue J, Okamoto H. Correlation between positivity for immunoglobulin A antibodies and viraemia of swine hepatitis E virus observed among farm pigs in Japan. J Gen Virol. 2005;86:1807–1813. doi: 10.1099/vir.0.80909-0. [DOI] [PubMed] [Google Scholar]
- 28.Kasorndorkbua C, Opriessing T, Huang FF, et al. Infectious swine hepatitis E virus is present in pig manure storage facilities on United States farms, but evidence of water contamination is lacking. Appl Environ Microbiol. 2005;71:7831–7837. doi: 10.1128/AEM.71.12.7831-7837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Poel WHM, Verschoor F, van der Heide R, et al. Hepatitis E virus sequences in swine related to sequences in humans, the Netherlands. Emerg Infect Dis. 2001;7:970–976. doi: 10.3201/eid0706.010608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai L, Dai X, Meng J. Hepatitis E virus genotyping based on full-length genome and partial genomic regions. Virus Res. 2006;120:57–69. doi: 10.1016/j.virusres.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi M, Nishizawa T, Miyajima H, et al. Swine hepatitis E virus strains in Japan form four phylogenetic clusters comparable with those of Japanese isolates of human hepatitis E virus. J Gen Virol. 2003;84:851–862. doi: 10.1099/vir.0.18918-0. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka Y, Takahashi K, Orito E, et al. Molecular tracing of Japan-indigenous hepatitis E viruses. J Gen Virol. 2006;87:949–954. doi: 10.1099/vir.0.81661-0. [DOI] [PubMed] [Google Scholar]
- 33.Tokita H, Harada H, Gotanda Y, Takahashi M, Nishizawa T, Okamoto H. Molecular and serological characterization of sporadic acute hepatitis E in a Japanese patient infected with a genotype III hepatitis E virus in 1993. J Gen Virol. 2003;84:421–427. doi: 10.1099/vir.0.18802-0. [DOI] [PubMed] [Google Scholar]
- 34.Nishizawa T, Takahashi M, Mizuo H, Miyajima H, Gotanda Y, Okamoto H. Characterization of Japanese swine and human hepatitis E virus isolates of genotype IV with 99% identity over the entire genome. J Gen Virol. 2003;84:1245–1251. doi: 10.1099/vir.0.19052-0. [DOI] [PubMed] [Google Scholar]