Abstract
Although the complete DNA sequences of several microbial genomes are now available, nearly 40% of the putative genes lack identifiable functions. Comprehensive screens and selections for identifying functional classes of genes are needed to convert sequence data into meaningful biological information. One particularly significant group of bacterial genes consists of those that are essential for growth or viability. Here, we describe a simple system for performing transposon mutagenesis on naturally transformable organisms along with a technique to rapidly identify essential or conditionally essential DNA segments. We show the general utility of this approach by applying it to two human pathogens, Haemophilus influenzae and Streptococcus pneumoniae, in which we detected known essential genes and assigned essentiality to several ORFs of unknown function.
Identification of essential genes that have no defined function provides a starting point for uncovering novel and important biological processes in microorganisms. In addition, because all conventional antibiotics target the products of essential genes, it is likely that the discovery of new essential gene products will have a significant impact on antimicrobial drug development. Essential gene products traditionally have been identified through the isolation of conditional lethal mutants (1) or by transposon mutagenesis in the presence of a complementing wild-type allele (balanced lethality) (2, 3). However, such approaches are laborious because they require isolation or construction of individual mutants on a gene-by-gene basis. These methods also are limited to species with well developed genetic systems and, therefore, cannot be applied readily to several microorganisms whose genomes have been sequenced recently (4–6).
We have developed a method, termed “GAMBIT” (“genomic analysis and mapping by in vitro transposition”), that identifies essential genes through the application of extended-length PCR, in vitro transposition, transformation, and genetic footprinting. Two naturally competent bacterial species, Haemophilus influenzae and Streptococcus pneumoniae, were chosen for evaluation of this approach. GAMBIT analysis of ≈50 kilobases of H. influenzae DNA and 10 kilobases of S. pneumoniae DNA confirmed the essential nature of nine of nine known essential genes. Of a total of 13 conserved hypothetical genes analyzed in these two organisms, 4 were found to putatively encode essential functions based on GAMBIT analysis. Thus, application of GAMBIT to these regions predicts that approximately one-third of all conserved hypothetical genes may encode functions essential for bacterial growth or viability.
METHODS
Transposon Mutagenesis.
H. influenzae Rd strain (American Type Culture Collection no. 9008) (7), the gift of Andrew Wright (Tufts University), was grown on Brain Heart Infusion medium supplemented with 5% Levinthal’s base (BXV) (8) or on MIc medium (9). S. pneumoniae (strain Rx1) (10) was grown on tryptic soy agar supplemented with 5% defibrinated sheep blood. Minitransposons were constructed that contained the inverted repeats of the Himar1 transposon and ≈100 bp of Himar1 transposon sequence flanking either a kanamycin resistance gene (11) for H. influenzae or a chloramphenicol resistance gene (12) for S. pneumoniae. Transposition reactions were performed by using purified Himar1 transposase as described (13). Targets for transposition were either chromosomal DNA or PCR products. PCR of ≈10-kilobase chromosomal regions was performed by using Taq polymerase (Takara Shuzo, Kyoto) and Pfu polymerase (Stratagene) at a 10:1 ratio, 100 pmol of primers, and 30 cycles of amplification (30 sec denaturation at 95°C, 30 sec annealing at 62°C, and 5 min extension at 68°C with 15 seconds added to the extension time for each cycle). Gaps in transposition products were repaired with T4 DNA polymerase and nucleotides followed by T4 DNA ligase with ATP (New England Biolabs) (14). Repaired transposition products were transformed into H. influenzae as described (15) and into S. pneumoniae as described (16) by using the synthetic 17-aa residue competence-inducing peptide CSP-1 for competence induction. Potential S. pneumoniae ORFs were analyzed for homology by using the gap-blast program (17). Mutants were evaluated by Southern blot analysis (14).
Genetic Footprinting.
Genetic footprinting was carried out as described (18) by using a transposon-specific primer (5′-CCGGGGACTTATCAGCCAACC-3′) and primers specific to each chromosomal region designed by using the chromosomal sequence from The Institute for Genomic Research (sequences available on request). PCR was performed by using the above protocol. Products were analyzed by gel electrophoresis on 0.8% agarose gels. To select for and against mutants in thyA, mutants first were plated on BXV and then were replated onto either MIc or BXV containing 5 μg/ml of trimethoprim. Plasmid pSecA, which contains the Escherichia coli secA gene, was constructed by cloning the BamHI fragment from pT7secA (19), the gift of Carol Kumamoto (Tufts University), into the BglII site of the E. coli–H. influenzae shuttle plasmid pGJB103 (15), the gift of Gerard Barcak (University of Maryland). Primers used in Fig. 3 lie within or close to the following loci: (a) HI0449 (primer in lane 1 hybridizes 114 bp 5′ of the primer in lane 2), (b) HI1658, (c) HI0911, (d) HI0905, (e) HI0461, (f) same primer as in (c), and (g) HI0456.
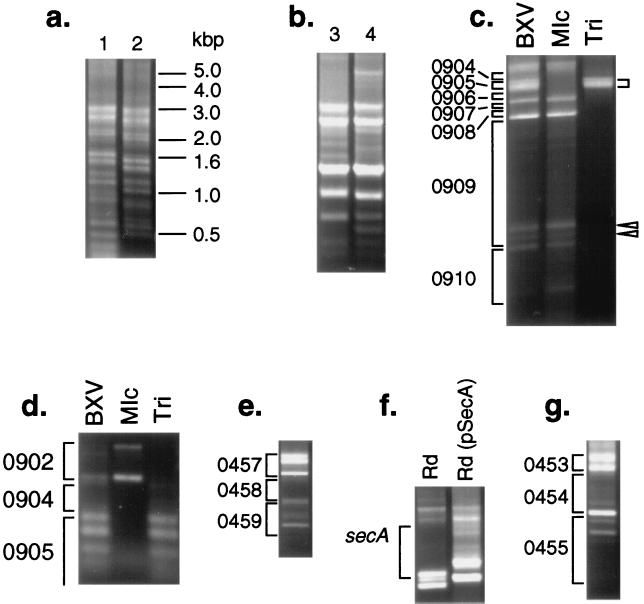
Figure 3.
Genetic footprinting of H. influenzae mutant pools. Genetic footprinting was carried out by using a Himar1-specific primer and a chromosomal primer. In a, the positions of molecular weight standards are indicated; other panels are labeled with locus names by HI number. In c and d, cells were selected on BXV, MIc, or BXV containing trimethoprim (“Tri”). In f, in vitro mutagenesis of a chromosomal fragment that included the secA gene was performed, and the mutagenized DNA was transformed into both wild-type H. influenzae and an H. influenzae strain containing pSecA.
RESULTS
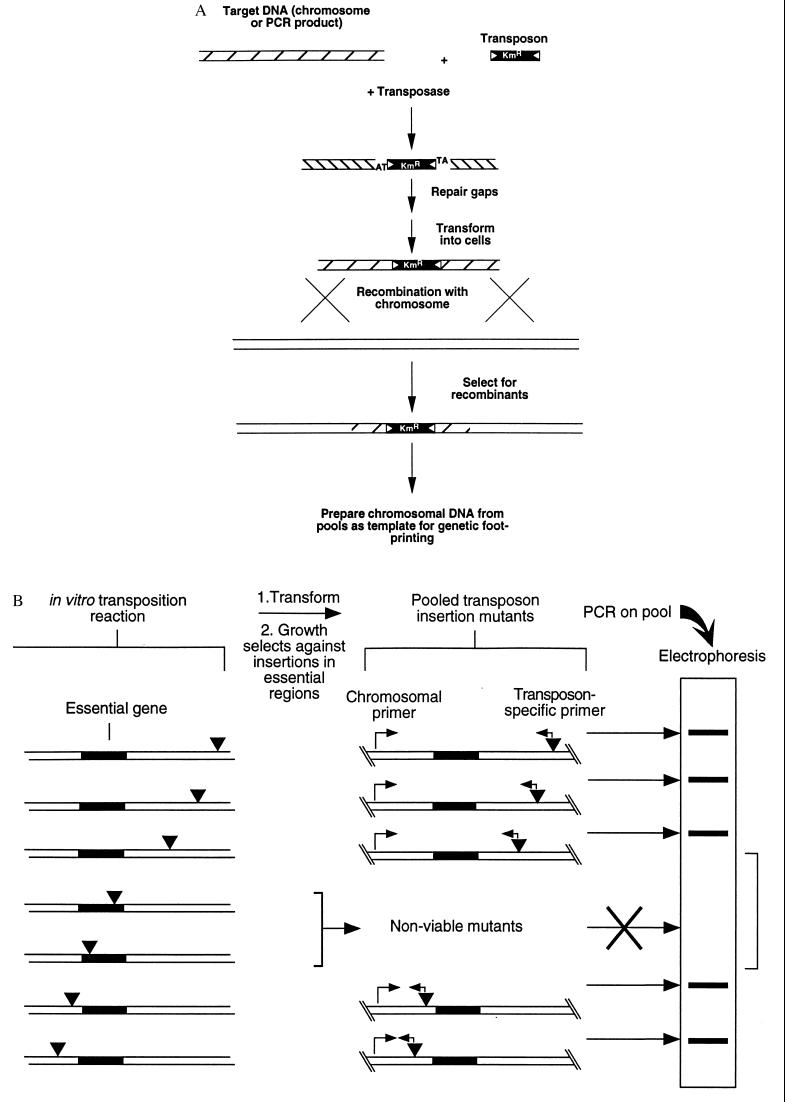
The GAMBIT approach is outlined schematically as two steps in Fig. 1 a and b. The first step involves efficient in vitro transposition mutagenesis and recombination onto the chromosome. The second step maps the genomic location of each transposon insertion in a pool of mutants by genetic footprinting.
Figure 1.
Schematic diagram of the two steps required for GAMBIT (a) Strategy for producing chromosomal mutations by using in vitro transposon mutagenesis. (b) Genetic footprinting for detection of essential genes. Target DNA mutagenized in vitro with the Himar1 transposon was introduced into bacteria by transformation and homologous recombination. Recombinants were selected for drug resistance encoded by the transposon, and insertions in essential genes were lost from the pool during growth. PCR with primers that hybridize to the transposon and to specific chromosomal sites yielded a product corresponding to each mutation in the pool. DNA regions containing no insertions yielded a blank region on electrophoresis gels.
In Vitro Transposon Mutagenesis.
To use GAMBIT, it was necessary to develop an in vitro mutagenesis protocol that could be used on purified chromosomal DNA derived from a naturally competent bacterial species. We chose H. influenzae and S. pneumoniae, both of which are transformable, and the mariner-family transposon Himar1, originally isolated from the horn fly, Haematobia irritans (13). Although other transposons have been shown to function in vitro (20, 21), Himar1 offers two practical advantages. First, a single protein mediates efficient Himar1 transposition in vitro and does not require cellular cofactors. Second, under the conditions we used, Himar1 shows very little insertion site specificity, requiring only the dinucleotide TA in the target sequence [and even this minor site specificity can be easily altered by using different reaction conditions (13)].
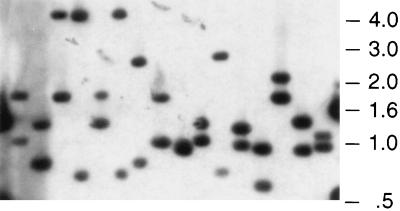
Chromosomal DNA isolated from H. influenzae and S. pneumoniae was mutagenized with the Himar1 transposase and an artificial minitransposon containing genes encoding resistance to either kanamycin (magellan1) or chloramphenicol (magellan2). Insertion of the transposon produces a short single-stranded gap on either end of the insertion site (13). Because natural competence in H. influenzae and S. pneumoniae requires a single-stranded DNA intermediate, these gaps required repair (using a DNA polymerase and a DNA ligase) to produce the flanking DNA sequence required for recombination into the chromosome (Fig. 1a). The mutagenized DNA was transformed into bacteria, and cells that had acquired transposon insertions by homologous recombination were selected on the appropriate antibiotic-containing medium. Using this method, we were able to produce mutant libraries with ≈9,000 H. influenzae mutants and ≈100,000 S. pneumoniae mutants, indicating, as predicted, that this approach is equally effective in Gram-negative and Gram-positive bacteria. AseI-digested DNA from individual H. influenzae transposon mutants was transferred to a Southern blot and was probed with magellan1 DNA. Because AseI cleaves magellan1 only once, these two fragments correspond to chromosomal junction fragments. Thus, each analyzed mutant contained a single transposon insertion, and Himar1 inserted at diverse chromosomal sites (Fig. 2).
Figure 2.
Southern blot analysis of H. influenzae transposon mutants. Genomic DNA was isolated from 16 individual mutants and was digested with AseI, which cleaves once within magellan1. Digested DNA was subjected to agarose gel electrophoresis, was transferred to nitrocellulose, and was hybridized with a probe composed solely of magellan1 minitransposon-derived DNA.
Development of the GAMBIT Technique.
Although mutant libraries such as those described above are quite useful for obtaining a given mutant, GAMBIT demands a greater degree of saturation of mutations to provide a high-density insertion map of a given chromosomal region. To perform such highly saturated mutagenesis, we targeted specific genomic segments for transposition by purifying these via extended-length PCR. In brief, specific oligonucleotide primers were synthesized and were used to amplify selected ≈10-kilobase regions of the chromosome. The resulting PCR products were purified and were used as targets for in vitro Himar1 transposon mutagenesis. Each mutagenized pool of DNA was transformed into competent bacteria and was plated on rich medium containing an appropriate antibiotic, resulting in libraries of ≈400–800 mutants, all of which contained insertions within the target chromosomal segment. The position of each of these insertion mutations with respect to any given PCR primer, designed from genome sequence data, then could be assessed by genetic footprinting (18) conducted on the entire pool of mutants by using a primer that hybridizes to the transposon and another primer that hybridizes to a specified location in the chromosome (Fig. 1b). After amplification, products were analyzed by agarose gel electrophoresis. Each band on the agarose gel represents a transposon insertion a given distance from the chromosomal primer site. Insertions into regions that produce significant growth defects then are represented by areas of decreased intensity on the footprinting gel (Fig. 1b). Note that either one of the two primers used for amplifying a genomic segment also can be used to analyze mutations within that segment by genetic footprinting.
Fig. 3a, lane 1 shows agarose gel electrophoresis of the PCR products obtained from a region of the H. influenzae chromosome chosen for GAMBIT analysis. Areas of the gel corresponding to DNA regions that carry many Himar1 insertions contain many bands; blank regions on the gel, on the other hand, correspond to segments of the chromosome that are devoid of Himar1 insertions. That the banding pattern seen in Fig. 3a, lane 1 reflects an accurate assessment of the position of insertion mutations within the targeted segment can be shown by simply moving the chromosomal primer by 114 bp (Fig. 3a, lane 2). Bands and blank regions on the gel are shifted down in migration by a distance corresponding to ≈114 bases. In addition, sequencing of several gel-purified bands demonstrated that they were in the predicted loci (data not shown). GAMBIT footprinting results are quite reproducible. When two independent insertion libraries were created for a given region, the pattern exhibited only minor differences, and the blank regions were unchanged (Fig. 3b, lane 3 vs. lane 4).
Fig. 3c demonstrates the use of GAMBIT to examine essential genes in the chromosome region containing a homologue of the E. coli gene thyA, which encodes thymidylate synthetase. Mutation of the thyA gene prevents growth on minimal medium lacking thymidine but confers resistance to trimethoprim (22). Thus, this gene provided us with the opportunity to test directly the fidelity of the system because mutations in thyA can be selected both positively and negatively. A primer that hybridizes 3′ to the H. influenzae secA gene, 5,159 bp from the thyA gene, was used as a chromosomal primer. When libraries selected on rich medium were analyzed by genetic footprinting, the region corresponding to the thyA gene (indicated by brackets on the right in Fig. 3c) contained multiple bands. When the analysis was performed on the same mutant pool plated on a defined medium lacking thymidine, the thyA region PCR products were no longer seen. Because thyA mutants are resistant to the antibiotic trimethoprim, selection of the same pool on a medium containing trimethoprim and thymidine followed by PCR analysis yielded products only in the thyA region, confirming the identity of the bands seen in this region of the gel. Analysis of the same mutant pool with a primer that hybridizes close to the thyA gene demonstrates that the bands seen in the lane labeled “Tri” in Fig. 3c can be resolved into a series of bands that correspond to multiple Himar1 inserts distributed within the thyA gene (Fig. 3d).
We found several regions with a decreased number and intensity of PCR products. Some regions contained no detectable PCR products. For example, no bands could be seen in the region in H. influenzae corresponding to an ORF with a high degree of similarity to the E. coli gene surA (Fig. 3e). In E. coli, this gene is required for colony formation (23), and, thus, it is not surprising that insertions in surA were undetectable. Another group of regions were identified that were largely devoid of insertions but that did contain a few insertions, usually in specific reproducible locations. For example, the H. influenzae homologue of the E. coli secA gene (which encodes a portion of the preprotein translocase required for protein secretion) contained two clear insertions near the predicted 3′ end of the gene (Fig. 3c, open arrowheads). This finding is consistent with the previous observation that C-terminal truncations in the E. coli SecA protein do not prevent survival or growth (24).
We tested whether the distribution of Himar1 insertions revealed by GAMBIT analysis reflects the essential nature of a given gene or simply site specificity of the transposon. As discussed above, no insertions could be found in the first 75% of the secA gene. However, when GAMBIT was performed on the same region in a strain complemented with E. coli secA, numerous transposon insertions could be found throughout the gene (Fig. 3f). These data provide strong evidence that gaps in the distribution of Himar1 insertions can be attributed confidently to the presence of an essential DNA sequence.
Detection of Candidate Essential Genes by GAMBIT.
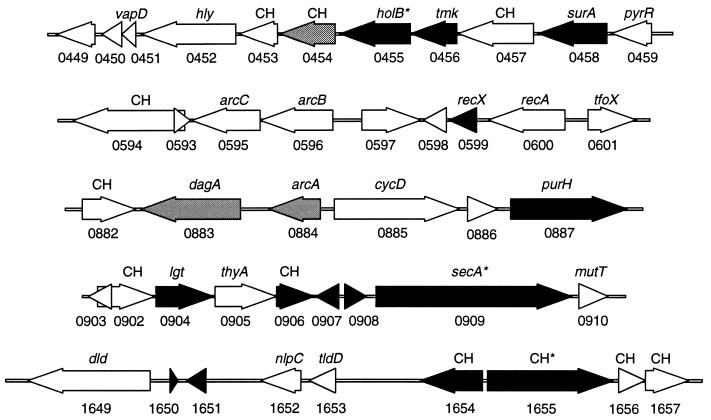
Using this method, we studied five genomic segments in H. influenzae and two in S. pneumoniae. We identified several candidate genes required for growth or viability (Fig. 4 and Table 1). Some of these genes are known to be essential in other organisms, including secA, surA, tmk (25), and lgt (26). Other genes have no known function. To facilitate future genomic analysis, we propose to name genes whose only known functions have been determined by GAMBIT analysis “peg,” for “putative essential gene.” Thus, peg1655 would correspond to ORF HI1655.
Figure 4.
Essential ORFs of H. influenzae. Five chromosomal segments are shown. ORFs with essential functions are shown in black, ORFs that are nonessential are shown in white, and ORFs in which mutations produce growth attenuation are shown in gray. The direction of transcription for each ORF is shown along with the TIGR designation below the ORF and the closest homologue above the ORF. Conserved hypothetical ORFs of unknown function are designated CH. [*, Essential ORFs that can sustain a highly limited number of discrete insertions (<2/kbp).]
Table 1.
S. pneumoniae essential genes
| S. pneumoniae ORF* | Position† | Essential‡ | Similarity (gap-blast E-value) |
|---|---|---|---|
| Conserved hypothetical | 840–2174 | No | Archaeoglobus fulgidus, hypothetical protein, AF0170, (1e–47) |
| Unknown | 3051–3866 | No | None |
| rbfA | 4109–4459 | Yes | Bacillus subtilis ribosome-: binding factor A, P32731, (4e–20) |
| IF-2 | 4710–7586 | Yes | H. influenzae translation, initiation factor IF-2, P44323, (1e–153) |
| L7AE | 7603–7902 | Yes | Enterococcus faecium, probable ribosomal protein in L7AE family, P55768, (6e–23) |
| nusA | 8210–9346 | Yes | B. subtilis NusA, Z99112, (3e–96) |
| p15A | 9390–9860 | No | B. subtilis P15A homolog, unknown function P32726, (2e–27) |
| ytmQ | 9995–10630 | No | B. subtilis YtmQ, unknown function, Z99119, (5e–73) |
PCR Primers used to amplify the 11,266 bp corresponding to contig 4151 of TIGR S. pneumoniae genomic sequence release 112197 and are Forward 5′-CTTTCTGTAAAATGTGGGATTCAA-3′ and Reverse 5′-AATTATTATGGAGTCGTCGTTTGG-3′.
S. pneumoniae orf designations are based on matches giving the highest gap-blast score.
Positions are given with respect to the first base of the Forward primer.
Essential regions as defined in the text.
The major power of GAMBIT is its ability to interrogate specific regions or, by scanning a large series of regions, entire genomes for the presence of essential genes or loci. Mutants reduced in growth, however, also can be detected. Our analysis did, in fact, detect regions with partial reductions of band intensity, suggesting that mutants with insertions in these regions had reduced growth rates but remained viable. For example, among the genes we studied were three genes of unknown function that had been hypothesized to be members of the minimal gene set required by all bacteria (27). Two of these [HI0454 (see Fig. 3g) and HI1654] apparently did cause growth attenuation when disrupted. GAMBIT analysis of HI0454 yielded detectable bands that were reduced in intensity whereas HI1654 yielded no detectable bands. The third (HI0597), however, proved to be nonessential in H. influenzae under the conditions used here.
DISCUSSION
GAMBIT provides a powerful system for identifying genes required for growth or survival. It is likely that some of the essential genes identified by this screen represent previously unidentified components of basic known biological processes such as gene expression, cell division, DNA replication, or protein translocation. It is also possible that GAMBIT will identify fundamentally new biological processes that have remained undiscovered solely because mutations in these essential genes are, by definition, usually lethal. From a practical standpoint, the products of essential genes represent an important set of potential new targets for antimicrobial drugs.
The GAMBIT approach should prove equally useful for identifying genes required for growth or viability under conditions that are more stringent than the rich in vitro media used here. For example, GAMBIT should allow systematic identification of the genes required by pathogenic organisms to grow and survive within a host. Although GAMBIT is applicable to genome scale analysis, it also can be targeted to specific DNA elements or regions of interest such as phages or pathogenicity islands. It is particularly well-suited to the analysis of naturally competent organisms (a group that includes important human pathogens belonging to the genera Haemophilus, Streptococcus, Helicobacter, Neisseria, Campylobacter, and Bacillus). It is also apparent that, with the use of allelic replacement vectors or efficient linear DNA transformation methods, GAMBIT should be adaptable to other bacteria and microorganisms.
In this report, we have used GAMBIT to investigate the essential nature of several genes postulated to be part of the minimal complement of genes needed to sustain life (27). This proposed set was derived from a comparison of the smallest currently sequenced genome capable of encoding a cell, that of Mycoplasma genitalium, and the highly divergent H. influenzae genome. Of seven genes from the proposed minimal gene set examined by our assay (HI0454, HI0456, HI0597, HI0600, HI0905, HI0909, and HI1654), three were essential for growth on rich medium (HI0456, HI0909, and HI1654). These results highlight the concept that a minimal gene set must be defined in terms of the environments encountered by the cell and the diverse strategies that cells use to survive. It is likely that, if the remaining four nonessential genes do constitute part of a minimal gene set, then they are functionally redundant with other genes or are needed for aspects of the bacterial life-cycle that are not represented by growth on rich medium. Such conditions may include environmental alterations affecting oxygen tension, osmolarity, pH, or nutrient availability. Alternatively, some genes may play essential roles only under unusual growth states such as prolonged stationary phase. It is also possible that these genes are required specifically for adaptation to conditions encountered during infection of the human host, the primary niche of both H. influenzae and M. genitalium.
Acknowledgments
We thank E. Hansen, L. Cope, G. Barcak, N. Judson, and members of the Mekalanos laboratory for helpful discussion. This work was supported in part by National Institutes of Health Grants AI02137 (to E.J.R) and AI26289 (to J.J.M.) and Pew Scholars Program Grant P0168SC (to A.C.). B.J.A. was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship, DRG-1371.
References
- 1.Harris S D, Cheng J, Pugh T A, Pringle J R. J Mol Biol. 1992;225:53–65. doi: 10.1016/0022-2836(92)91025-k. [DOI] [PubMed] [Google Scholar]
- 2.Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Nature (London) 1996;383:829–832. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 3.Murphy C K, Stewart E J, Beckwith J. Gene. 1995;155:1–7. doi: 10.1016/0378-1119(94)00920-n. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 6.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 7.Reidl J, Mekalanos J J. J Exp Med. 1996;183:621–629. doi: 10.1084/jem.183.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander H. In: Bacterial and Mycotic Infections of Man. Dubos R, Hirsch J, editors. Philadelphia: Lippincott; 1965. pp. 724–741. [Google Scholar]
- 9.Herriott R M, Meyer E M, Vogt M. J Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoemaker N B, Guild W R. Mol Gen Genet. 1974;128:283–290. doi: 10.1007/BF00268516. [DOI] [PubMed] [Google Scholar]
- 11.Alexeyev M F, Shokolenko I N, Croughan T P. Gene. 1995;160:63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- 12.Claverys J P, Dintilhac A, Pestova E V, Martin B, Morrison D A. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 13.Lampe D J, Churchill M E, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual, Second Edition. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Barcack G J, Chandler M S, Redfield R J, Tomb J-F. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 16.Havarstein L S, Coomaraswamy G, Morrison D A. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh I R, Crowley R A, Brown P O. Proc Natl Acad Sci USA. 1997;94:1304–1309. doi: 10.1073/pnas.94.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt M G, Oliver D B. J Bacteriol. 1989;171:643–649. doi: 10.1128/jb.171.2.643-649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuuchi K. Cell. 1983;35:785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- 21.Gwinn M L, Stellwagen A E, Craig N L, Tomb J F, Smith H O. J Bacteriol. 1997;179:7315–7320. doi: 10.1128/jb.179.23.7315-7320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertino J B, Stacey K A. Biochem J. 1966;101:32C–33C. doi: 10.1042/bj1010032c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tormo A, Almiron M, Kolter R. J Bacteriol. 1990;172:4339–4347. doi: 10.1128/jb.172.8.4339-4347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajapandi T, Oliver D. Biochem Biophys Res Commun. 1994;200:1477–1483. doi: 10.1006/bbrc.1994.1617. [DOI] [PubMed] [Google Scholar]
- 25.Reynes J P, Tiraby M, Baron M, Drocourt D, Tiraby G. J Bacteriol. 1996;178:2804–2812. doi: 10.1128/jb.178.10.2804-2812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan K, Sankaran K, Williams M G, Aldea M, Rudd K E, Kushner S R, Wu H C. J Bacteriol. 1995;177:1879–1882. doi: 10.1128/jb.177.7.1879-1882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mushegian A R, Koonin E V. Proc Natl Acad Sci USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]