Charcot–Marie–Tooth (CMT) disease is the most common hereditary neuropathy. CMT falls into two main forms: the demyelinating CMT type 1 with decreased nerve conduction velocities and the axonal CMT type 2. CMT2 is further subtyped by linkage analysis into >10 loci, with eight genes identified.
Recently, mutations in the mitochondrial fusion protein 2 (MFN2) gene were reported in families with CMT2A1 and additional mutations have been detected in other studies, bringing to 42 the total number of different MFN2 mutations described thus far.2,3,4
In the current study, we report a novel MFN2 mutation shared by two apparently unrelated CMT2 families originating from the same area in Southern Italy.
Vertical transmission and male‐to‐male inheritance were documented in both families, indicating that CMT2 segregates as an autosomal dominant trait. In family 1, 14 affected individuals were identified in four generations (three deceased before the study). After giving informed consent, eight affected individuals and 12 unaffected family members were examined by neurologists and enrolled in the genetic study. All affected individuals showed bilateral pes cavus, lower extremity wasting and steppage gait; only two of eight affected family members (IV‐6, IV‐7) complained of leg pain. Electrophysiological examination revealed decreased compound motor action potential (CMAP) and sensory action potential (SAP) amplitudes and mildly slowed motor and sensory nerve conductions that were consistent with axonal neuropathy. Age at onset was very variable and ranged from the first decade (IV‐6 and IV‐7) to the fifth decade (III‐5 and III‐7). Generally, the affected subjects belonging to the last generation showed an early onset that appeared to correlate with a faster progression of the disease and a more severe phenotype.
In family 2, six subjects with CMT2 were reported in three generations. Two (mother and son) signed the informed consent to be enrolled in the clinical and genetic study.
The proband (indicated by an arrow in fig 1) noticed pes cavus and walking difficulties at age 10 years. On examination at age 33 years, he showed bilateral pes cavus, steppage gait, distal muscle weakness and wasting (mild in the upper limbs and moderate in the lower limbs), reduced to absent deep tendon reflexes and mild sensory loss in both feet. He also had postural upper limb tremor. Nerve conduction studies showed reduced amplitudes of CMAPs and SAPs; nerve conduction velocities were preserved in the upper limbs and mildly slowed in the lower limbs. His mother showed a less severe phenotype. On examination at age 61 years, she had slight pes cavus and mild steppage gait; wasting and weakness in distal muscle was minor in the upper limbs but clear in the lower limbs, ankle jerks were absent and there was foot sensory loss. On nerve conduction studies, CMAP and SAP amplitudes were slightly decreased to normal in the upper limbs and reduced in the lower limbs; nerve conduction velocities were mildly decreased only in the lower limbs.
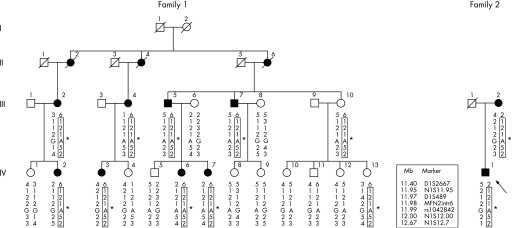
Figure 1 Pedigrees of the CMT2A families. Markers D1S489, MFN2intr6 and SNP rs1042842 are located, respectively, in intron 2, intron 6 and exon 19 of MFN2, whereas D1S2667, N1S11.95, N1S12.00 and N1S12.7 map in the immediate vicinity of the gene. Markers N1S11.95, MFN2intr6 and N1S12.7 are microsatellite sequences starting respectively at positions 11954818, 11981194 and 12006003 of the chromosome 1 assembly (UCSC hg18, NCBI Build 36.1). Boxes indicate the at risk haplotype cosegregating with the p.A383V (*) mutation in both families.
Mutational screening of the MFN2 gene was performed by denaturing high performance liquid chromatography (Transgenomic WAVE system).
In the index cases of both families, denaturing high performance liquid chromatography analysis, followed by direct sequencing of the PCR product of exon 11, revealed a heterozygous C>T transition at position 1148 (c.1148C>T) leading to the p.A383V amino acid substitution. Analysis of the other family members confirmed the presence of the mutation in all CMT2 affected subjects and in two unaffected subjects of family 1 (III‐10 and IV‐13). Neurological and EMG examinations of these subjects, 70 years and 36 years old, respectively, revealed no clinical signs or symptoms or any electrophysiological motor or sensory nerve impairment. The possibility of a common polymorphism was excluded as the mutation was not detected in 440 control chromosomes.
The presence of the same mutations in unrelated cases may be ascribed to two different events, namely a founder effect or the presence of a mutational hot spot. In order to explore these hypotheses, seven polymorphic markers (three intragenic and four close to MFN2) have been analysed on chromosome 1p36.22. In family 1, an at risk haplotype segregates in all of the affected subjects. Interestingly, the affected subjects of family 2 share the same at risk alleles for six out of seven markers (fig 1) thus indicating that our finding is more likely the result of a founder effect rather than a mutational hot spot. In addition, the fact that both families originate from the same geographic area supports the hypothesis that the mutation may have been inherited from a common ancestor.
In common with previously reported MFN2 mutations, CMT2 phenotype associated with the p.A383V substitution demonstrates consistent variability in expressivity ranging from late onset with mild neuropathy to early onset with more severe phenotype. In addition, the mutation was detected in two asymptomatic subjects, thus supporting the incomplete penetrance of MFN2 mutations, as already reported by Lawson and collaborators.5
The clinical variability and the incomplete penetrance underline the importance of other factors in defining the phenotype. Indeed, the presence of two non‐penetrant cases in the same family branch corroborate the importance of the genetic background suggesting that an interacting gene(s) may modulate the CMT2A phenotype or protect it against pathological mutations.
Mitofusin 2 (MFN2) is a mitochondrial transmembrane GTPase which is located on the outer mitochondrial membrane and regulates the mitochondrial network architecture by fusion of mitochondria. Most of the mutations previously reported are within or immediately upstream of the GTPase domain (residues 103–261) or within the two coiled coil domains (residues 408–433 and 724–752, respectively). The mutation p.A383V described here is located on exon 11 of the MFN2 gene. Nine additional mutations have been identified in this exon2,3,4,5 and two of them (p.H361Y and p.R364W) have been reported in families affected by axonal CMT associated with optic atrophy. In our patients no visual impairments were observed.
Interestingly, all of the mutations identified in exon 11 involve amino acids highly conserved across different species. In particular, the alanine at position 383 is conserved in both mouse Mfn2 and the cognate protein mitofusin 1 (Mfn1). Together, these findings suggest that, even if exon 11 is not located in a functionally characterised domain, the region it encodes plays a crucial role for the function or stability of the protein. From this standpoint, it remains to be established whether the mutation exerts its noxious effect by a dominant negative mechanism or haploinsufficiency. Further investigations, including the study of heterozygous MFN2 knock‐in mouse, are necessary to clarify the function of normal and mutated MFN2 gene.
Acknowledgements
The financial support of Telethon‐UILDM (grants GUP04009 to FT and MM, and GUP02169 to DP) and Fondazione Pierfranco e Luisa Mariani (grant R‐05‐44 to FT and AQ) is gratefully acknowledged.
Footnotes
Competing interests: None.
References
- 1.Züchner S, Mersiyanova I V, Muglia M.et al Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot–Marie–Tooth neuropathy type 2A. Nat Genet 200436449–451. [DOI] [PubMed] [Google Scholar]
- 2.Verhoeven K, Claeys K G, Zuchner S.et al MFN2 mutation distribution and genotype/phenotype correlation in Charcot–Marie–Tooth type 2. Brain 20061292093–2102. [DOI] [PubMed] [Google Scholar]
- 3.Chung K W, Kim S B, Park K D.et al Early onset severe and late‐onset mild Charcot–Marie–Tooth disease with mitofusin 2 (MFN2) mutations. Brain 20061292103–2118. [DOI] [PubMed] [Google Scholar]
- 4.Engelfried K, Vorgerd M, Hagedorn M.et al Charcot–Marie–Tooth neuropathy type 2A: novel mutations in the mitofusin 2 gene (MFN2). BMC Med Genet 2006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawson V H, Graham B V, Flanigan K M. Clinical and electrophysiologic features of CMT2A with mutations in the mitofusin 2 gene. Neurology 200565197–204. [DOI] [PubMed] [Google Scholar]