Abstract
Viable cells of Micrococcus luteus secrete a factor, which promotes the resuscitation and growth of dormant, nongrowing cells of the same organism. The resuscitation-promoting factor (Rpf) is a protein, which has been purified to homogeneity. In picomolar concentrations, it increases the viable cell count of dormant M. luteus cultures at least 100-fold and can also stimulate the growth of viable cells. Rpf also stimulates the growth of several other high G+C Gram-positive organisms, including Mycobacterium avium, Mycobacterium bovis (BCG), Mycobacterium kansasii, Mycobacterium smegmatis, and Mycobacterium tuberculosis. Similar genes are widely distributed among high G+C Gram-positive bacteria; genome sequencing has uncovered examples in Mycobacterium leprae and Mb. tuberculosis and others have been detected by hybridization in Mb. smegmatis, Corynebacterium glutamicum, and Streptomyces spp. The mycobacterial gene products may provide different targets for the detection and control of these important pathogens. This report is thus a description of a proteinaceous autocrine or paracrine bacterial growth factor or cytokine.
The essential role of cytokines in controlling the activation, growth, and proliferation of eukaryotic cells is now widely recognized (1). These proteinaceous cell-signaling molecules include many growth factors that are widely distributed among vertebrates, and probably also invertebrates (2). Similar growth factors have also recently been found in unicellular organisms such as ciliates (3–6). In prokaryotic organisms, intercellular signaling usually involves small metabolites (e.g., N-acyl homoserine lactones) or peptides (7–16). Although specific interactions have been documented between vertebrate cytokines and prokaryotes (17–20), there are no known examples of autocrine or paracrine growth factors produced by prokaryotic microorganisms.
The number of observable microbial cells in a natural sample often exceeds the number that can be cultured therefrom by orders of magnitude (21–23). It is not known in general whether such nonculturable (and often noncultured) cells are dead, are killed by our media, or are in a dormant state from which we could, in principle, resuscitate them with appropriate growth factors (24). After growth to stationary phase and starvation in spent growth medium, cells of the nonsporulating, Gram-positive bacterium Micrococcus luteus can enter a dormant state in which they can persist for at least 7 months. Whereas exponentially growing cultures have a viability of ≈100%, as estimated by comparing colony forming units (cfu) on agar plates with the total cell count determined microscopically, such dormant cultures can exhibit a viability of less than 10−4 (25). The viable count of this type of culture as measured by the Most Probable Number (MPN) method also corresponds to the number of cfu. However, in the presence of sterile (filtered) culture supernatant from the late logarithmic phase of batch growth, resuscitation occurs and the viable count by MPN increases by several orders of magnitude to a value approaching the total count (26). These and other (27, 28) experiments showed that growing bacteria produce a pheromonal resuscitation-promoting factor (Rpf).
We describe herein the isolation and characterization of the Rpf, which is a ≈16–17 kDa protein. The gene encoding the Rpf has been isolated, sequenced, and expressed in Escherichia coli. Similar genes are of widespread occurrence among the high G+C Gram-positive bacteria, including streptomycetes, corynebacteria, and mycobacteria. These findings, together with our demonstration that the purified Rpf from M. luteus stimulates the growth of several mycobacteria, including Mb. tuberculosis, have significant implications for detection and treatment of, as well as protection against, mycobacterial infections.
MATERIALS AND METHODS
Organisms and Media.
M. luteus NCIMB 13267 (previously described as “Fleming strain 2665”) was grown aerobically at 30°C in conical flasks in lactate minimal medium (LMM) containing l-lactate as described (25, 28). When the culture had reached stationary phase, agitation was continued at 30°C for up to 2 months. Cultures were then held aerobically at room temperature without agitation for periods up to a further 3 months. The initial viability of these cultures at this point (measured by comparing the plate count with the microscopic count) was less than 10−3. Mycobacterium smegmatis (“fast” strain) was obtained from All-Russia’s State Institute for Control and Standardization of Veterinary Preparations (Moscow) and was grown in either Sauton medium (29) or nutrient broth E (LabM), which contains beef extract (1 g/liter), yeast extract (2 g/liter), peptone (5 g/liter), and NaCl (5 g/liter). Overnight precultures were washed several times by centrifugation and used to inoculate cultures to an initial density of 2.102 cells/ml. Mycobacterium tuberculosis H37Rv, Mycobacterium bovis (BCG), Mycobacterium avium, and Mycobacterium kansasii were obtained from the Central Institute for Scientific Research on Tuberculosis (Moscow), and Mb. tuberculosis H37Ra (avirulent strain) was obtained from the All-Russia’s Centre for Applied Microbiology (Moscow). Cultures in Sauton medium (29) were inoculated with cells from 4- to 6-week-old cultures to an initial density (total count) of 106 cells/ml [2.5 × 105 cells/ml for Mb. bovis (BCG)], and incubated without shaking at 37°C.
M. luteus Spent Medium Preparation.
Supernatant was obtained after the centrifugation of late logarithmic phase M. luteus cultures (200–1,000 ml) grown in LMM or LMM in which lactate was replaced by succinate plus 0.01% yeast extract from which macromolecules had been removed by dialysis. The inoculum consisted of 2% (vol/vol) of cells grown in rich medium (nutrient broth E, LabM) and then washed in LMM lacking lactate. The supernatants were passed through a 0.22-μm filter (Whatman) before use.
M. luteus Cell Viability by Plating.
Plates consisting of 1.3% nutrient broth E (LabM) or LMM were used. Dilutions were made in quadruplicate with centrifuged and autoclaved spent medium taken from starved cultures. Plates were incubated at 30°C for 3–5 days.
M. luteus Cell Viability by MPN.
The MPN assay was performed in a Bioscreen C optical growth analyzer (Labsystems, Finland) by using LMM supplemented with 0.5% lactate and 0.05% of yeast extract as a resuscitation medium. Dilutions of starved cells were made as described (27). Ten microliters of each dilution (5–10 replicates) were added to wells containing 200 μl of either LMM supplemented with 0.5% lactate and 0.05% yeast extract or the same medium with the fraction being tested (2–20 μl). Growth (optical density) was monitored using a 600-nm filter. Plates were incubated at 30°C with intensive continuous shaking. The overall measurement period was 120 h, each well being measured hourly. Fractions obtained after chromatography were dialyzed against elution buffer 2 (see below), diluted in resuscitation medium in various proportions (1:10, 1:100, 1:500, 1:1000, 1:5,000, and 1:10,000), and filtered through 0.22-μm Gelman filters before testing. MPN values were calculated by using published tables (28).
Total Cell Counts.
Unstained cells were counted with a phase-contrast microscope and an improved Neubauer counting chamber (Fisons, UK) (28). In long-term experiments with mycobacteria, organisms were stained with Ziehl-Neelsen reagent (52) before counting.
Chromatography.
Prewetted DEAE cellulose was added to culture supernatant (1:10 vol/vol) and incubated at 4°C for 1 h with slow stirring. The cellulose was loaded into a column and washed with 5 volumes of buffer 1 (10 mM Tris⋅HCl, pH 7.4/1 mM EDTA/1 mM DTT/10% (vol/vol) glycerol) containing 10 mM KCl. The column was eluted stepwise with 2–3 bed volumes of 0.3 M KCl in buffer 1. The fraction obtained was slowly diluted with buffer 1 on ice to give a final KCl concentration of 0.08 M. Forty column volumes of this fraction were then loaded onto a DEAE-Sepharose fast flow column (1 part of Sepharose preequilibrated with buffer 1 containing 0.08 M KCl). The column was washed with 5 bed volumes of buffer 1 containing 0.08 M KCl and eluted stepwise with 3 volumes of 0.25 M KCl in buffer 1. The fraction obtained was again slowly diluted with buffer 1 on ice to a final KCl concentration of 0.08 M, filtered through a 0.22 μm Gelman filter, and loaded onto a Mono Q column (model HR5/5, prepacked, Pharmacia) equilibrated with buffer 2 (10 mM Tris⋅HCl, pH 7.4/10% glycerol) containing 0.08 M KCl. The Mono Q column was eluted with a linear gradient from 0.08 M to 0.28 M KCl in buffer 2 (total volume 20 ml). The flow rate was 1 ml/min and 1 ml fractions were collected. All manipulations except the Mono Q chromatography step were performed at 4°C. The fractions obtained were dialyzed against buffer 2 (dialysis is important for the retention of activity) and stored at 4°C for up to 5 days without loss of activity. For prolonged storage at −70°C, fractions were dialyzed in the same way and glycerol added to a final concentration of 20–30% (wt/vol). The protein content in purified preparations was estimated from the predicted molar extinction coefficient of the deduced gene product (see below; E280 nm, 1 cm = 46,470).
Trypsin Treatment.
Trypsin was added to the active, dialyzed fraction obtained from the Mono Q column and diluted with LMM supplemented with 0.5% wt/vol lactate and 0.05% yeast extract (1:100) to final concentration of 50 μg/ml). The mixture was incubated for 30 min at 37°C and then the reaction was stopped by adding trypsin inhibitor (100 μg/ml). In control experiments trypsin inhibitor was added to the mixture before incubation.
SDS/PAGE.
SDS/PAGE was performed according to Laemmli (30). Chromatographic fractions were dialyzed against 10 mM Tris⋅HCl, (pH 7.4) for 4–5 h, vacuum-dried (Speedvac, 1.5 h), dissolved in sample buffer (Sigma, S-3401), loaded onto a 15% (wt/vol) acrylamide gel, and run at a constant voltage of 200 V. The gel was stained with colloidal Coomassie G (Sigma).
Chemicals.
Nutrient broth E, yeast extract and agar were obtained from Lab M, whilst l-lactate (Li salt), succinate, trypsin, soya bean trypsin inhibitor and DEAE-Sepharose fast flow were obtained from Sigma. DEAE cellulose DE52 was obtained from Whatman, and Mono Q from Pharmacia. Other chemicals were of analytical grade and were obtained from Sigma or BDH Chemicals (Poole, U.K.).
DNA Manipulations.
Protein microsequence data from the N terminus (ATVDTWDRLAEexSNGTxD) and an internal peptide (VGGEGYPHQASK) obtained from the purified Rpf was used to design two oligonucleotides, A1 (5′-GCSACSGTSGACACSTGGGACCGSCTSGCSGAG-3′) and A2 (5′-GCYTGRTGIGGRTAICCYTCICC-3′) (underlined residues were employed in primer design). Taq polymerase was employed under standard conditions to amplify (4 min at 94°C followed by 30 cycles of: 30 s at 94°C, 30 s at 60°C, 90 s at 72°C) a 147-bp PCR product from M. luteus DNA with these primers. The PCR product was labeled with digoxygenin and used as a probe for Southern hybridizations. A strongly hybridizing 1.4 kbp SmaI fragment was detected, cloned in pMTL20 (31) and established in E. coli strain XL-2 blue (Stratagene). Recombinant plasmids carrying the fragment in both possible orientations were detected by hybridization, confirmed by PCR by using primers A1 and A2, and 1,135 bp of the insert in one of them was manually sequenced on both strands by using the dideoxy chain termination method. The DNA sequence was confirmed by cycle sequencing with fluorescent dye terminators (Applied Biosystems PRISM).
Standard procedures were employed to isolate DNA from M. luteus and Mb. smegmatis. Streptomyces rimosus DNA was kindly supplied by D. Hranueli (Pliva Research Institute, Zagreb). Southern hybridizations with Mb. smegmatis and S. rimosus DNA were initially carried out under nonstringent conditions (0.5 SSC, 37°C). Stringent conditions (0.1 SSC, 65°C) were subsequently employed for screening an ordered cosmid library (32) of Streptomyces coelicolor A3(2) DNA.
Expression of Rpf in E. coli.
Two primers (5′-GTCAGAATTCATATGGCCACCGTGGACACCTGGG-3′) and (5′-TGACGGATCCTATTAGGCCTGCGGCAGGACGAG-3′) (EcoRI, NdeI, and BamHI restriction sites underlined) were employed to amplify (5 cycles of 30 s at 94°C, 30 s at 60°C, 30 s at 72°C, followed by 15 cycles of 30 s at 94°C, 60 s at 72°C) the Rpf coding sequence (i.e., lacking the signal sequence) from the cloned 1.4-kbp SmaI fragment of genomic DNA. It was first established in E. coli DH5α as a 567-bp EcoRI–BamHI fragment in pMTL20 (31) and then excised as a 562-bp NdeI–BamHI fragment, inserted into pET19b (Novagen) and re-established in E. coli DH5α. The sequence of the PCR product and vector-insert junction in this plasmid, denoted pRPF1, was verified. Rpf was expressed from pRPF1 after transforming it into E. coli HSM174(DE3). The protein, containing a His10-tag at the N terminus, was isolated by sonicating bacteria (previously grown to an OD600 nm = 0.9 and induced with 0.4 mM isopropyl-β-d-thiogalactoside for 4 h) in binding buffer (5 mM imidazole, pH 7.9/0.5 M NaCl/20 mM Tris⋅HCl/8 M urea). After low speed centrifugation, a Ni2+-chelation column (Ni2+-coordinated iminodiacetic acid immobilized on Sepharose 6B), was loaded with the supernatant, washed with 20 vol binding buffer, 20 vol binding buffer containing 100 mM imidazole, and then eluted with 10 vol binding buffer containing 0.5 M imidazole. Additional purification was achieved by Mono Q column chromatography (see above, save that the salt gradient was from 0.1–1 M NaCl). Monoclonal anti-(polyHis) antibodies (Sigma, clone His-1) were employed for immunoblot analysis of fractions subjected to SDS/PAGE and electroblotted using standard methods. Fractions were dialyzed against buffer 2 and assayed for biological activity as indicated above.
RESULTS
Characterization of Rpf from Culture Supernatants.
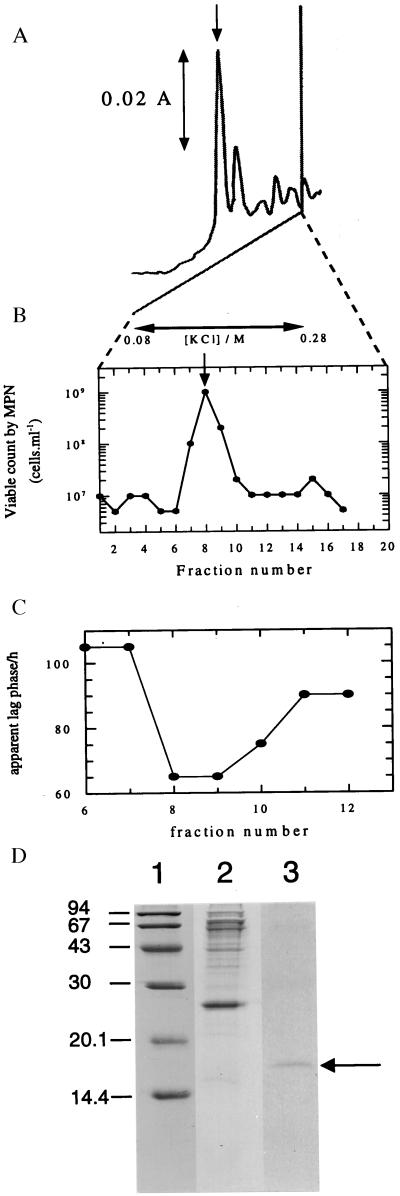
The proteinaceous nature of Rpf in M. luteus culture supernatants was inferred from the results of preliminary resuscitation experiments with dormant bacteria in which it was established that the activity was heat-labile, nondialyzable, and trypsin-sensitive. It was purified to homogeneity in several steps (see Materials and Methods), the final stage of which is shown in Fig. 1A. The peak of resuscitation and apparent lag phase-reducing activity (Fig. 1 B and C) corresponded to a protein with an apparent molecular mass of 16–17 kDa, as estimated electrophoretically (Fig. 1D), and 16–19 kDa, as estimated by Sephacryl gel filtration (not shown).
Figure 1.
Purification and assay of Rpf (A) Elution profile of the resuscitation activity. Fractions eluted from the DEAE-Sepharose column (see Materials and Methods) with 0.25 M KCl were applied to a Mono Q column that was developed with a 20 ml linear gradient from 0.08 to 0.28 M KCl in 10 mM Tris⋅HCl buffer containing 10% glycerol, pH 7.4. The A280 of the eluate was monitored continuously. (B) Resuscitation activity. 10 μl of a diluted suspension of starved cells (cfu 3 × 106 cells/ml, total count of unstained cells 5 × 109 cells/ml) was added to 200 μl of LMM supplemented with 0.5% wt/vol lactate and 0.05% yeast extract containing 2 μl of each fraction in 5–10 replicates in the Bioscreen instrument. For details see Materials and Methods. (C) Reduction of the apparent lag phase of viable cells. 10 μl of a diluted suspension of viable, stationary phase cells (viable count 20 cells) was added to 200 μl of LMM supplemented with 0.5% wt/vol l-lactate and containing 2 μl of each fraction (from a different experiment to that shown in A and B) in 5–10 replicates in the Bioscreen instrument. The apparent lag phase was estimated by extrapolating the exponential growth line to the abscissa. The detection limit of the instrument is approx. 107 cells/ml. (D) SDS/PAGE profile of fractions after DEAE-cellulose and Mono Q chromatography. The molecular mass (kDa) markers (Pharmacia) were phosphorylase B (94), BSA (67), ovalbumin (43), carbonic anhydrase (30), soya bean trypsin inhibitor (20.1), and lactalbumin (14.4). Lanes: 1, markers; 2, fraction from DEAE-cellulose column; 3, purifed preparation (fraction number 8 from the Mono Q column).
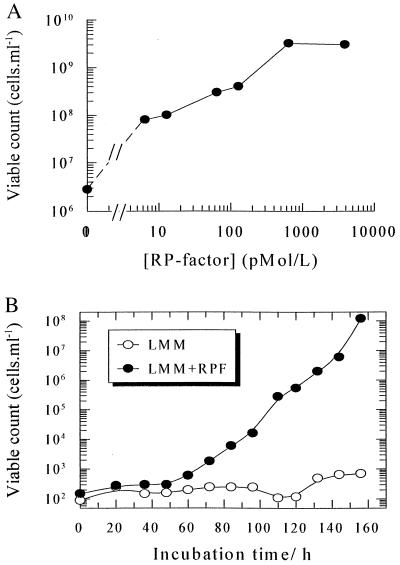
The purified protein lost biological activity after boiling or treatment with trypsin and activity was retained by a 12-kDa cutoff filter (not shown). Rpf was active in the picomolar range in increasing the number of culturable M. luteus cells from dormant populations by three orders of magnitude (Fig. 2A) and in stimulating the growth of washed cells of this organism in batch culture (Fig. 2B). Whereas unwashed cells proliferate normally in liquid LMM (25, 28) the proliferation of washed cells in this medium appeared to depend absolutely on added Rpf over the 160 h duration of the experiment (the prolonged lag phase in this experiment results from the use of a small inoculum of intensively washed cells and a minimal medium). In view of the low concentrations necessary for activity, a trivial nutritional role for (proteolytic degradation products of) Rpf may be discounted. Moreover, in resuscitation experiments Rpf is active in the presence of yeast extract. Rpf is not visible as a distinct band in crude culture supernatants, nor after partial purification using a DEAE cellulose column (Fig. 1D), in which the major species is a ca. 27 kDa protein that may be equivalent to the major 30–32 kDa species found in supernatants of Mb. tuberculosis (33, 34).
Figure 2.
Effect of purified Rpf on M. luteus. (A) Resuscitation of dormant cells with different concentrations of Rpf. 10 μl of a diluted suspension of starved cells (cfu 3 × 106 cells/ml, total count 5 × 109 cells/ml) was added to 200 μl of LMM supplemented with 0.5% wt/vol l-lactate and 0.05% yeast extract and Rpf at the concentrations shown in 5–10 replicates in the Bioscreen instrument. For details see Materials and Methods. (B) Growth of washed cells. Stationary phase cells of M. luteus grown in LMM were washed 5 times by suspension and centrifugation in LMM from which lactate had been omitted. Bacteria were finally suspended in the same medium by repeatedly passing them through a syringe, diluted, and inoculated into a 20 ml flask with LMM or LMM in the presence of Rpf (31 pmol/l). The initial cell density was ca. 102 viable cells per ml and incubation was at 30°C with intensive shaking. Growth was monitored by plating 0.1 ml samples on plates containing nutrient broth E solidified with agar.
Characterization of the Gene Encoding Rpf from M. luteus.
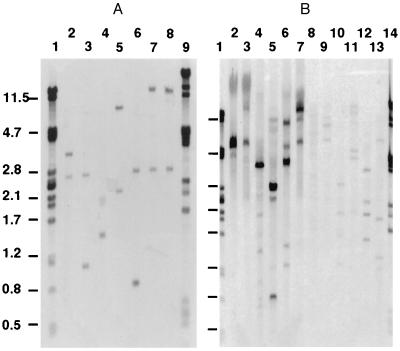
By using primers A1 and A2, designed from protein microsequence data (see Materials and Methods), a 147-bp fragment was PCR-amplified from M. luteus DNA. This fragment was cloned and sequenced and when used as a hybridization probe, it detected a ≈1.4-kbp SmaI genomic restriction fragment as well as two larger, more weakly hybridizing bands (see Fig. 4A, lane 4). The 1.4 kbp fragment was cloned and sequencing revealed that the original PCR product was derived from a gene, denoted rpf, capable of encoding a 220-residue product. The predicted gene product (Fig. 3) commences with a typical 38-residue Gram-positive bacterial signal sequence (35), and the protein sequence immediately after the consensus AQA↓ AT signal peptidase cleavage site corresponds to that of the 25 N-terminal residues of Rpf purified from culture supernatants. After removal of the signal sequence, the predicted gene product has a molecular mass of 19,148.5 Da, which is larger than the measured size of Rpf (see above). Codon usage is similar to that of other M. luteus genes sequenced to date, with a very strong preference for codons ending in G or C throughout.
Figure 4.
Detection of genes similar to rpf in M. luteus, Mb. smegmatis, and S. rimosus. The 147-bp PCR product obtained with oligonucleotides A1 and A2 was used as hybridization probe. (A) Lane1, λPstI; lanes 2–8, M. luteus DNA digested with XhoI, StuI, SmaI, PvuII, PstI, KpnI, and BamHI, respectively, lane 9, λPvuII. (B) Lane 1, λPstI, lanes 2–7, S. rimosus DNA digested with XhoI, StuI, SmaI, PvuII, PstI, and BamHI; lanes 8–13, Mb. smegmatis DNA digested with XhoI, StuI, SmaI, PvuII, PstI, and BamHI; lane 14, λPvuII.
Figure 3.
Mb. tuberculosis and Mb. leprae contain genes whose products are similar to Rpf. Multiple sequence alignment of M. luteus Rpf (Z96935) with predicted gene products from Mb. tuberculosis g2052146 (MTCI237.26), g2791490 (MTV008.06c), g1655671 (MTCY253.32), g2225976 (MTCY180.34), e1254009 (MTV043.60c), and Mb. leprae g2440090 (MLCB57.05c), MSGB38COS (L01095, nucleotides 12,292–12,759). Conserved blocks are in uppercase, residues conserved or conservatively substituted in five or more sequences are in boldface and predicted signal sequences (35) are underlined.
Genes Similar to Rpf Have Been Found in Mycobacteria.
Database searches revealed that the N-terminal region of the secreted form of the M. luteus rpf gene product is substantially similar to the predicted products of seven other genes, five from Mb. tuberculosis and two from Mb. leprae (Fig. 3). One of the predicted gene products in Mb. tuberculosis (g2052146) has a prokaryotic membrane lipoprotein lipid attachment site and all of the others appear to have secretory signal sequences (35), suggesting that they may perform a similar biological function to that of the secreted M. luteus protein. However, these hypothetical mycobacterial proteins have been uncovered solely by DNA sequencing. Their biological functions are currently unknown.
Genes Similar to Rpf Are Found in Other Groups of High G+C Gram-Positive Bacteria.
Genes similar to rpf appear to be widely distributed among those Gram-positive bacteria whose DNA has a high G+C content. Southern hybridization and/or PCR experiments with primers A1 and A2 (not shown) revealed that a second similar gene is present in M. luteus (Fig. 4A) and that similar genes are also detectable in Mb. smegmatis, Mb. bovis (BCG), Corynebacterium glutamicum, and several species of Streptomyces (Fig. 4B and unpublished data). The number of similar genes detected, as deduced from the number of hybridizing bands, varies from one (possibly more) in S. rimosus to four in Mb. smegmatis. Two cosmids from the ordered library of S. coelicolor A3(2) DNA (32) contain genes that hybridize under stringent conditions with the 147-bp PCR product and they are being characterized currently. Genes encoding products comparably similar to Rpf are not present in any of the other organisms whose genomes have been completely or almost completely sequenced to date (Aquifex aeolicus, Archaeoglobus fulvidus, Bacillus subtilis, Borrelia burgdorferi, E. coli, Haemophilus influenzae, Helicobacter pylori, Methanococcus jannaschii, Methanococcus thermoautotrophicum, Mycoplasma genitalium, Mycoplasma pneumoniae, Saccharomyces cerevisiae, Staphylococcus aureus, Streptococcus pneumoniae, Synechocystis PCC6803).
Rpf from M. luteus Stimulates the Growth of Mycobacteria.
In addition to (i) promoting resuscitation of dormant cells, (ii) stimulating growth of washed viable cells and (iii) shortening the apparent lag phase of M. luteus cells in batch culture, purified Rpf also stimulated the growth of both rapidly growing and slow-growing mycobacteria. When added to intensively washed cells of Mb. smegmatis, growth occurred in Sauton medium after 20–24 h, whereas the control lacking Rpf showed no growth after 6 days (Table 1). Moreover, trypsinized Rpf had no such activity (data not shown). With unwashed Mb. smegmatis cells growing in nutrient broth E, the maximum optical density attained was doubled in the presence of Rpf (not shown). Experiments with slow-growing mycobacteria showed that Rpf stimulates the growth of Mb. tuberculosis, Mb. avium, Mb. bovis (BCG), and Mb. kansasii (summarized in Table 1).
Table 1.
Purified M. luteus Rpf stimulates growth of mycobacteria
| Organism | Bacterial growth*
|
|
|---|---|---|
| Rpf omitted | Rpf added | |
| Mb. tuberculosis H37Ra | 1.3 ± 1.9 (5) | 110 ± 32 (5) |
| Mb. tuberculosis H37Rv | 1.5 ± 2 (4) | 45 ± 28 (4) |
| Mb. avium | 0 (3) | >300 (3) |
| Mb. bovis (BCG) | 0 (5) | 54 ± 38 (5) |
| Mb. smegmatis† | 0 (8) | 225 ± 44 (8) |
| Mb. kansasii | 2.5 ± 2.5 (3) | 90 ± 77 (3) |
Growth was estimated microscopically (magnification, ×600) after 14 days of incubation; ca. 50 μl of each culture was fixed, stained using Ziehl-Neelsen reagent and counted. Values in the body of the table are average numbers of cells in a microscope field (10–20 fields counted) ±SD with the number of independent determinations in parentheses. Rpf (after elution from the Mono Q column and dialysis) was used at a concentration of ca. 40 pmol/liter; activity was lost after trypsin treatment, heating (autoclaving), or filtration through a 12-kDa cutoff membrane.
Washed cells of Mb. smegmatis were used for this experiment.
Analysis of Recombinant Rpf.
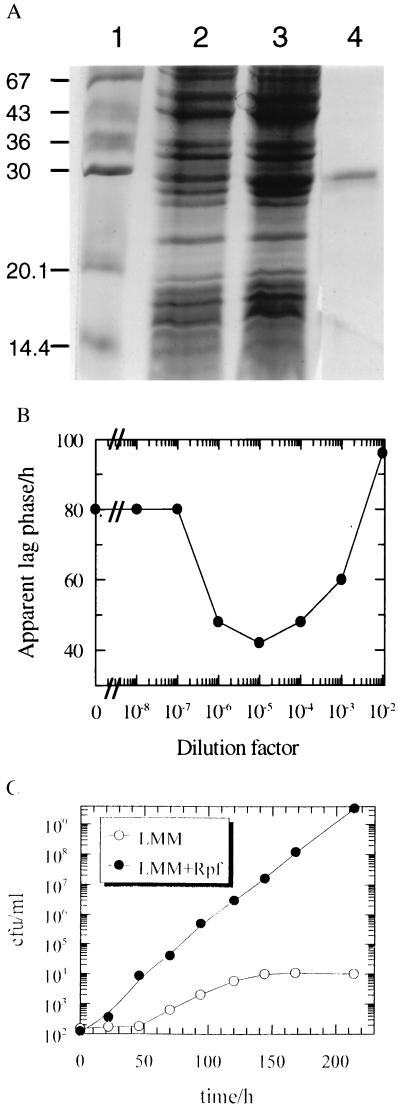
The coding sequence corresponding to the secreted form of Rpf, starting at residue A39, was inserted into pET19b to generate plasmid pRPF1 (see Materials and Methods). Extracts of isopropyl-β-d-thiogalactoside-induced E. coli strain HSM174(DE3) containing pRPF1 were challenged with a poly-His antibody. A strong signal was associated with a protein (apparent size, 29 kDa; predicted size, 22 kDa) that was eluted from the affinity column by 0.5 M imidazole followed by Mono Q purification (Fig. 5A). The histidine-tagged protein from HSM174(DE3) reduced the apparent lag phase of viable cells of M. luteus at picomolar concentrations, whereas the control (material eluted from the same column under the same conditions when an extract from cells containing plasmid vector only was applied) showed no activity (Fig. 5B). Finally, purified recombinant-Rpf strongly stimulated the growth of washed cells of M. luteus (Fig. 5C). The association of biological activity with the recombinant protein, produced in E. coli containing pRPF, and the absence of biological activity in the isogenic control containing pET19b, demonstrates unequivocally that the active molecule is indeed a product of the rpf gene.
Figure 5.
Purification and activity of recombinant Rpf. (A) Purification of his-tagged Rpf. Rpf was expressed in E. coli HSM174(DE3) and purified as described in Materials and Methods. SDS/PAGE profile of fractions after Ni2+-chelation chromatography. The molecular mass (kDa) markers (Sigma) were BSA (67), ovalbumin (43), glyceraldehyde 3-phosphate dehydrogenase (36), carbonic anhydrase (30), soya bean trypsin inhibitor (20.1), and lactalbumin (14.4). Lanes: 1, markers; 2, crude extract from E. coli containing pET19b vector; 3, crude extract from E. coli containing pRPF1; 4, purifed recombinant Rpf. (B) Reduction of the apparent lag phase of viable cells of M. luteus by purifed recombinant Rpf. For experimental details see the legend for Fig. 1C. A dilution factor of 100 corresponds to 33 μg Rpf/ml. (C) Stimulation of the growth of washed cells. Stationary phase cells of M. luteus grown in LMM were washed 5 times by suspension and centrifugation in LMM from which lactate had been omitted. Bacteria were finally suspended in the same medium by repeatedly passing them through a syringe, diluted, and inoculated into a 20 ml flask with LMM or LMM in the presence of Rpf (230 pmol/liter). The initial cell density was ca. 102 viable cells per ml and incubation was at 30°C with intensive shaking. Growth was monitored by plating 0.1-ml samples on plates containing nutrient broth E solidified with agar.
DISCUSSION
A variety of different autocrine chemical compounds (pheromones, ref. 36) produced as secondary metabolites are responsible for eliciting and coordinating the differentiation of prokaryotes during processes such as sporulation, conjugation and the expression of virulence (for reviews see refs. 9, 10, and 15). The important properties of such molecules in this context, which discriminates them from nutrients, are that (i) they are produced by the organisms themselves, (ii) they are active at very low concentrations, and (iii) unless they are generated from prohormones their metabolism is not necessary for activity (although they may be degraded subsequently). The chemical nature of these pheromonal compounds varies widely: those exploited by Gram-negative organisms tend to be of low molecular weight, particularly N-acyl homoserine lactone derivatives (7–11), whilst a number of Gram-positive organisms use proteins and polypeptides (9, 10, 12–16, 37, 38).
The results described herein document the existence of a protein, denoted Rpf, which is secreted by M. luteus and which stimulates growth and cell division when added in picomolar concentrations to minimal media. Although a number of mammalian hormones act as growth factors for unicellular organisms such as ciliates (4, 5), and can be potent stimulators of bacterial growth (18, 20), to our knowledge this is the first characterization of an autocrine or paracrine factor that stimulates the resuscitation of truly dormant bacteria. The protein has the properties of a cytokine, because it also stimulates the growth of viable cells, and is likely to be involved in the normal control of cell multiplication. The extent to which the observed cross-reactivity may permit the resuscitation of other organisms, and thus the question of inter-species specificity, remains to be evaluated. Given that the number of observable microbial cells in a natural sample often exceeds the number that can be cultured therefrom by orders of magnitude (21–23, 39), the current findings are consistent with the view (20) that to permit their regrowth many of these may in fact merely require the addition of such a cytokine, whose potency, and potential susceptibility to degradation in the environment, has delayed their earlier recognition.
By analogy with other protein signaling systems found in bacteria such as S. aureus (9), B. subtilis (14), and S. pneumoniae (40, 41), as well as higher organisms (many prohormones in animals, systemin (42, 43) in plants), it is possible that the proximate signaling molecule is derived from the secreted form of the rpf gene product by proteolytic cleavage. Significantly, the secreted form of the predicted gene product appears to be larger than Rpf purified from culture supernatants. Because the N-terminal amino acid sequence of Rpf was identical to that of the predicted gene product, Rpf is presumably generated by C-terminal processing. The recombinant protein expressed in E. coli also has growth-promoting activity, but it remains to be established whether Rpf is further modified to generate the proximate signaling molecule.
Mb. tuberculosis is a very significant re-emerging pathogen (44–46) that shows latency (i.e., dormancy) in vivo (47–50). It is tempting to speculate that resuscitation and growth of this organism (and possibly also Mb. leprae) may be controlled, in part at least, by members of a family of secreted Rpf-like proteins, which function as autocrine and/or paracrine growth factors. Four of them are secreted, whereas the fifth appears to be a membrane-anchored protein. This protein family may show functional redundancy. Alternatively, as in many animal cell systems (51), the different signaling molecules may be produced under different conditions or for different specific purposes. For example, the membrane-anchored protein (g2052146) may be responsible for juxtacrine signaling and/or signal perception, whereas the protein with the alanine+proline-rich C-terminal extension, comprising a complex family of repeating units (e1254009), may play a role in autocrine signaling by binding to a specific molecule located at the cell surface, and the smallest protein lacking any additional domain (g1655671) may function as a paracrine signaling molecule. Further experimental work will be required to explore these hypotheses, which may lead, in the short term, to substantially improved laboratory methods for the detection and cultivation of these organisms and, in the longer term, to therapeutic strategies and vaccines for preventing their growth in vivo.
Acknowledgments
We thank Drs. Andrew Berry, Nick Harpham, Mustak Kaderbhai, Igor Moshkov, Galina Novikova, and Obolbek Turapov for their help with chromatography, Drs. Jeff Keen and Arthur Moir for protein microsequencing, Dr. Tim Langdon and Miss Katrin Jennert for their help with automated cycle sequencing, Dr. Daslav Hranueli for providing S. rimosus DNA and Prof. V. I. Golyshevskaya for help with the cultivation of mycobacteria. We thank the Royal Society under the terms of the Royal Society/Russian Academy of Sciences Joint Project scheme, the Russian Foundation for Basic Research and the WHO Global Programme for Vaccines and Immunization, for financial support.
Note Added in Proof
The Rpf homologues detected in Mb. tuberculosis H37Rv and described in Fig. 3 have now been given the following gene designations following the publication of the finished genome sequence of this organism: Rv1884c = g2225976 (MTCY180.34), Rv0867c = e1254009 (MTV043.60c), Rv1009 = g2052146 (MTC1237.26), Rv2389c = g1655671 (MTCY253.32), and Rv2450c = g2791490 (MTV008.06c) (53). Further details are available via the Sanger Centre’s Website at http://www.sanger.ac.uk/Projects/M_tuberculosis/.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: Rpf, resuscitation-promoting factor; LMM, lactate minimal medium; cfu, colony-forming units; MPN, most probable number.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Z96935).
References
- 1.Callard R, Gearing A. The Cytokine Facts Book. London: Academic; 1994. [Google Scholar]
- 2.Ottaviani E, Franchini A, Kletsas D, Franceschi C. Ital J Zool. 1996;63:317–323. [Google Scholar]
- 3.Tanabe H, Nishi N, Takagi Y, Wada F, Akamatsu I, Kaji K. Biochem Biophys Res Commun. 1990;170:786–792. doi: 10.1016/0006-291x(90)92160-2. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen L, Christensen S T, Schousboe P, Wheatley D N. FEMS Microbiol Lett. 1996;137:123–128. doi: 10.1016/0378-1097(96)00053-5. [DOI] [PubMed] [Google Scholar]
- 5.Christensen S T, Leick V, Rasmussen L, Wheatley D N. Int Rev Cytol. 1998;177:181–253. doi: 10.1016/s0074-7696(08)62233-0. [DOI] [PubMed] [Google Scholar]
- 6.Vallesi A, Giuli G, Bradshaw R A, Luporini P. Nature (London) 1995;376:522–524. doi: 10.1038/376522a0. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser D, Losick R. Cell. 1993;73:873–885. doi: 10.1016/0092-8674(93)90268-u. [DOI] [PubMed] [Google Scholar]
- 8.Swift S, Bainton N J, Winson M K. Trends Microbiol. 1994;2:193–198. doi: 10.1016/0966-842x(94)90110-q. [DOI] [PubMed] [Google Scholar]
- 9.Ji G Y, Beavis R C, Novick R P. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kell D B, Kaprelyants A S, Grafen A. Trends Ecol Evol. 1995;10:126–129. doi: 10.1016/s0169-5347(00)89013-8. [DOI] [PubMed] [Google Scholar]
- 11.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 12.Solomon J M, Magnuson R, Srivastava A, Grossman A D. Genes Dev. 1995;9:547–558. doi: 10.1101/gad.9.5.547. [DOI] [PubMed] [Google Scholar]
- 13.Solomon J M, Grossman A D. Trends Genet. 1996;4:150–155. doi: 10.1016/0168-9525(96)10014-7. [DOI] [PubMed] [Google Scholar]
- 14.Solomon J M, Lazazzera B A, Grossman A D. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua C, Winans S, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 16.Nodwell J R, McGovern K, Losick R. Mol Microbiol. 1996;22:881–893. doi: 10.1046/j.1365-2958.1996.01540.x. [DOI] [PubMed] [Google Scholar]
- 17.LeRoith D, Roberts C, Lesniak M A, Roth J. Experientia. 1986;42:782–788. doi: 10.1007/BF01941525. [DOI] [PubMed] [Google Scholar]
- 18.Denis M, Campbell D, Gregg E O. Res Microbiol. 1991;142:979–983. doi: 10.1016/0923-2508(91)90008-x. [DOI] [PubMed] [Google Scholar]
- 19.Lenard J. Trends Biochem Sci. 1992;17:147–150. doi: 10.1016/0968-0004(92)90323-2. [DOI] [PubMed] [Google Scholar]
- 20.Kaprelyants A S, Kell D B. Trends Microbiol. 1996;4:237–242. doi: 10.1016/0966-842X(96)10035-4. [DOI] [PubMed] [Google Scholar]
- 21.Amann R I, Ludwig W, Schleifer K H. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davey H M, Kell D B. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torsvik V, Sorheim R, Goksoyr J. J Ind Microbiol. 1996;17:170–178. [Google Scholar]
- 24.Kaprelyants A S, Gottschal J C, Kell D B. FEMS Microbiol Rev. 1993;104:271–286. doi: 10.1111/j.1574-6968.1993.tb05871.x. [DOI] [PubMed] [Google Scholar]
- 25.Kaprelyants A S, Kell D B. Appl Env Microbiol. 1993;59:3187–3196. doi: 10.1128/aem.59.10.3187-3196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaprelyants A S, Mukamolova G V, Kell D B. FEMS Microbiol Lett. 1994;115:347–352. [Google Scholar]
- 27.Votyakova T V, Kaprelyants A S, Kell D B. Appl Environ Microbiol. 1994;60:3284–3291. doi: 10.1128/aem.60.9.3284-3291.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaprelyants A S, Kell D B. J Appl Bacteriol. 1992;72:410–422. [Google Scholar]
- 29.Connell N D. Methods Cell Biol. 1994;45:107–125. doi: 10.1016/s0091-679x(08)61848-8. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Chambers S P, Prior S E, Barstow D A, Minton N P. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 32.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 33.Young D B, Kaufmann S H E, Hermans P W M, Thole J E R. Mol Microbiol. 1992;6:133–145. doi: 10.1111/j.1365-2958.1992.tb01994.x. [DOI] [PubMed] [Google Scholar]
- 34.Harth G, Lee B Y, Wang J, Clemens D L, Horwitz M A. Infect Immun. 1996;64:3038–3047. doi: 10.1128/iai.64.8.3038-3047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen H, Engelbrecht J S B, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Stephens K. CRC Crit Rev Microbiol. 1986;13:309–334. doi: 10.3109/10408418609108741. [DOI] [PubMed] [Google Scholar]
- 37.Dunny G M, Leonard B A B. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 38.Kleerebezem M, Quadri L E N, Kuipers O P, de Vos W M. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 39.DeLong E F. Trends Biotechnol. 1997;15:203–207. doi: 10.1016/S0167-7799(97)01044-5. [DOI] [PubMed] [Google Scholar]
- 40.Håvarstein L S, Coomaraswamy G, Morrison D A. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Q, Campbell E A, Naughton A M, Johnson S, Masure H R. Mol Microbiol. 1997;23:683–692. doi: 10.1046/j.1365-2958.1997.2481617.x. [DOI] [PubMed] [Google Scholar]
- 42.McGurl B, Pearce G, Orozco Cardenas M, Ryan C A. Science. 1992;255:1570–1573. doi: 10.1126/science.1549783. [DOI] [PubMed] [Google Scholar]
- 43.Bergey D R, Hoi G A, Ryan C A. Proc Natl Acad Sci USA. 1996;93:12053–12058. doi: 10.1073/pnas.93.22.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bloom B R, Murray C J L. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 45.Bloom B R. Tuberculosis: Pathogenesis, protection and control. Washington, D.C.: Am. Soc. Microbiol.; 1994. [Google Scholar]
- 46.Duncan K. Exp Op Ther Pat. 1998;8:137–142. [Google Scholar]
- 47.Antia R, Koella J C, Perrot V. Proc R Soc London B. 1996;263:257–263. doi: 10.1098/rspb.1996.0040. [DOI] [PubMed] [Google Scholar]
- 48.Gangadharam P R J. Tubercle Lung Dis. 1995;76:477–479. doi: 10.1016/0962-8479(95)90521-9. [DOI] [PubMed] [Google Scholar]
- 49.Wayne L G. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 50.Young D B, Duncan K. Annu Rev Microbiol. 1995;49:641–673. doi: 10.1146/annurev.mi.49.100195.003233. [DOI] [PubMed] [Google Scholar]
- 51.Heath J K. Growth Factors. Oxford: IRL; 1993. [Google Scholar]
- 52.Murray R G E, Doetsch R N, Robinow C F. In: Methods for General and Molecular Bacteriology. Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Washington, DC: Am. Soc. Microbiol.; 1994. p. 33. [Google Scholar]
- 53.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]