Abstract
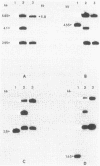
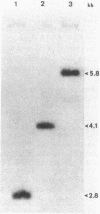
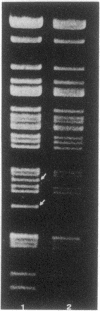
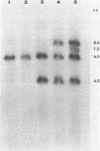
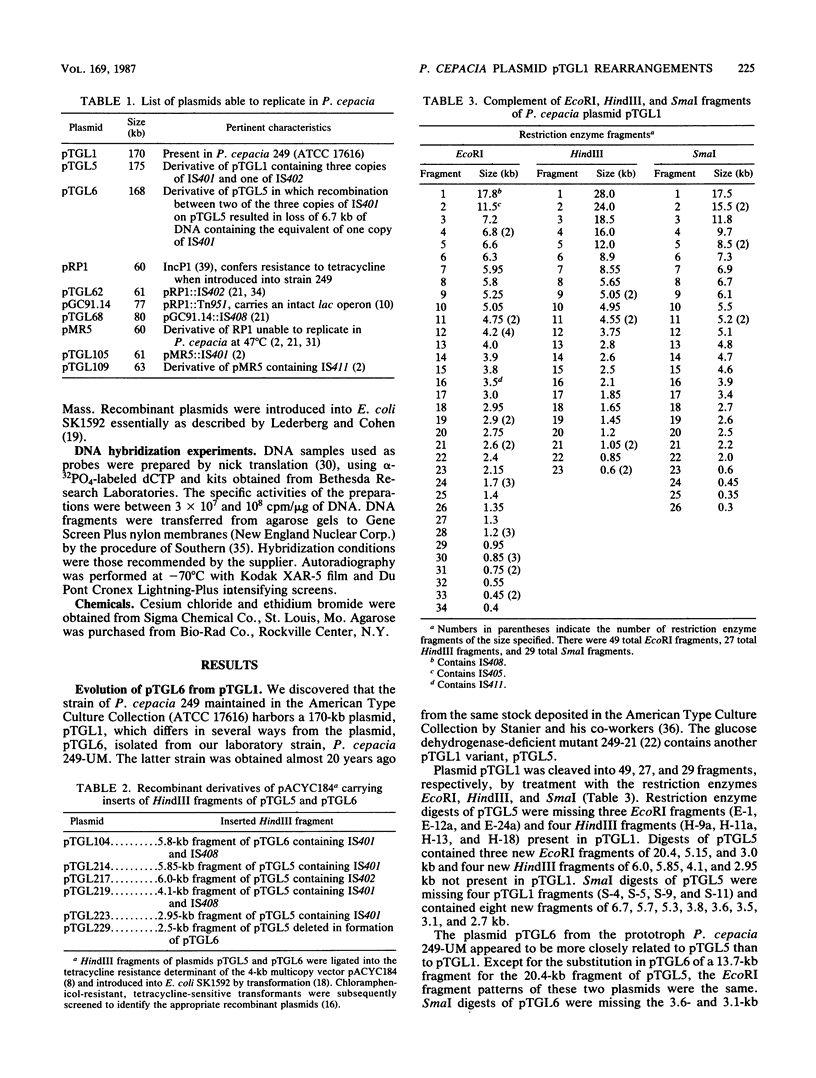
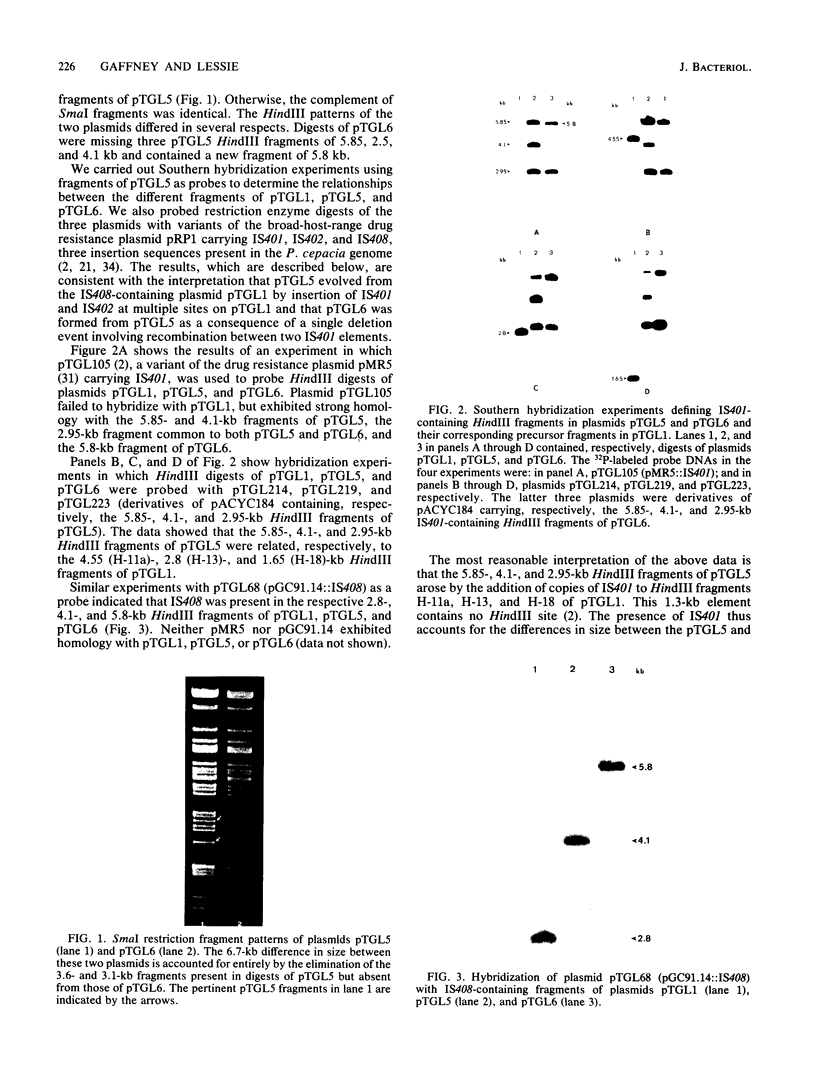
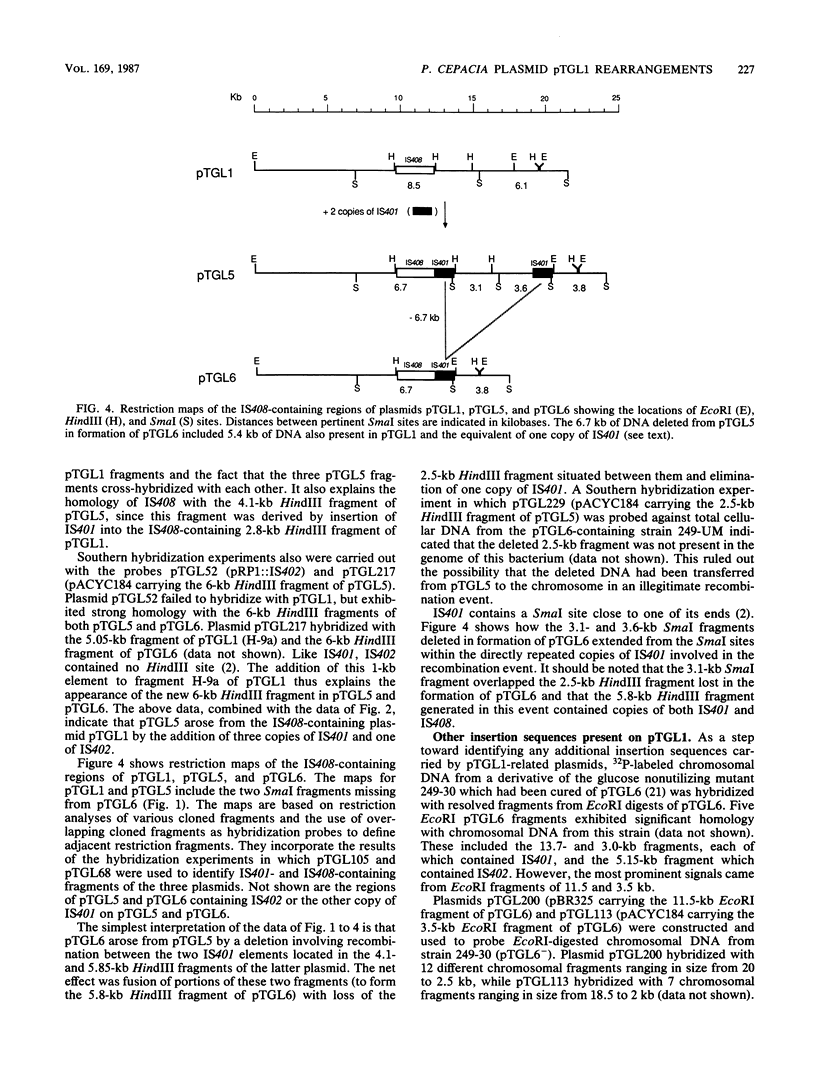
Pseudomonas cepacia 249 (ATCC 17616) harbors a 170-kilobase (kb) plasmid designated pTGL1. We identified three insertion sequences, IS405, IS408, and IS411, on this plasmid. Various prototrophic and auxotrophic derivatives in our collection contained variants of pTGL1 formed by accretion and deletion of other elements. Plasmid pTGL6, the variant in one prototroph, evolved from pTGL1 by the addition of three copies of IS401 (1.3 kb) and one of IS402 (1 kb), to generate pTGL5, and recombination between two of the copies of IS401 on pTGL5 to form pTGL6. The latter event entailed loss of one copy of IS401 and an additional 5.4 kb of plasmid DNA. Derivatives of the broad-host-range plasmid pRP1 carrying the above insertion sequences and recombinant plasmids carrying fragments of plasmids pTGL6 and pTGL5 were used as probes to ascertain the extent of reiteration of the various elements in the P. cepacia genome. The data indicate a high frequency of genomic rearrangements which presumably contributes to the extraordinary adaptability of this bacterium.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballard R. W., Palleroni N. J., Doudoroff M., Stanier R. Y., Mandel M. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970 Feb;60(2):199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- Barsomian G., Lessie T. G. Replicon fusions promoted by insertion sequences on Pseudomonas cepacia plasmid pTGL6. Mol Gen Genet. 1986 Aug;204(2):273–280. doi: 10.1007/BF00425509. [DOI] [PubMed] [Google Scholar]
- Beckman W., Gaffney T., Lessie T. G. Correlation between auxotrophy and plasmid alteration in mutant strains of Pseudomonas cepacia. J Bacteriol. 1982 Mar;149(3):1154–1158. doi: 10.1128/jb.149.3.1154-1158.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman W., Lessie T. G. Response of Pseudomonas cepacia to beta-Lactam antibiotics: utilization of penicillin G as the carbon source. J Bacteriol. 1979 Dec;140(3):1126–1128. doi: 10.1128/jb.140.3.1126-1128.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J. O., Atherly A. G. Induced plasmid-genome rearrangements in Rhizobium japonicum. J Bacteriol. 1984 Jan;157(1):218–224. doi: 10.1128/jb.157.1.218-224.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Kosuge T. Transposable element that causes mutations in a plant pathogenic Pseudomonas sp. J Bacteriol. 1983 Jun;154(3):1162–1167. doi: 10.1128/jb.154.3.1162-1167.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G., Ghosal D., Saedler H. Multiple integration sites for the lactose transposon Tn 951 on plasmid RP 1 and establishment of a coordinate system for Tn 951. Mol Gen Genet. 1979 Jan 5;168(1):61–67. doi: 10.1007/BF00267934. [DOI] [PubMed] [Google Scholar]
- Curiale M. S., Mills D. Integration and partial excision of a cryptic plasmid in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1982 Nov;152(2):797–802. doi: 10.1128/jb.152.2.797-802.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier T. C., Nester E. W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976 Dec;76(2):431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- DasSarma S., RajBhandary U. L., Khorana H. G. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2201–2205. doi: 10.1073/pnas.80.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederer G. M., Matsen J. M. Colonization and infection with Pseudomonas cepacia. J Infect Dis. 1972 Jun;125(6):613–618. doi: 10.1093/infdis/125.6.613. [DOI] [PubMed] [Google Scholar]
- Heckman J. E., RajBhandary U. L. Organization of tRNA and rRNA genes in N. crassa mitochondria: intervening sequence in the large rRNA gene and strand distribution of the RNA genes. Cell. 1979 Jul;17(3):583–595. doi: 10.1016/0092-8674(79)90266-6. [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbane J. J., Chatterjee D. K., Karns J. S., Kellogg S. T., Chakrabarty A. M. Biodegradation of 2,4,5-trichlorophenoxyacetic acid by a pure culture of Pseudomonas cepacia. Appl Environ Microbiol. 1982 Jul;44(1):72–78. doi: 10.1128/aem.44.1.72-78.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. N., Lessie T. G. Purification and characterization of the two 6-phosphogluconate dehydrogenase species from Pseudomonas multivorans. J Bacteriol. 1974 Dec;120(3):1043–1057. doi: 10.1128/jb.120.3.1043-1057.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessie T. G., Berka T., Zamanigian S. Pseudomonas cepacia mutants blocked in the direct oxidative pathway of glucose degradation. J Bacteriol. 1979 Jul;139(1):323–325. doi: 10.1128/jb.139.1.323-325.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano F. E., Kaplan S. Plasmid rearrangements in the photosynthetic bacterium Rhodopseudomonas sphaeroides. J Bacteriol. 1984 Jun;158(3):1094–1103. doi: 10.1128/jb.158.3.1094-1103.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Weidinger G., Goebel W. Genetic variability in Halobacterium halobium. J Bacteriol. 1981 Jan;145(1):375–381. doi: 10.1128/jb.145.1.375-381.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priefer U. B., Burkardt H. J., Klipp W., Pühler A. ISR1: an insertion element isolated from the soil bacterium Rhizobium lupini. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):87–91. doi: 10.1101/sqb.1981.045.01.016. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinson M. K., Bennett P. M., Falkow S., Dodd H. M. Isolation of a temperature-sensitive derivative of RP1. Plasmid. 1980 May;3(3):343–347. doi: 10.1016/0147-619x(80)90047-5. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Long S. R., Meade H. M., van den Bos R. C., Ausubel F. M. ISRm1: A Rhizobium meliloti insertion sequence that transposes preferentially into nitrogen fixation genes. J Mol Appl Genet. 1982;1(5):405–418. [PubMed] [Google Scholar]
- Sapienza C., Rose M. R., Doolittle W. F. High-frequency genomic rearrangements involving archaebacterial repeat sequence elements. Nature. 1982 Sep 9;299(5879):182–185. doi: 10.1038/299182a0. [DOI] [PubMed] [Google Scholar]
- Scordilis G. E., Ree H., Lessie T. G. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J Bacteriol. 1987 Jan;169(1):8–13. doi: 10.1128/jb.169.1.8-13.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Szabo L. J., Mills D. Integration and excision of pMC7105 in Pseudomonas syringae pv. phaseolicola: involvement of repetitive sequences. J Bacteriol. 1984 Mar;157(3):821–827. doi: 10.1128/jb.157.3.821-827.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Stalker D. M., Helinski D. R. Replication and incompatibility properties of segments of the origin region of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):1–7. doi: 10.1007/BF00338996. [DOI] [PubMed] [Google Scholar]