Abstract
Interferon (IFN) consensus sequence-binding protein/IFN regulatory factor 8 (IRF8) is a transcription factor that regulates the differentiation and function of macrophages, granulocytes, and dendritic cells through activation or repression of target genes. Although IRF8 is also expressed in lymphocytes, its roles in B cell and T cell maturation or function are ill defined, and few transcriptional targets are known. Gene expression profiling of human tonsillar B cells and mouse B cell lymphomas showed that IRF8 transcripts were expressed at highest levels in centroblasts, either from secondary lymphoid tissue or transformed cells. In addition, staining for IRF8 was most intense in tonsillar germinal center (GC) dark-zone centroblasts. To discover B cell genes regulated by IRF8, we transfected purified primary tonsillar B cells with enhanced green fluorescent protein–tagged IRF8, generated small interfering RNA knockdowns of IRF8 expression in a mouse B cell lymphoma cell line, and examined the effects of a null mutation of IRF8 on B cells. Each approach identified activation-induced cytidine deaminase (AICDA) and BCL6 as targets of transcriptional activation. Chromatin immunoprecipitation studies demonstrated in vivo occupancy of 5′ sequences of both genes by IRF8 protein. These results suggest previously unappreciated roles for IRF8 in the transcriptional regulation of B cell GC reactions that include direct regulation of AICDA and BCL6.
The response of peripheral B cells to T-dependent antigen involves a highly orchestrated series of changes. These are manifested by successive alterations in responsiveness to external signals, anatomic relocalization with passage through the germinal center (GC), modifications in Ig genes by somatic hypermutation (SHM) and class switch recombination (CSR), and the assumption of alternative fates as long-lived memory cells or Ig-secreting plasma cells (1). This multistep process of differentiation is governed by an interlocking chain of transcription factors, some of which function as “master regulators” of developmental decision points, each with its own set of target genes (2–4).
Among these factors, activation of NF-κB downstream of CD40 is required for initiation of the GC reaction (5) and induction of activation-induced cytidine deaminase (AICDA) (6), a protein required for SHM and CSR. BCL6 is also required for the formation of GC, where it acts in a B cell–autonomous manner to repress genes involved in the control of lymphocyte activation and cell cycle progression, allowing rapid cell proliferation (4). BCL6 also arrests further development by acting in concert with PAX5 (BSAP) to repress genes involved in plasmacytic differentiation (2). These genes include PRDM1 (BLIMP1) (2), a direct target of BCL6, and XBP1 (7), a target of PAX5. Commitment to plasmacytic differentiation appears to initiate within the GC, marked by the appearance of cells with an incomplete plasma cell phenotype. These cells are PNA+ in the mouse but do not express BCL6 or PAX5; instead, they are positive for CD138 (Syndecan), BLIMP1, and a member of the IFN regulatory factor (IRF) family, IRF4. The importance of carefully regulated expression of these genes to normal B cell differentiation is indicated by the fact that aberrant expression of NF-κB, BCL6, BLIMP1, PAX5, and IRF4 as a result of chromosomal translocations is associated with a variety of human non-Hodgkin's lymphomas (8).
In this paper we report that another member of the IRF family of transcription factors, IRF8 (also known as IFN consensus sequence-binding protein [ICSBP]) (9), is differentially expressed during normal B cell activation, with peak levels in GC dark-zone centroblasts and lowest levels in Ig-secreting populations of plasma cells and plasmablasts. IRF8 is a hematopoietic cell–specific member of the IRF family of transcription factors (9–11). It is expressed constitutively in macrophages, B cells, and some DCs and at low levels in resting T cells and granulocytes. Studies of mice with a null mutation of the gene (IRF8−/−) (12) showed that IRF8 controls myeloid differentiation by promoting macrophage development while inhibiting granulocyte differentiation (12–14). IRF8 also regulates the maturation and function of subsets of DCs and Langerhans cells, thus providing a critical link between innate and acquired immunity (15–17).
IRF8 functions as a transcriptional activator or repressor through the formation of different DNA-binding heterocomplexes with a series of proteins including IRF family members (IRF1 and -2) (18) and Ets family members (PU.1 and TEL) (19, 20), as well as E47 (19), NFATc1 (21), Trip-15/CSN2 (22), and Miz-1 (23). As part of these complexes, IRF8 can exhibit direct binding to target DNA sequences, including the IFN-stimulated response element and the Ets/IRF composite element (EICE) (10, 11), as well as a newly recognized consensus sequence comprising an IRF/Ets composite element distinct from EICE (24); however, IRF8 is also recruited to IFN-γ activation and isolated Ets sites in a nonbinding capacity through protein interactions (10, 25). It is noteworthy that these associations do not necessarily take place in all mammalian cells, as a functional association of IRF8 with Miz-1 occurred in hematopoietic but not NIH3T3 cells (23), whereas interactions of IRF8 and IRF4 capable of repressing expression of the gene ISG-15 (G1P1) were functional in macrophages but not B cells (26).
Targets of transcriptional regulation by IRF8 and its partners have been studied in macrophages, granulocytic cells, and DCs. They include a series of genes involved in innate and adaptive immunity including IL-12p40, IL-18, toll-like receptor 4, Dab2, gp91-phox, and IL-1β, as well as those regulating apoptosis (BCL-Xl and BCL2) (10, 11, 24–27). In addition, a series of studies has implicated IRF8 in the development of human chronic myelogenous leukemia (27–31), where it functions as a tumor suppressor gene that modulates the transforming activity of the BCR/ABL protooncogene in vivo (30) and in vitro (31).
In contrast to this rapidly expanding understanding of the contributions of IRF8 to myeloid development, function, and transformation, very little is known about the role played by this gene in lymphoid cells. Roles in B cell differentiation and function were suggested by the findings that IRF8−/− mice (12) have increased numbers of splenic and LN B cells. Mice deficient in both IRF8 and IRF4, a second hematopoietic cell–specific IRF family member (32, 33), exhibited a block in B cell development at the pre-B to B transition, a defect not seen in mice singly deficient in these factors (34). This phenotype was associated with a striking reduction in the rearrangement of Ig L chain genes. A likely role for IRF8 in regulating Ig L chain expression is also indicated by the findings that PU.1, E47, and Spi-B recruit IRF8 and IRF4 to EICE in the Igκ 3′ and λ enhancers (35–37). The possibility that regulated expression of IRF8 might be involved in later events in B cell differentiation was further suggested by the finding that the gene most stringently down-regulated during the maturation of human B cells from CD19+ tonsillar B cells to plasma cells was IRF8 (38).
The current studies of IRF8 expression and transcriptional regulation in B cell lineage cells were prompted by parallel analyses of expression profiles exhibited by different classes of mouse B cell lineage lymphomas (39, 40) and subsets of human primary tonsillar B cells (41, 42). These studies showed that the gene that best distinguished mouse centroblastic (CB) diffuse large B cell lymphomas from other lymphoma classes was IRF8 and that IRF8 was expressed at higher levels in tonsillar GC centroblasts than in less or more mature primary human B cells. In addition, we found that IRF8 regulates the expression of genes critically involved in GC including AICDA and BCL6, which were shown to be direct transcriptional targets by promoter reporter and chromatin immunoprecipitation (ChIP) analyses.
RESULTS
High-level expression of IRF8 is a distinguishing feature of mouse CB lymphomas
We used a 70mer oligonucleotide array interrogating 6,800 genes to establish expression profiles for 10 classes of mouse B cell lineage lymphomas (43, 44). A t test was used to identify genes that distinguished each subset from all the others. IRF8 was the gene that best distinguished the histologically defined CB subset of diffuse large B cell lymphomas (unpublished data). Analyses of the relative expression of IRF8 transcripts among the lymphoma classes showed that the highest levels were seen in CB and the other GC-derived lymphomas, with transcript levels in CB being around sixfold higher than in the lowest-expressing class, plasmacytomas (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20051450/DC1). These results paralleled earlier studies of normal human primary B cells that showed that the levels of IRF8 transcripts were ∼10-fold higher in total tonsillar CD19+ B cells than in BM plasma cells (38). This suggested that the pattern of IRF8 expression seen with mouse lymphoma subsets was likely to reflect differences related to the differentiative state of the cell of origin rather than a mechanistic contribution to transformation.
IRF8 expression is modulated during the human GC reaction
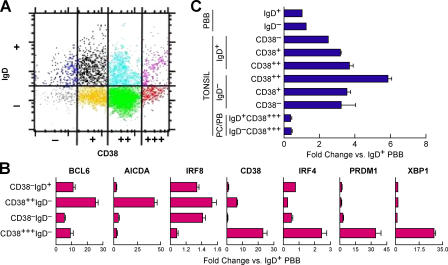
To evaluate a possible relationship between levels of IRF8 expression and stages in normal B cell differentiation, we turned to studies of human primary tonsillar B cells. Previous studies demonstrated that eight subsets of CD19+ B cells can be distinguished by flow cytometry using antibodies to IgD and CD38 (Fig. 1 A) (41, 42): IgD+ preswitch populations (CD38−IgD+, CD38+IgD+, and CD382+IgD+), CD382+IgD− (GC dark-zone centroblasts), CD38+IgD− (GC light-zone centrocytes), CD38−IgD−, CD38+++IgD+ (plasmablasts), and CD38+++IgD− (plasma cells).
Figure 1.
Expression of IRF8 transcripts in human tonsillar and PBBs. (A) Subsets of tonsillar B cells as defined by expression of IgD and CD38 were purified by flow cytometric sorting. RNA prepared from sorted cells belonging to each fraction was used to quantitate IRF8 expression. (B) Quantitation of transcripts of the indicated genes as determined by array analyses of purified tonsillar and peripheral B cell subsets. Data for each gene were normalized to levels in IgD+ PBBs. (C) qPCR analyses of IRF8 transcripts in sorted PBB (IgD+ and IgD−) and in purified subsets of tonsillar B cells. Results were normalized to transcript levels in IgD+ PBB. PB, plasmablasts; PC, plasma cells. Data are expressed as means ± 1 SEM compared with IgD+ PBB for two (B) or three to four (C) independent experiments.
Gene arrays were used to quantitate transcripts in primary human tonsillar and peripheral blood B cell (PBB) subsets (Fig. 1 B). We monitored levels of transcripts for genes known to change in expression in association with GC passage and subsequent maturation to memory and plasma cells: BCL6, IRF4, AICDA, CD38, XBP1, and PRDM1 (BLIMP1) (Fig. 1 B). Transcripts for BCL6 and AICDA were highest in CB and decreased in memory and plasma cells. In addition, transcripts for CD38, IRF4, PRDM1, and XBP1 were highest in plasma cells. These results are completely in keeping with earlier studies of tonsillar B cell subsets using microarrays or serial analysis of gene expression (45, 46), validating the methods used to subset the tonsillar B cell populations. Notably, transcripts for IRF8 were highest in CD38++IgD− centroblasts.
RNA prepared from the sorted tonsillar subsets and purified IgD+ and IgD− PBB was then examined by quantitative RT-PCR (qPCR) for expression of IRF8 transcripts, with results normalized against levels in sorted IgD+ PBB (Fig. 1 C). Of note, transcripts were higher in nonsecreting B cells in activated secondary lymphoid tissue than in circulating B cells. The results also showed that IRF8 transcripts were highest in centroblasts (6-fold over PBB), lower in pre- and postswitch populations (2–4-fold), and lowest in Ig-secreting cells (∼0.5-fold).
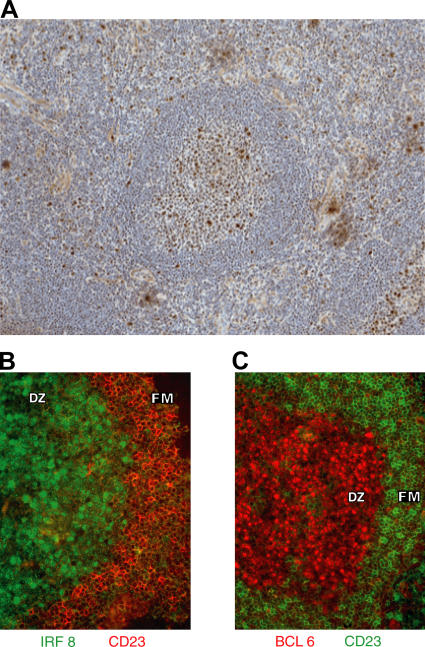
We next evaluated expression of some of these genes at the protein level using immunohistochemistry to study frozen sections of human tonsil. As expected, IRF8 was expressed in cells of the myeloid lineage with high levels in extrafollicular elements including macrophages, monocytes, granulocytes, and DCs (Fig. 2 A). Cells of the follicular mantle zone were mostly negative except for a minor population of small lymphoid cells. In contrast, all GC were positive, with large centroblast-like cells staining most intensely. We used two-color studies to examine expression of CD23, a marker of follicular mantle cells, BCL6, which is expressed most highly in centroblasts, and IRF8 (Fig. 2, B and C). Within GC, antibodies to IRF8 showed intense nuclear staining of cells in the dark zone. This reactivity colocalized with nuclear staining for BCL6. In addition, IRF8+ cells were almost all positive for Ki-67, a marker for the highly proliferative population of centroblasts (not depicted).
Figure 2.
Immunohistochemical staining of frozen sections from tonsil. (A) Staining for IRF8 at an original magnification of 10. (B and C) Staining on serial sections for IRF8 and CD23 (B) and BCL6 and CD23 (C) at an original magnification of 20. Areas comprising the GC dark zone (DZ) and follicular mantle zone (FM) are indicated. Results are representative of three experiments.
IRF8 regulates the expression of genes involved in the GC reaction
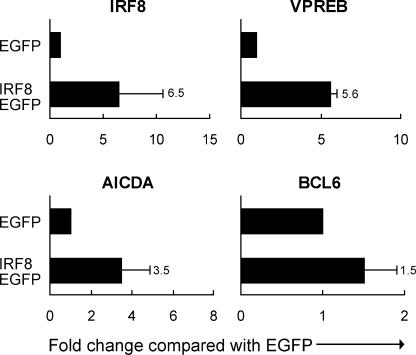
We applied three approaches to identifying genes regulated by IRF8 in B cell lineage cells. First, we transfected primary CD19+ tonsillar B cells with a construct expressing an enhanced GFP (EGFP)–tagged WT mouse IRF8. Because IRF8 positively regulates its own expression (10), this allowed us to monitor human IRF8 expression from the endogenous locus using species-specific primers. We also used a second construct that expressed only EGFP. The cells expressing either EGFP alone or EGFP-tagged IRF8 were sorted into EGFP+ and EGFP− fractions (Fig. S2, http://www.jem.org/cgi/content/full/jem.20051450/DC1, and the transcript levels for various genes were assessed by qPCR. Of note, FACS analyses of EGFP levels at 18 h after transfection showed that IRF8 was equally well expressed in the eight subsets of tonsillar B cells (unpublished data).
These studies showed that human IRF8 transcripts in the EGFP+ population expressing mouse IRF8 were increased an average of 6.5-fold over the levels in cells expressing EGFP alone (Fig. 3), confirming the predicted autoregulation of the gene and validating the experimental system. Expression of two known IRF8 targets, VPREB (34) and NF1 (not depicted) (47), was increased 5.6- and 2.4-fold, respectively. Further analyses showed that cells with enhanced expression of IRF8 had increased levels of transcripts for mature B cell/GC-related genes AICDA and BCL6. These experiments suggested that AICDA and BCL6 are transcriptionally regulated by IRF8, either as direct or indirect targets.
Figure 3.
Transcript levels measured by qPCR for genes in total tonsillar B cells transfected with EGFP-tagged WT IRF8 or EGFP only and sorted 18 h later for EGFP+ and EGFP cells. Data are from independent studies (three for VPREB and nine for IRF8, AICDA, and BCL6). Numbers indicate the fold change represented by the closed bars. Values indicate SD.
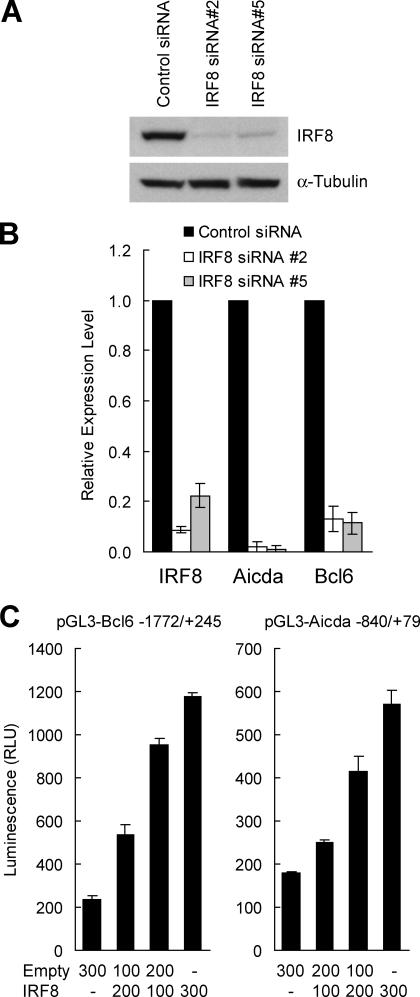
In a second approach to discovering genes regulated by IRF8, we used small interfering RNA (siRNA) to knock down IRF8 expression in the mouse follicular lymphoma–derived B cell line NFS-202. The cells were infected with a retrovirus that expressed puromycin and either of two siRNA for IRF8 or a nonreactive siRNA. After selection for puromycin resistance, cloned cells were assayed for levels of IRF8 transcripts by qPCR. We selected two independent clones, one expressing siRNA #2 and one expressing siRNA #5, in which Irf8 transcripts were reduced to levels 10% or less of those expressed by control cells when normalized to levels of β-actin or Gapdh (unpublished data). By immunoblotting, the siRNA construct reduced IRF8 protein to 10% or less of control levels with α-tubulin serving as a loading control (Fig. 4 A).
Figure 4.
Effect of IRF8 on expression of BCL6 and AICDA. (A) Western blot analyses of IRF8 and α-tubulin expression in NFS-202 cells expressing a negative siRNA and clones expressing either suppressive siRNA #2 or #5. (B) qPCR analyses of gene transcripts in comparisons of NFS-202 cells with an inactive siRNA (closed bar) or with active siRNA #2 and #5 (open and shaded bars, respectively). Results obtained with cells with IRF8 siRNAs are normalized to the values for cells with the control siRNA. Results are representative of three experiments. (C) Luciferase reporter assays of Bcl6 and Aicda promoter sequences. HeLa cells were cotransfected with 800 ng pGL3-Bcl6 or pGL3-Aicda reporter vector, 0–300 ng pCDNA3.1-IRF8 or pCDNA3.1 empty expression vector, and 50 ng pRL-SV40 reporter vector. Luciferase activities were measured after 22 h, and transfection efficiency was normalized with values of Renilla luciferase activities. Results are representative of three experiments. Values in B and C indicate SD.
RNA prepared from these clones was examined by qPCR. Transcripts for Aicda and Bcl6 were strikingly reduced (Fig. 4 B), reinforcing the suggestion that IRF8 functions as a transcriptional activator for both genes. To investigate the possibility that IRF8 directly regulates Aicda and Bcl6 gene expression, we cloned putative promoter fragments flanking the start sites of both genes and placed them upstream of a luciferase reporter gene. Transient expression of the reporters in HeLa cells with increasing amounts of an IRF8 expression plasmid showed a greater than fivefold increase in activity of the Bcl6 reporter and a greater than threefold increase for the Aicda reporter (Fig. 4 C).
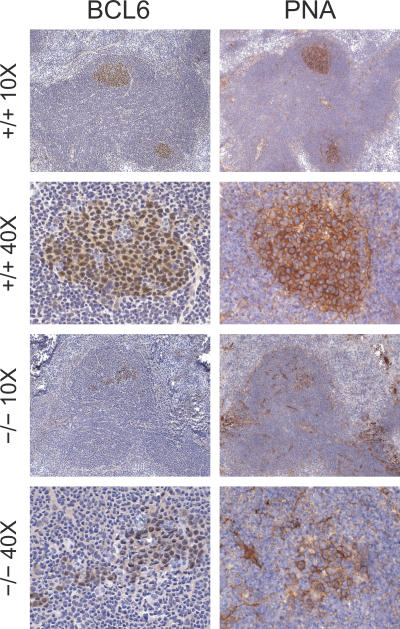
As a third approach to understanding genes regulated by IRF8 in B cells, we examined mice homozygous for a null mutation of the gene (−/−) or WT animals (+/+) (12). Histologic studies of spleens and LN from young −/− mice showed the previously described accumulations of granulocytes, macrophages, and pseudo-Gaucher cells in association with lymphadenopathy and splenomegaly. GC were readily identified in lymph nodes and spleens of both +/+ and −/− mice. Those of +/+ mice were compact and well defined, with centrocytes and centroblasts exhibiting characteristic nuclear staining for BCL6 and surface staining for PNA (Fig. 5). GC of −/− mice exhibited comparable staining of the B cell population but were usually less well organized. Some, as shown in Fig. 5, had populations of BCL6+PNA+ cells trailing away from a more dense collection of cells like those seen in +/+ GC. Others had substantial numbers of small BCL6− lymphocytes localized in small clusters and dispersed throughout a loosened structure of BCL6+PNA+ cells along with scattered apoptotic bodies not seen in +/+ GC (unpublished data).
Figure 5.
Immunohistochemical studies of LN from WT (+/+) and IRF8-deficient (−/−) mice. Frozen serial sections were stained with anti-BCL6 or PNA and counterstained with hematoxylin. Original magnifications are indicated.
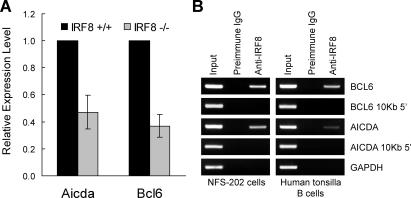
To determine how these histologic features related to expression of GC-associated genes, RNA prepared from purified splenic B cells of +/+ and −/− mice was analyzed by qPCR for levels of transcripts of Aicda and Bcl6 normalized to transcript levels for Gapdh. The results (Fig. 6 A) showed that, compared with levels in +/+ B cells, Aicda and Bcl6 transcripts were reduced approximately two- and threefold, respectively. These findings support the conclusion that IRF8 is involved in the transcriptional regulation of Bcl6 and Aicda and showed that reduced expression of these genes was associated with poorly organized GC in mutant mice.
Figure 6.
Relations of IRF8 to expression of BCL6 and AICDA. (A) RNA prepared from splenic B cells from IRF8-deficient and WT mice was tested by qPCR for expression of Bcl6 and Aicda. Data for IRF8−/− B cells are normalized to levels in +/+ B cells and indicate the means ± SEM for four mice. (B) IRF8 is present at the promoter regions of BCL6 and AICDA in vivo. ChIP analyses were performed with mouse NFS-202 cells or purified human tonsillar B cells. Protein–DNA complexes were immunoprecipitated by addition of antibody to IRF8 and analyzed by PCR for the presence of BCL6 and AICDA promoter sequences. Data are representative of three experiments.
IRF8 is bound to the endogenous AICDA and BCL6 promoters in mouse and human B cells
We performed ChIP analyses to determine whether IRF8 protein was bound in vivo to promoters of genes that it appeared to regulate. As likely positive targets, we selected regions in the promoters of the mouse and human BCL6 and AICDA genes. Protein from the mouse NFS-202 cell line or human tonsillar B cells was cross-linked to DNA using formaldehyde, and immunoprecipitations were performed with anti-IRF8 or control antibodies. After cross-linking was reversed, the DNA was amplified with species- and gene-specific primers for the promoter regions of AICDA and BCL6. Primer pairs for AICDA and BCL6 amplified DNA products of the expected sizes from the total input DNA at specific locations in the 5′ sequences but not at 10-kb sites 5′ of these targets (Fig. 6 B). Notably, bands were observed only with anti-IRF8–precipitated DNA. These data indicated that IRF8 bound the endogenous promoters of AICDA and BCL6 in a mouse GC B cell–derived cell line and in human tonsillar B cells.
DISCUSSION
The GC reaction is the hallmark feature of the B cell response to T cell–dependent antigen. Studies using DNA microarray and SAGE analyses, primarily of human cells, have identified large numbers of genes that are differentially expressed as cells move into and through the GC, maturing from the naive state to CB, to centrocytes, and then to memory or plasma cells as they exit (38, 45, 46, 48, 49). A substantial number of these genes are transcription factors. Among them are a series of “master regulators”—PAX5, BCL6, PRDM1, and XBP1—situated at critical developmental decision points and forming a transcriptional regulatory cascade in which they function in a hierarchic as well as a combinatorial fashion to govern the differentiation of B cells involved in the GC reaction (2–4, 7). The transcriptional functions of these master regulators are complex, involving both gene activation and repression as well as mutual antagonism.
The data from this study identify the transcription factor, IRF8, as another important regulator of the GC program of molecular differentiation by demonstrating two striking and previously unappreciated roles in B cell biology, the direct transcriptional activation of both BCL6 and AICDA, critical determinants of GC development, and function and effects on GC organization. By activating BCL6, a known repressor of PRDM1, IRF8 may also function to prevent GC B cells from initiating a plasma cell differentiation program. Studies of mice homozygous for a null allele of IRF8 showing that transcripts for both Bcl6 and Aicda were reduced approximately two- to threefold in isolated B cells strongly support a direct role for IRF8 in regulating their expression.
Neither BCL6 nor AICDA was previously identified as a transcriptional target of IRF8. BCL6 is absolutely required for GC formation, and control of expression is known to be regulated at multiple levels (50–52). Although transcripts can be detected at low levels in several tissues, protein expression is limited to lymphocytes (53). Within the B cell lineage, BCL6 is expressed only in mature B cells of the GC. In B cells, protein levels are modulated by phosphorylation and ubiquitination, resulting in proteasomal degradation, whereas acetylation negatively regulates BCL6 function (51, 52). Transcriptional down-regulation of Bcl6 appears to be required for B cells to exit the GC, and our experiments indicate that this may be caused by synchronous reductions in the expression of IRF8 and activation of BLIMP1, a transcriptional repressor of Bcl6 (2).
AICDA is a critical determinant of GC formation and function. Deficiencies in AICDA in humans and mice result in markedly impaired CSR and SHM and, in contrast to mice deficient in BCL6, very large GC (54, 55). AICDA transcripts are detected only in peripheral lymphoid tissues, where they are almost exclusively associated with GC (56). AICDA can be induced in resting B cells by several exogenous stimuli including IL-4, CD40, TGF-β, or LPS (56, 57), BLys or APRIL (58), or IL-21 (59) alone or in different combinations. Our data strongly suggest that IRF8 is downstream of the signaling cascades initiated by these agents in B cells and plays a role in regulating expression of AICDA in the GC. Earlier studies showed that the levels of switched Ig classes in activated cells from AICDA+/− and AICDA+/+ mice were comparable, indicating that a 50% reduction in AICDA expression had no major effect on CSR (6). The fact that AICDA transcripts were ∼37% of normal in B cells from the KO may explain why levels of switched Ig classes were close to normal in IRF8−/− mice (60).
Immunohistochemical studies demonstrated that both IRF8 and BCL6 are expressed in GC B cells localized to the dark zone. In contrast, expression of the other lymphoid-restricted member of the IRF family, IRF4, is limited to a small population of centrocytic B cells in the light zone, as well as a few plasmablasts and plasma cells (53). Expression of IRF4 in B cells is regulated primarily by pathways known to drive lymphocyte activation rather than by IFN, which is the case for most other IRF family members. Stimuli provided by IL-4, CD40, and Ig cross-linking received by cells during selection in the GC not only activate IRF4 but also repress BCL6, providing a likely explanation for the contrasting expression patterns of these two genes (61). Whether down-regulation of IRF8 or altered expression of its binding partners plays a role in down-regulating the expression of BCL6 is currently not known but is worth considering. A mechanistic basis for the contrasting patterns of IRF8 and IRF4 expression has not been defined, but the promoter regions of the two genes are apparently regulated by distinct members of the STAT family (STAT1 in the case of IRF8 and STAT6 for IRF4) (62). The nature of the signals that induce the expression and activity of IRF8 in the tonsil is currently unknown but of great interest.
The approaches used to define transcriptional targets of IRF8 all have limitations. First, the knockdown studies were based on the use of a mouse cell line derived from a lymphoma with associated uncertainties about changes in gene expression patterns that might have occurred during the process of transformation or on adaptation to tissue culture. In addition, the use of stable siRNA knockdowns makes it difficult to determine which effects of IRF8 on gene expression are primary or secondary.
Second, splenic B cells from the IRF8 KO mice developed in an abnormal cellular environment caused by extensive myeloid proliferation and a T helper type 2 cell–based cytokine milieu deficient in IL-12 and IFN-γ (12). Although IRF8 has not been shown to play a role in the development or function of follicular DCs, it has profound effects on subsets of DCs and Langerhans cells (15–17). Irregularities in the organization of follicles and GC in secondary lymphoid tissue of IRF8-null mice may well be caused by IRF8-dependent changes in the development and function of follicular DCs.
Third, the studies of total tonsillar B cells for effects of IRF8 overexpression were performed 18 h after transfection. This again leaves open the issue of primary versus secondary targets of IRF8 regulation. Finally, there are several differences between the immunology of mice and humans (63, 64), and conclusions drawn from cross-species comparisons must be made with caution. Nonetheless, the studies of IRF8-deficient mice, the ChIP experiments, and the transcriptional reporter assays all argue that BCL6 and AICDA are primary targets of IRF8 in both species.
In conclusion, our experiments identify IRF8 as a novel component of the regulatory cascade that moves B cells into and through the GC reaction. Increased levels of IRF8 in GC B cells appear to play a role in fostering the GC response by up-regulating BCL6 and AICDA, and down-regulation of expression may contribute to GC B cell entry into the plasma cell differentiation program. Because GC can form in IRF8-deficient mice, IRF8 appears to function as a rheostat that modulates B cell differentiation and function during the GC reaction rather than as a master regulator in the mode of BCL6 or PRDM1.
MATERIALS AND METHODS
Mice, lymphomas, cell lines, and microarray analyses of gene expression.
C57BL/6-Irf8−/− mice described previously (12) were studied at 6–8 wk of age under a protocol (Lip-4) approved by the National Institute of Allergy and Infectious Diseases (NIAID) Animal Care and Use Committee. Splenic B cells were purified by negative selection using standard techniques and were (95% pure. The NFS-202 cell line was cultured from the second generation scid passage of a spontaneous follicular B cell lymphoma from an NFS.V+ mouse. Plasmacytoma cell lines were provided by M. Potter (National Institutes of Health [NIH], Bethesda, MD). The origins and characteristics of primary B cell–lineage lymphomas from NFS.V+ congenic, B6.λ-MYC, SJL-β2m−/−, and BALB/c-gld/gld mice, and the techniques used for transcriptional profiling of the lymphomas using oligonucleotide arrays, were detailed previously (39, 40). Microarray data is available under accession no. GSE1908.
Negative selection of human primary tonsillar B cells, transfection, sorting, and analyses of gene expression.
For negative selection of B cells, tonsillar mononuclear cells were prepared (41, 42) and were stained on ice with a StemSep human B cell–negative selection cocktail containing a mixture of dextran cross-linked to mAb specific for glycophorin A, CD2, CD3, CD14, CD16, and CD56, followed by exposure to a magnetic coil covalently linked to antidextran mAb (StemCell Technologies, Inc.). Polystyrene tubes (12 × 75 mm) containing the suspension were placed into purple EasySep magnets (Stem Cell Technologies). After 5 min at room temperature, the suspension of purified B cells was decanted into collection tubes. The cells were pelleted, resuspended in PBS 1% BSA, and counted.
For transfections, negatively selected tonsillar B cells were diluted to 5 × 106 ml, separated into 1-ml aliquots, and pelleted. The supernatant was removed, and the cells were resuspended in 100 μl of nucleofector solution (Amaxa Biosystems). 5 μg of plasmid expressing EGFP or IRF8 tagged with EGFP (17), both gifts from K. Ozato (NIH, Bethesda, MD) was added to each tube and run on the U-15 program of the Amaxa nucleofector machine. 2 ml of room-temperature RPMI medium (Life Technologies) supplemented with 200 U/ml penicillin G, 10 μg/ml gentamycin, and 10% FCS was added, and cells were placed in a 12-well plate to incubate at 37(C overnight.
For sorting of EGFP+ and EGFP− populations, cells were harvested and resuspended in RPMI medium (Life Technologies) supplemented with 200 U/ml penicillin G, 10 μg/ml gentamycin, and 10% FCS before using a cell sorter (MoFlo; DakoCytomation). For analysis, cells were stained with PE–Cy7 anti–human CD38 (HD37 mIgG1; Becton Dickinson) and A647 anti–human IgD (IA6-1 mIgG2a; BD Biosciences); the A647 conjugation kit was purchased from Invitrogen. PE–Cy7 and A647-conjugated isotype-matched mAbs were used as controls.
Affymetrix arrays were used to profile gene expression in human tonsillar B cell subsets. RNA was prepared from tonsillar B cell subsets purified as described previously (41). We studied RNA from three experiments for centroblasts, plasma cells, and CD38− IgD+ cells and from four experiments for CD38−IgD− cells. The purified RNA was reverse transcribed into cDNA (Invitrogen). The template cDNA was purified for amplification and in vitro transcription reaction to cRNA (BioArray High Yield RNA transcript labeling kit [T7]; Enzo Life Sciences). cRNA was biotin labeled, purified, and hybridized to HG-U133A Affymetrix Genechips according to the Affymetrix protocol (available at http://www.affymetrix.com). Genechips were scanned on a high-resolution Affymetrix scanner using GCOS software (version 1.2). Microarray data were log2 transformed. Relative gene expression of tonsillar B cell subsets was determined by comparison with levels in IgD+ PBBs.
siRNA knockdown of IRF8 expression.
The mammalian expression vector pSUPER.retro.puromycin (OligoEngine) was used for expression of siRNA in NFS-202 cells. To generate the pSR.puro-IRF8 #2 and pSR.puro-IRF8 #5, the pSUPER.retro.puromycin vector was digested with Bgl II and Hind III and the annealed oligos (IRF8 #2, 5′-gatccccACCACCACCTGCCTTGAAGttcaagagaCTTCAAGGCAGGTGGTGGTtttt tggaaa-3′ and 5′-agcttttccaaaaaACCACCACCTGCCTTGAAGtctcttgaaCTTCAAGGCAGGTGGTGGTggg-3′; IRF8 #5, 5′-gatccccACTCATTCTGGTGCAGGTAttcaagagaTACCTGCACCAGAATGAGTtttttggaaa-3′ and 5′-agcttttccaaaaaACTCATTCTGGTGCAGGTAtctcttgaaTACCTGCACCAGAATGAGTggg-3′) were ligated into the vector. The 19-nucleotide IRF8 target sequences are indicated in capitals in the oligonucleotide sequences. A control vector (pSR.puro-negative) was constructed using 64-nucleotide sequences (5′-gatccccTGTAGATGGGTACGCGCTCttcaagagaGAGCGCGTACCCATCTACAtttttggaaa-3′ and 5′-agcttttccaaaaaTGTAGATGGGTACGCGCTCtctcttgaaGAGCGCGTACCCATCTACAggg-3′) with no substantial homology to any mammalian gene sequence and thus serves as a nonsilencing control. The constructs were transformed into DH5α-competent cells (Invitrogen) according to the manufacturer's instructions. Positives were confirmed by sequencing.
Retroviral infection and transfection of cell lines.
Ecotropic retroviral supernatants were produced by transfection of phoenix packaging cells by a reagent (LipofectAMINE 2000; Invitrogen) according to the manufacturer's instructions. After transfection (48 h), the tissue culture medium was filtered through a 0.45-μm filter, and the viral supernatant was used for infection of NFS-202 cells after addition of 4 μg/ml polybrene. Cells were infected for 24 h and allowed to recover for 24 h with fresh medium. Infected cells were selected with 4 μg/ml puromycin for 72 h and cloned for 2 mo. Cloned cells were assayed for levels of IRF8 transcripts by qPCR.
Luciferase reporter assays.
The 5′(flanking regions of Bcl6 (−1772/+245) and Aicda (−840/+79) were PCR amplified from genomic DNA extracted from NFS-202 cells and cloned into the pGL3–Basic Firefly luciferase reporter vector (Promega). (1 corresponds to the first nucleotide of exon 1 of the gene. A murine IRF8 expression vector (pCDNA3.1-IRF8) was provided by K. Ozato. The 800-ng reporter plasmid was used for transfection of HeLa cells grown in 12-well plates. The 50-ng pRL-SV40 Renilla luciferase vector (Promega) was used as an internal control. To test dosage effects, 100, 200, and 300 ng of pCDNA3.1-IRF8 expression vector were adjusted to a total 300 ng of plasmid with pCDNA3.1 empty vector. Luciferase activities were measured 22 h after transfection using the dual-luciferase reporter assay kit (Promega) according to the manufacturer's protocol. All samples were tested in triplicate.
Western blotting.
Total cell lysates were prepared in lysis buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 0.2 mM EDTA, 0.1% Nonidet P-40, 1 mM DTT, 10% glycerol) supplemented with protease inhibitor cocktail solution (Pierce Chemical Co.). Lysates were cleared by centrifugation at 14,000 g for 20 min at 4(C, and the protein contents were then determined by the BCA protein assay kit (Pierce Chemical Co.). For Western blotting, 20 μg of protein per lane was separated on a NuPage 10% Bis-Tris gel (Invitrogen) and transferred to a polyvinylidene difluoride membrane. After blocking with a 5% skim milk solution, the blot was incubated with anti-IRF8 antibody (Santa Cruz Biotechnology, Inc.) or anti–α-tubulin antibody (Sigma-Aldrich). These antibodies were detected with horseradish peroxidase–conjugated donkey anti–rabbit or anti–mouse secondary antibody (GE Healthcare) and developed by Super Signal detection kit (Pierce Chemical Co.) according to the manufacturer's instructions.
qPCR.
Total RNA was isolated using a modified version of the RNA extraction protocol detailed elsewhere (available at http://research.nhgri.nih.gov/microarray). In brief, total RNA was collected using the initial steps of the TRIzol protocol (Invitrogen). After collecting the aqueous supernatant, the RNeasy column coupled with DNase set–based (QIAGEN) protocol was followed. Reverse transcription was performed using 2 μg total RNA, 25 μg/ml oligo (dT)12-18 primers, and 200 units of SuperScript II reverse transcriptase (Invitrogen). For qPCR, 2 ng cDNA was amplified using SYBR Green PCR Master Mix (Applied Biosystems) and 0.33 μM each of forward and reverse primers on the ABI PRISM 7900HT sequence detector system (Applied Biosystems) with published techniques (43). Primers for qPCR were designed using the Primer Express software (Applied Biosystems) and synthesized at MWG-Biotec (Tables S1 and S2, available at http://www.jem.org/cgi/content/full/jem.20051450/DC1). All samples were tested in triplicate, and mean values were used for quantification. Analysis was performed using software (SDS version 2.1; Applied Biosystems) according to the manufacturer's instructions. Samples were normalized using the housekeeping gene GAPDH for human studies and β-actin for mouse. Results for human cells are expressed as the relative fold increase compared with normal IgD+ PBBs. Assays of peripheral blood and tonsillar B cells were run in separate plates, compared with β-actin as a control, and normalized to IgD+ PBBs.
ChIP assays.
ChIP assays were performed by using the ChIP assay kit (Upstate Biotechnology) according to the manufacturer's protocol. 4 × 106 cells were cross-linked with 1% formaldehyde for 10 min at room temperature before ChIP assays. The chromatin was immunoprecipitated with goat anti-IRF8 Ab or preimmune goat IgG (Santa Cruz Biotechnology, Inc.) was used as a control Ab. A 3-μl aliquot out of the 60-μl solution of DNA recovered from each immunoprecipitate was used for PCR, and the products were analyzed on a 2% agarose gel after 36 cycles of amplification. Input DNA (1:400) was used as a control. The following primers were used for PCR: mouse Bcl6 promoter (5′-GTGCCTAATACTCTAGCTGGAAGGAG-3′ and 5′-GCTCGGCCTCTGGAATTCT-3′), mouse Bcl6 10-kb 5′ region (5′-CCAAAGTTCTGGAATGCCCA-3′ and 5′-CAGCATATCCGTGCATGTGC-3′), human BCL6 promoter (5′-AAGTGCAGGAGAGACACACTTCAG-3′ and 5′-CATATGTAACAATCCCAGCCCC-3′), human BCL6 10-kb 5′ region (5′-TATTGTAATTTACTTAATCATTCTTCATCCAA-3′ and 5′-GCAGACCCTTTGACCCAGAG-3′), mouse Aicda promoter (5′-ACATGGTGGCTTTCAACCG-3′ and 5′-GCATCCAGAGAGTGAACTTTAGCC-3′), mouse Aicda 10-kb 5′ region (5′-TTCCTATGGCATGTGTACGGC-3′ and 5′-AACACTCTTCGGGCCAATGA-3′), human AICDA promoter (5′-TAGCATTGCATCCCTAGCACC-3′ and 5′-TGGTCTATTAAAGATTTTATTTCTCTCTCCT-3′), human AICDA 10-kb 5′ region (5′-GTCTCTACTGAAAATACAAAAAAATTGGCT-3′ and 5′-ACTGCAACCTCTGCCTCCC-3′), mouse Gapdh promoter (5′-CACCCTGGCATTTTCTTCCA-3′ and 5′-GACCCAGAGACCTGAATGCTG-3′), and human GAPDH promoter (5′-GCCTGAGCAGTCCGGTGT-3′ and 5′-GATCGGTGCTGGTTCCCA-3′).
Online supplemental material.
Fig. S1 shows the relative expression of IRF8 in mouse lymphomas and plasmacytomas. Fig. S2 depicts flow cytometric analyses of CD19+ tonsillar B cells transfected with vectors expressing EGFP only or WT IRF8 tagged with EGFP at 18 h after infection. Tables S1 and S2 show mouse and human primers, respectively, for qPCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20051450/DC1.
Supplemental Material
Acknowledgments
We thank Drs. Janet W. Hartley and Torgny N. Fredrickson for their assistance in classification of the mouse lymphomas and thoughtful input during the course of this work. We thank Drs. A. Martinez and M. Raffeld for sharing unpublished information. We are grateful to the Ozato laboratory for the generous sharing of reagents and ideas. We thank James Simone and Derek Hewgill of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) Flow Cytometry Facility for multiparameter sorting. We thank NIAID intramural editor Brenda Rae Marshall for assistance.
This work was supported by the Divisions of Intramural Research of the NIAID and of the NIAMS at the NIH. R. Slota is the recipient of the NIH Undergraduate Scholar Program (UGSP) award and is currently an NIH-UGSP scholar.
The authors have no conflicting financial interests.
Abbreviations used: AICDA, activation-induced cytidine deaminase; CB, centroblastic; ChIP, chromatin immunoprecipitation; CSR, class switch recombination; EGFP, enhanced GFP; EICE, Ets/IRF composite element; GC, germinal center; ICSBP, IFN consensus sequence-binding protein; IRF, IFN regulatory factor; PBB, peripheral blood B cell; qPCR, quantitative RT-PCR; SHM, somatic hypermutation; siRNA, small interfering RNA.
C.H. Lee and M. Melchers contributed equally to this work.
References
- 1.MacLennan, I.C.M., A. Gulbransonjudge, K.M. Toellner, M. Casamayorpalleja, E. Chan, D.M.Y. Sze, S.A. Luther, and H.A. Orbea. 1997. The changing preference of T and B cells for partners as T-dependent antibody responses develop. Immunol. Rev. 156:53–66. [DOI] [PubMed] [Google Scholar]
- 2.Lin, K.I., C. Tunyaplin, and K. Calame. 2003. Transcriptional regulatory cascades controlling plasma cell differentiation. Immunol. Rev. 194:19–28. [DOI] [PubMed] [Google Scholar]
- 3.Busslinger, M. 2004. Transcriptional control of early B cell development. Annu. Rev. Immunol. 22:55–79. [DOI] [PubMed] [Google Scholar]
- 4.Shaffer, A.L., X. Yu, Y.S. He, J. Boldrick, E.P. Chan, and L.M. Staudt. 2000. Bcl-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 13:199–212. [DOI] [PubMed] [Google Scholar]
- 5.Quezada, S.A., L.Z. Jarvinen, E.E. Lind, and R.J. Noelle. 2004. CD40/CD154 interactions at the interface of tolerance and immunity. Annu. Rev. Immunol. 22:307–328. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 7.Reimold, A.M., N.N. Iwakoshi, J. Manis, P. Vallabhajosyula, E. Szomolanyi-Tsuda, E.M. Gravallese, D. Friend, M.J. Grusby, F. Alt, and L.H. Glimcher. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 412:300–307. [DOI] [PubMed] [Google Scholar]
- 8.Kuppers, R., and R. Dalla-Favera. 2001. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 20:5580–5594. [DOI] [PubMed] [Google Scholar]
- 9.Driggers, P.H., D.L. Ennist, S.L. Gleason, W.-H. Mak, M.S. Marks, B.-Z. Levi, J.R. Flanagan, E. Appella, and K. Ozato. 1990. An interferon γ-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc. Natl. Acad. Sci. USA. 87:3743–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura, T., and K. Ozato. 2002. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J. Interferon Cytokine Res. 22:145–152. [DOI] [PubMed] [Google Scholar]
- 11.Levi, B.Z., S. Hashmueli, M. Gleit-Kielmanowicz, A. Azriel, and D. Meraro. 2002. ICSBP/IRF-8 transactivation: a tale of protein-protein interaction. J. Interferon Cytokine Res. 22:153–160. [DOI] [PubMed] [Google Scholar]
- 12.Holtschke, T., J. Lohler, Y. Kanno, T. Fehr, N. Giese, F. Rosenbauer, J. Lou, K.P. Knobeloch, L. Gabriele, J.F. Waring, et al. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 87:307–317. [DOI] [PubMed] [Google Scholar]
- 13.Tamura, T., T. Nagamura-Inoue, Z. Shmeltzer, T. Kuwata, and K. Ozato. 2000. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 13:155–165. [DOI] [PubMed] [Google Scholar]
- 14.Tsujimura, H., T. Nagamura-Inoue, T. Tamura, and K. Ozato. 2002. IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J. Immunol. 169:1261–1269. [DOI] [PubMed] [Google Scholar]
- 15.Schiavoni, G., F. Mattei, P. Sestili, P. Borghi, M. Venditti, H.C. Morse, F. Belardelli, and L. Gabriele. 2002. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med. 196:1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aliberti, J., O. Schulz, D.J. Pennington, H. Tsujimura, C.R.E. Sousa, K. Ozato, and A. Sher. 2003. Essential role for ICSBP in the in vivo development of murine CD8α+ dendritic cells. Blood. 101:305–310. [DOI] [PubMed] [Google Scholar]
- 17.Tsujimura, H., T. Tamura, C. Gongora, J. Aliberti, C.R.E. Sousa, A. Sher, and K. Ozato. 2003. ICSBP/IRF-8 retrovirus transduction rescues dendritic cell development in vitro. Blood. 101:961–969. [DOI] [PubMed] [Google Scholar]
- 18.Bovolenta, C., P.H. Driggers, M.S. Marks, J.A. Medin, A.D. Politis, S.N. Vogel, D.E. Levy, K. Sakaguchi, E. Appella, J.E. Coligan, and K. Ozato. 1994. Molecular interactions between interferon consensus sequence binding-protein and members of the interferon regulatory factor family. Proc. Natl. Acad. Sci. USA. 91:5046–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brass, A.L., E. Kehrli, C.F. Eisenbeis, U. Storb, and H. Singh. 1996. Pip, a lymphoid-restricted IRF, contains regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU. 1. Genes Dev. 10:2335–2347. [DOI] [PubMed] [Google Scholar]
- 20.Kuwata, T., C. Gongora, Y. Kanno, K. Sakaguchi, T. Tamura, T. Kanno, V. Basrur, R. Martinez, E. Appella, T. Golub, and K. Ozato. 2002. Gamma interferon triggers interaction between ICSBP (IRF-8) and Tel, recruiting the histone deacetylase HDAC3 to the interferon-responsive element. Mol. Cell. Biol. 22:7439–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu, C., K. Rao, H.B. Xiong, K. Gagnidze, F.L. Li, C. Horvath, and S. Plevy. 2003. Activation of the murine interleukin-12 p40 promoter by functional interactions between NFAT and ICSBP. J. Biol. Chem. 278:39372–39382. [DOI] [PubMed] [Google Scholar]
- 22.Cohen, H., A. Azriel, T. Cohen, D. Meraro, S. Hashmueli, D. Bech-Otschir, R. Kraft, W. Dubiel, and B.Z. Levi. 2000. Interaction between interferon consensus sequence-binding protein and Cop9/signalosome subunit CSN2 (Trip15)—a possible link between interferon regulatory factor signaling and the Cop9/signalosome. J. Biol. Chem. 275:39081–39089. [DOI] [PubMed] [Google Scholar]
- 23.Alter-Koltunoff, M., S. Ehrlich, N. Dror, A. Azriel, M. Eilers, H. Hauser, H. Bowen, C.H. Barton, T. Tamura, K. Ozato, and B.Z. Levi. 2003. NRAMP1-mediated innate resistance to intraphagosomal pathogens Is regulated by IRF-8, PU.1, And Miz- 1. J. Biol. Chem. 278:44025–44032. [DOI] [PubMed] [Google Scholar]
- 24.Tamura, T., P. Thotakura, T.S. Tanaka, M.S.H. Ko, and K. Ozato. 2005. Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood. 106:1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Contursi, C., I.M. Wang, L. Gabriele, M. Gadina, J. O'Shea, H.C. Morse, and K. Ozato. 2000. IFN consensus binding protein potentiates STAT1-dependent activation of IFN-γ-responsive promoters in macrophages. Proc. Natl. Acad. Sci. USA. 97:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenbauer, F., J.F. Waring, J. Foerster, M. Wietstruk, D. Philipp, and I. Horak. 1999. Interferon consensus sequence binding protein and interferon regulatory factor-4/Pip form a complex that represses the expression of the interferon-stimulated gene-15 in macrophages. Blood. 94:4274–4281. [PubMed] [Google Scholar]
- 27.Burchert, A., D. Cai, L.C. Hofbauer, M.K.R. Samuelsson, E.P. Slater, J. Duyster, M. Ritter, A. Hochhaus, R. Muller, M. Eilers, et al. 2004. Interferon consensus sequence binding protein (ICSBP; IRF-8) antagonizes BCR/ABL and down-regulates Bcl-2. Blood. 103:3480–3489. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt, M., S. Nagel, J. Proba, C. Thiede, M. Ritter, J.F. Waring, F. Rosenbauer, D. Huhn, B. Wittig, I. Horak, and A. Neubauer. 1998. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood. 91:22–29. [PubMed] [Google Scholar]
- 29.Schmidt, M., A. Hochhaus, A. Nitsche, R. Hehlmann, and A. Neubauer. 2001. Expression of nuclear transcription factor interferon consensus sequence binding protein in chronic myeloid leukemia correlates with pretreatment risk features and cytogenetic response to interferon-α. Blood. 97:3648–3650. [DOI] [PubMed] [Google Scholar]
- 30.Hao, S.X., and R.B. Ren. 2000. Expression of interferon consensus sequence binding protein (ICSBP) is downregulated in BCR-ABL-induced murine chronic myelogenous leukemia-like disease, and forced coexpression of ICSBP inhibits BCR-ABL-induced myeloproliferative disorder. Mol. Cell. Biol. 20:1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura, T., H.J. Kong, C. Tunyaplin, H. Tsujimura, K. Calame, and K. Ozato. 2003. ICSBP/IRF-8 inhibits mitogenic activity of p210 Bcr/Abl in differentiating myeloid progenitor cells. Blood. 102:4547–4554. [DOI] [PubMed] [Google Scholar]
- 32.Eisenbeis, C.F., H. Singh, and U. Storb. 1995. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9:1377–1387. [DOI] [PubMed] [Google Scholar]
- 33.Mittrucker, H.W., T. Matsuyama, A. Grossman, T.M. Kundig, J. Potter, A. Shahinian, A. Wakeham, B. Patterson, P.S. Ohashi, and T.W. Mak. 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 275:540–543. [DOI] [PubMed] [Google Scholar]
- 34.Lu, R., K.L. Medina, D.W. Lancki, and H. Singh. 2003. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 17:1703–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pongubala, J.M.R., S. Nagulapalli, M.J. Klemsz, S.R. McKercher, R.A. Maki, and M.L. Atchison. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol. Cell. Biol. 12:368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenbeis, C.F., H. Singh, and U. Storb. 1993. Pu.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda 2-4 enhancer. Mol. Cell. Biol. 13:6452–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagulapalli, S., and M.L. Atchison. 1998. Transcription factor PIP can enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol. Cell. Biol. 18:4639–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan, F., E.M. Tian, K. Bumm, R. Smith, B. Barlogie, and J. Shaughnessy Jr. 2003. Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood. 101:1128–1140. [DOI] [PubMed] [Google Scholar]
- 39.Morse, H.C., III, T. McCarty, C.F. Qi, T.A. Torrey, Z.N. Naghashfar, S.K. Chattopadhyay, T.N. Fredrickson, and J.W. Hartley. 2003. B lymphoid neoplasms of mice: characteristics of naturally occurring and engineered diseases and relationships to human disorders. Adv. Immunol. 81:97–121. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, J.Q., C. Okumura, T. McCarty, T.A. Torrey, M.S. Shin, C.H. Lee, M. Hori, Z. Naghashfar, D.C. Roopenian, H.C. Morse III, and W.F. Davidson. 2004. Evidence for selective transformation of autoreactive immature plasma cells in mice deficient in Fasl. J. Exp. Med. 200:1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grammer, A.C., R. Fischer, O. Lee, X. Zhang, and P.E. Lipsky. 2004. Flow cytometric assessment of the signaling status of human B lymphocytes from normal and autoimmune individuals. Arthritis Res. Ther. 6:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grammer, A.C., R.D. McFarland, J. Heaney, B.F. Darnell, and P.E. Lipsky. 1999. Expression, regulation, and function of B cell-expressed CD154 in germinal centers. J. Immunol. 163:4150–4159. [PubMed] [Google Scholar]
- 43.Shin, M.S., T.N. Fredrickson, J.W. Hartley, T. Suzuki, K. Agaki, and H.C. Morse III. 2004. High-throughput retroviral tagging for identification of genes involved in initiation and progression of mouse splenic marginal zone lymphomas. Cancer Res. 64:4419–4427. [DOI] [PubMed] [Google Scholar]
- 44.Morse, H.C., M.R. Anver, T.N. Fredrickson, D.C. Haines, A.W. Harris, N.L. Harris, E.S. Jaffe, S.C. Kogan, I.C.M. MacLennan, P.K. Pattengale, and J.M. Ward. 2002. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 100:246–258. [DOI] [PubMed] [Google Scholar]
- 45.Feldhahn, N., I. Schwering, S. Lee, M. Wartenberg, F. Klein, H. Wang, G.L. Zhou, S.M. Wang, J.D. Rowley, J. Hescheler, et al. 2002. Silencing of B cell receptor signals in human naive B cells. J. Exp. Med. 196:1291–1305. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Klein, U., Y.H. Tu, G.A. Stolovitzky, J.L. Keller, J. Haddad, V. Miljkovic, G. Cattoretti, A. Califano, and R. Dalla-Favera. 2003. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. USA. 100:2639–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, C., G. Saberwal, Y.F. Lu, L.C. Platanias, and E.A. Eklund. 2004. The interferon consensus sequence-binding protein activates transcription of the gene encoding neurofibromin 1. J. Biol. Chem. 279:50874–50885. [DOI] [PubMed] [Google Scholar]
- 48.Tarte, K., J. De Vos, T. Thykjaer, F. Zhan, G. Fiol, V. Costes, T. Reme, E. Legouffe, J.-F. Rossi, J. Shaughnessy Jr., et al. 2002. Generation of polyclonal plasmablasts from peripheral blood B cells: a normal counterpart of malignant plasmablasts. Blood. 100:1113–1122. [PubMed] [Google Scholar]
- 49.Tarte, K., F. Zhan, J. De Vos, B. Klein, and J. Shaughnessy. 2003. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 102:592–600. [DOI] [PubMed] [Google Scholar]
- 50.Allman, D., A. Jain, A. Dent, R.R. Maile, T. Selvaggi, M.R. Kehry, and L.M. Staudt. 1996. BCL-6 expression during B-cell activation. Blood. 87:5257–5268. [PubMed] [Google Scholar]
- 51.Niu, H., B.H. Ye, and R. Dalla-Favera. 1998. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 12:1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bereshchenko, O.R., W. Gu, and R. Dalla-Favera. 2002. Acetylation inactivates the transcriptional repressor BCL 6. Nat. Genet. 32:606–613. [DOI] [PubMed] [Google Scholar]
- 53.Falini, B., M. Fizzotti, A. Pucciarini, B. Bigerna, T. Marafioti, M. Gambacorta, R. Pacini, C. Alunni, L. Natali-Tanci, B. Ugolini, et al. 2000. A monoclonal antibody (Mum1p) detects expression of the Mum 1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 95:2084–2092. [PubMed] [Google Scholar]
- 54.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (Aid), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 55.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Lagelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 56.Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. [DOI] [PubMed] [Google Scholar]
- 57.Dedeoglu, F., B. Horwitz, J. Chaudhuri, F.W. Alt, and R.S. Geha. 2004. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NF-κB. Int. Immunol. 16:395–404. [DOI] [PubMed] [Google Scholar]
- 58.Litinskiy, M.B., B. Nardelli, D.M. Hilbert, B. He, A. Schaffer, P. Casali, and A. Cerutti. 2002. DCs induce CD40-independent immunoglobulin class switching through BLys and April. Nat. Immunol. 3:822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozaki, K., R. Spolski, R. Ettinger, H.P. Kim, G. Wang, C.F. Qi, P. Hwu, D.J. Shaffer, S. Akilesh, D.C. Roopenian, et al. 2004. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl 6. J. Immunol. 173:5361–5371. [DOI] [PubMed] [Google Scholar]
- 60.Giese, N.A., L. Gabriele, T.M. Doherty, D.M. Klinman, L. Tadesse-Heath, C. Contursi, S.L. Epstein, and H.C. Morse III. 1997. Interferon (IFN) consensus sequence binding-protein, a transcription factor of the IFN regulatory factor family, regulates immune responses in vivo through control of interleukin 12 expression. J. Exp. Med. 186:1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pernis, A.B. 2002. The role of IRF-4 in B and T cell activation and differentiation. J. Interferon Cytokine Res. 22:111–120. [DOI] [PubMed] [Google Scholar]
- 62.Gupta, S., M. Jiang, A. Anthony, and A.B. Pernis. 1999. Lineage-specific modulation of interleukin 4 signaling by interferon regulatory factor 4. J. Exp. Med. 190:1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gordon, C.J., G. Grafton, P.M. Wood, M. Larche, and R.J. Armitage. 2001. Modelling the human immune response: can mice be trusted? Curr. Opin. Pharmacol. 1:431–438. [DOI] [PubMed] [Google Scholar]
- 64.Mestas, J., and C.C.W. Hughes. 2004. Of mice and not men: differences between mouse and human immunology. J. Immunol. 172:2731–2738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.