Abstract
Giant axonal neuropathy (GAN; MIM 256850) is a severe childhood onset autosomal recessive sensorimotor neuropathy affecting both the peripheral nerves and the central nervous system. Bomont and colleagues identified a novel ubiquitously expressed gene they named Gigaxonin on chromosome 16q24 as the cause of GAN in a number of families. We analysed five families with GAN for mutations in the Gigaxonin gene and mutations were found in four families; three families had homozygous mutations, one had two compound heterozygous mutations and one family had no mutation identified. All families had the typical clinical features, kinky hair and nerve biopsy. We report some unusual clinical features associated with GAN and Gigaxonin mutations as well as confirm the heterogeneity in GAN and the identification of two families with manifesting carriers.
In 1972, Asbury and Gale1 and Berg and colleagues2 described the first case of giant axonal neuropathy (GAN) in a 6‐year‐old girl with kinky hair who had an age of onset of 3 years with progressive clumsiness, muscle weakness and atrophy, areflexia and sensory loss. Sural nerve biopsy revealed large axonal spheroids densely packed with neurofilaments in both myelinated and unmyelinated fibres. Since the original report that carefully defined GAN, many similar patients and families with this severe autosomal recessive motor and sensory neuropathy have been reported throughout the world.3
The majority of cases of GAN have an age of onset of approximately 3 years with developmental delay, patients have similar faces, usually kinky hair, an axonal neuropathy and CNS abnormalities.3,4 Sural nerve biopsy shows giant axons, a pathological feature that is also found in other neuropathies associated with toxins, such as acrylamide, iminodipropionitrile, methyl‐n‐butyl ketone and n‐hexane.3 Giant axons have also been described in a litter of German shepherd kinky haired dogs and other forms of Charcot–Marie–Tooth disease, including families with neurofilament light chain5 and KIAA19856 mutations.
The gene causing GAN was identified by Bomont and colleagues7 and is called Gigaxonin. They identified seven homozygous mutations and eight compound heterozygous mutations in families with GAN. Three typical families with GAN based on clinical and pathological families had no mutation in the Gigaxonin gene,8 suggesting possible heterogeneity. Mutations in the Gigaxonin gene have been identified in a number of other families.9,10,11,12,13
We have identified five families with GAN. Gigaxonin mutations were found in four families. Clinical and pathological details are described in these kindreds and we speculate on the genotype phenotype effect of these Gigaxonin mutations (table 1).
Table 1 Families with giant axonal neuropathy and Gigaxonin mutations identified.
| Family No | Age of onset | Diagnosis | Affected | Origin | Hair | Nerve biopsy |
|---|---|---|---|---|---|---|
| 1 | 3 y | GAN | 1 | England | Kinky | GAN (see fig 1) |
| 2 | II‐1: 4 y | GAN | 5 | Pakistan | Kinky | GAN |
| II‐5: 21 mo | ||||||
| 3 | II‐1: 5 y | GAN | 2, 2 possibly affected | Pakistan | Kinky | GAN |
| II‐2: 5 y | ||||||
| 4 | <2 y | GAN | 1 | Scotland | Kinky | GAN |
| 5 | 25 mo | GAN | 1 | Canada | Kinky | GAN |
| Family No | Exon/intron | Nucleotide change | Amino acid | Domain | Consanguinity | Mutation type |
|---|---|---|---|---|---|---|
| 1 (proband and mother) | Exon 5 | c.944 CCG to CTG | Pro 315 Leu | Kelch | No | Compound heterozygous |
| 1 (proband and father) | Exon 10 | 1553/1554 Del TT | Phe 518Fs | Kelch | No | Compound heterozygous |
| Probable NMD | ||||||
| 2 | Exon 1 | c.151 GCC to CCC | Ala 51 Pro | BTB | Yes | Homozygous |
| 3 | Exon 10 | c.1505 TGG to TAG | Trp 502 Stop | Kelch | Yes | Homozygous |
| Probable NMD | ||||||
| 4 | Exon 2 | c.213 TAT to TAA | Tyr 71 Stop | BTB | No | Homozygous |
| Probable NMD | ||||||
| 5 | No mutation identified in the coding region or flanking introns | No |
c., cDNA nucleotide numbering; GAN, Giant axonal neuropathy; Fs, frameshift; NMD, non‐sense mediated decay.
Methods
Ethics approval was obtained from the joint medical and ethics committee. DNA was extracted from blood samples obtained with informed consent. DNA from family No 5 was obtained from a cell pellet from the Montreal Children's Hospital Research Institute. The 11 Gigaxonin exons and flanking introns were sequenced, as previously described.7 Mutations were rechecked by sequencing in the proband and other family members. None of the mutations was identified in 100 control cases. The method and primer sequences are available on request.
Results
GAN families (table 1) were identified from England (family No 1), Pakistan (family Nos 2 and 3), Scotland (family No 4) and Canada (family No 5). Sequencing of the Gigaxonin gene revealed three homozygous mutations in the Gigaxonin gene in family Nos 2, 3 and 4. Family No 1 had two different compound heterozygous mutations (table 1). These conserved mutations were frameshift, non‐sense or missense and were not present in 100 mixed Caucasian or Asian controls.
No previous Pakistani, English or Scottish GAN families have been reported in the past. No mutations were identified in the Canadian GAN family; this family was clinically typical for GAN with severe childhood axonal neuropathy, age of onset in childhood and typical kinky hair. The fibroblasts from this case have been studied previously and found to have errors in the organisation of intermediate filaments.14
Clinical details
All four families with GAN were diagnosed prior to genetic studies. A clinical and electrophysiological, motor more than sensory, axonal neuropathy was present in all families with GAN, as were abnormal CNS findings. Family Nos 1 and 4 had particularly prominent cranial nerve abnormalities. Family No 2 had additional epilepsy and respiratory problems in the most severely affected sibling. Two cousins of the affected siblings in family No 2 also had a similar progressive neurological condition. One of the cousins had the distinctive hair and was found to be a heterozygous carrier of the mutation while the other had normal hair but died before testing was available. In family No 3, the most severely affected sibling had dysphagia, visual problems with dense optic atrophy, and problems with feeding and secretions. Family No 4 had an onset with delayed walking problems and developed early gastrointestinal reflux and regurgitation. Cranial nerve signs were prominent and she was hypotonic with brisk reflexes and scoliosis. MRI of the brain was carried out in family Nos 1, 2 and 4 and showed diffuse high T2 signal abnormalities.
The patient from family No 1 had an age of onset of 2–3 years and from then on developed progressive difficulties, with weakness of his feet and ankles and walking problems. His mother has a diagnosis of “mild multiple sclerosis”, diagnosed after problems in her legs and eyes. Her MRI was consistent with demyelination and was typical of multiple sclerosis, and lumbar puncture showed unmatched oligoclonal bands. His father was normal and there was no family history of consanguinity. At the age of 24 years the patient had blond frizzy hair that was different to his parents, and he walked with bilateral foot drop and an ataxic gait. He had a left ptosis, horizontal nystagmus in all directions, mild bilateral facial weakness and a cerebellar dysarthria. He had a kyphoscoliosis with weakness, wasting and sensory loss in his upper and lower limbs, distal more than proximal.
MRI of the brain showed symmetrical high T2 signal change within the medulla and dorsal pons but no supratentorial parenchymal lesions. Electromyography showed chronic denervation, and nerve conduction studies showed a moderately severe sensory motor axonal neuropathy. The father in family No 1 had normal nerve conduction studies, the mother had an absent response from the extensor digitorum brevis and rather small responses from the abductor pollicis brevis and abductor hallucis, suggesting a slightly patchy peripheral neuropathy. Nerve biopsy showed greatly swollen axons caused by accumulation of neurofilaments. The appearances were consistent with GAN (sural nerve biopsy from the proband shown in fig 1), as was the proband's tight kinky hair that was characteristic of GAN.
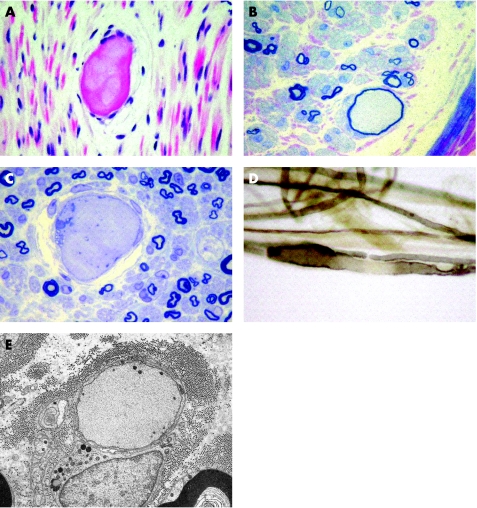
Figure 1 Sural nerve biopsy from the proband of family No 1. Images of paraffin sections (A), resin sections (B, C), teased fibres (D) and electron microscopy (E) of the sural nerve biopsy. (A) Axonal swellings show variable eosinophilia in haematoxylin and eosin stained paraffin sections. (B, C) At least one “giant” axon was present in each fascicle: some were surrounded by an attenuated myelin sheath (B) while others were de‐ or non‐myelinated (C). Teased fibres showed secondary demyelination in the region of the axonal swellings (D). Electron microscopy showed that the “giant” axons were filled with closely packed neurofilaments and organelles, and that unmyelinated axons may also be involved (E). Scale bar 20 μm (A–D) or 5 μm (E).
Neuropathology
Nerve biopsies were carried out in all five families and were consistent with the diagnosis of GAN. The most striking feature in the nerve biopsies was the abnormally enlarged axons with homogeneous axoplasm and thin or absent myelin sheath. There was no apparent reduction in the number of myelinated fibres (fig 1). Ultrastructural examination showed that the giant axons consisted of closely packed and irregularly oriented neurofilaments, with focal areas of increased density. Both myelinated and unmyelinated axons were affected. Aggregates of neurotubules with mitochondria, vesicles and dense bodies were sometimes confined to the centre or edges of the axoplasm (fig 1E, family No 1 sural nerve biopsy).
Discussion
We have reported four families with previously unidentified Gigaxonin mutations. The majority of Gigaxonin mutations are non‐sense or frameshift changes that lead to a premature termination codon. This was the case for three of the mutations identified here (table 1). One family with GAN (family No 5) did not have a Gigaxonin mutation. This confirms the genetic heterogeneity in GAN.
A number of studies have been carried out on the normal functional role and interactions of Gigaxonin and the consequences of mutations and features of null mice.15 Disruption of the Gigaxonin interacting genes (TBCB, MAP1B and MAP8) varied depending on the location of the mutation in Gigaxonin, suggesting that defects in Kelch repeats caused the greatest TBCB protein disruption compared with the BTB domain or null mutations that caused the greatest MAP1B and MAP8 protein disruption and toxic neuronal accumulation.15
The mutations in family Nos 1 and 3 caused a less severe phenotype and later age of onset as opposed to the more severe phenotype and earlier onset in family Nos 2 and 4. There are still relatively few mutations reported to draw significant genotype phenotype conclusions, but predicting complications is important for clinicians and families. Mutations in the BTB domain seem to cause an earlier age of onset, earlier age at loss of ambulation and/or death compared with mutations in the Kelch repeats or non‐domain areas. This is shown in family Nos 2 and 4 from this study and from other reported studies (family 1910) and the single case from Brockmann and colleagues.11 An unusual family (Tunisian family II) had a homozygous mutation in the BTB domain (R15S) but the phenotype was distinct, with a progressive multisystem degeneration with giant axons but lack of kinky hair.7
Two reports have suggested the possibility of manifesting heterozygous carriers.9,13 In our series of patients, likely manifesting carriers were identified in family Nos 1 and 2 where the carriers had a point mutation in the Gigaxonin gene, one in the BTB and one in the Kelch region. The reported manifesting carriers9,13 had a mutation in the Kelch domain (Arg201Stop) and one between the BTB and Kelch domains (Arg293Stop), suggesting that a Gigaxonin mutation may act as a risk factor for axon damage by either a haploinsufficiency or by causing a dominant negative effect and interfering with the normal allele in Gigaxonin carriers.
In summary, we have reported five novel Gigaxonin mutations and their associated clinical phenotypes. Genetic heterogeneity was confirmed and a genotype phenotype effect was suggested, based on the particular domain the mutation affected. We also identified a further two families who had manifesting heterozygous carriers. Further mutation reports and clinical analysis of heterozygous carriers will enable a more detailed evaluation to define the penetrance of Gigaxonin mutations.
Acknowledgements
We are grateful to the Medical Research Council (MRC) for their support. HH holds an MRC clinician scientist fellowship. We are also grateful to The Wellcome Trust and the Mason Medical Research Foundation. This work was undertaken at UCLH/UCL who received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme. We also thank the cell repository of neuromuscular diseases, Montreal Children's Hospital Research Institute and The Istituto G Gaslini, Genova. The families, CMT International and the National Organisation for Rare Disorders (NORD) are thanked for their continued interest and assistance with our work.
Abbreviations
GAN - giant axonal neuropathy
Footnotes
Competing interests: None.
References
- 1.Asbury A K, Gale M K. Giant axonal neuropathy—a unique case with segmental neurofilamentous masses. Acta Neuropathol (Berl) 197220237–247. [DOI] [PubMed] [Google Scholar]
- 2.Berg B O, Rosenberg S H, Asbury A K. Giant axonal neuropathy. Pediatrics 197249894–899. [PubMed] [Google Scholar]
- 3.Carpenter S, Karpati G, Andermann F.et al Giant axonal neuropathy. A clinically and morphologically distinct neurological disease. Arch Neuol 197431312–316. [DOI] [PubMed] [Google Scholar]
- 4.Treiber‐Held S, Budjarjo‐Welim H, Reimann D.et al Giant axonal neuropathy: a generalized disorder of intermediate filaments with longitudinal grooves in the hair. Neuropediatrics 19942589–93. [DOI] [PubMed] [Google Scholar]
- 5.Fabrizi G M, Cavallaro T, Angiari C.et al Giant axon and neurofilament accumulation in Charcot–Marie–Tooth disease type 2E. Neurology 2004621429–1431. [DOI] [PubMed] [Google Scholar]
- 6.Azzedine H, Ravise N, Verny C.et al Spine deformities in Charcot–Marie–Tooth 4C caused by SH3TC2 gene mutations. Neurology 200667602–606. [DOI] [PubMed] [Google Scholar]
- 7.Bomont P, Cavalier L, Blondeau F.et al The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet 200026370–374. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier L, BenHamida C, Amouri R.et al Giant axonal neuropathy locus refinement to a <590 kb critical interval. Eur J Hum Genet 20008527–534. [DOI] [PubMed] [Google Scholar]
- 9.Kuhlenbaumer G, Young P, Oberwittler C.et al Giant axonal neuropathy (GAN): case report and two novel mutations in the gigaxonin gene. Neurology 2002581273–1276. [DOI] [PubMed] [Google Scholar]
- 10.Bomont P, Ioos C, Yalcinkaya C.et al Identification of seven novel mutations in the GAN gene. Hum Mutat 200321446. [DOI] [PubMed] [Google Scholar]
- 11.Brockmann K, Pouwels P J, Dechent P.et al Cerebral proton magnetic resonance spectroscopy of a patient with giant axonal neuropathy. Brain Dev 20032545–50. [DOI] [PubMed] [Google Scholar]
- 12.Bruno C, Bertini E, Federico A.et al Clinical and molecular findings in patients with giant axonal neuropathy (GAN). Neurology 20046213–16. [DOI] [PubMed] [Google Scholar]
- 13.Demir E, Bomont P, Erdem S.et al Giant axonal neuropathy: clinical and genetic study in six cases. J Neurol Neurosurg Psychiatry 200576825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pena D J. Giant axonal neuropathy: An inborn error of organization of intermediate filaments. Muscle Nerve 19825166–172. [DOI] [PubMed] [Google Scholar]
- 15.Ding J, Allen E, Wang W.et al Gene targeting of GAN in mouse causes a toxic accumulation of microtubule‐associated protein 8 and impaired retrograde axonal transport. Hum Mol Genet 2006151451–1463. [DOI] [PubMed] [Google Scholar]