Abstract
Apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G), a cytidine deaminase, is a recently recognized innate intracellular protein with lethal activity against human immunodeficiency virus (HIV). Packaged into progeny virions, APOBEC3G enzymatic activity leads to HIV DNA degradation. As a counterattack, HIV virion infectivity factor (Vif) targets APOBEC3G for proteasomal proteolysis to exclude it from budding virions. Based on the ability of APOBEC3G to antagonize HIV infection, considerable interest hinges on elucidating its mechanism(s) of regulation. In this study, we provide the first evidence that an innate, endogenous host defense factor has the potential to promote APOBEC3G and rebuke the virus-mediated attempt to control its cellular host. We identify interferon (IFN)-α as a potent inducer of APOBEC3G to override HIV Vif neutralization of APOBEC3 proteins that pose a threat to efficient macrophage HIV replication. Our data provide a new dimension by which IFN-α mediates its antiviral activity and suggest a means to render the host nonpermissive for viral replication.
Identified as an HIV virion infectivity factor (Vif) target through subtracted cloning between permissive and nonpermissive cell lines to Vif-deficient HIV (1), cellular apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is packaged into new viral particles during viral synthesis to combat infectivity. APOBEC3G deaminates deoxycytidine to deoxyuridine of minus single–strand nascent viral DNA during reverse transcription, which results in hypermutation of plus-strand DNA. This enzymatic editing of HIV reverse transcripts leads to degradation of deaminated minus-strand DNA (2–4) and blockade of HIV replication, although nonenzymatic antiretroviral mechanisms for APOBEC3G have recently been invoked (5, 6). To circumvent this protective role of host cell APOBEC3G, Vif binds to APOBEC3G, targeting it for proteasome-dependent proteolysis (7–9) or alternative modes of inactivation (5), and in most cases, HIV Vif triumphs over cellular APOBEC3G. Despite the susceptibility of HIV to restriction by this species-specific innate and constitutively expressed path-way (9), little is known regarding the existence of regulatory mechanisms by which to embellish APOBEC3G and/or its related mammalian family of cytidine deaminases to destabilize retroviral genomes and potentially thwart infection.
IFN-α is a potent inhibitor of HIV infection in CCR5+CD4+ human macrophages (10, 11), which in addition to CD4+ T cells, represent major hosts of HIV (12). IFN-α exerts its antiviral activity through multiple mechanisms, including the RNA-dependent protein kinase (PKR)/eukaryotic initiation factor 2 α pathway, oligoadenylate synthetase/RNaseL pathway, and myxovirus resistance GTPase. Activated PKR phosphorylates multiple substrates, including itself, eukaryotic initiation factor 2 α, and IκBα to mediate not only antiviral activity, but also apoptosis and antiinflammatory actions (13, 14). IFN-α induction of an RNA-specific adenosine deaminase, which catalyzes adenosine to inosine in both viral RNA and cellular pre-mRNA to cause hypermutation, may also antagonize infection (13) and suggests a potential influence on other deaminases. Although the role of APOBEC3 proteins in macrophage HIV infection has received less emphasis than in T cells, we examined the possibility that IFN-α may influence APOBEC3G in association with its potent antiretroviral activity in these hosts. Using an in vitro primary human macrophage system, we define an IFN-specific up-regulation of APOBEC3G in association with inhibition of HIV, offering insight into reversing susceptibility to infection by manipulation of this family of innate resistance factors.
RESULTS AND DISCUSSION
IFN-α enhances APOBEC3G
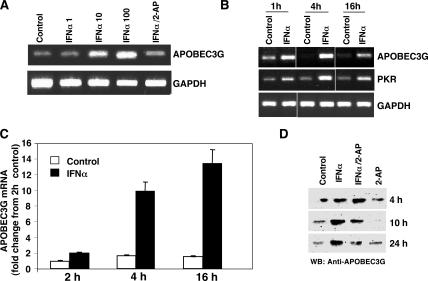
Human primary monocyte–derived macrophages express low levels of APOBEC3G (Fig. 1 A), consistent with a cell nonpermissive to HIV in the absence of Vif (1). To determine whether IFN-α directly influences APOBEC3G expression, macrophages were treated with IFN-α and by RT-PCR; a dose-dependent increase in APOBEC3G gene expression was detected (Fig. 1 A). A kinetic analysis revealed that a single treatment with 10 ng/ml IFN-α enhanced APOBEC3G gene expression within 1 h, which increased further at 4 h (Fig. 1 B). PKR gene expression and activation are prominent consequences of IFN activity (13), and a similar time-dependent pattern of gene expression was observed in macrophages treated with IFN-α for both APOBEC3G and PKR by RT-PCR (Fig. 1 B), and APOBEC3G expression was confirmed by real-time PCR (Fig. 1 C). A corresponding IFN-α–induced increase in APOBEC3G protein was evident in macrophage lysates by Western blot using a monoclonal anti-APOBEC3G antibody (Fig. 1 D). Thus, by both RNA and protein analyses, IFN-α was able to induce elevated APOBEC3G in primary macrophages. This IFN-α response was consistent, although variable baseline levels of APOBEC3G were donor dependent. Furthermore, 2-aminopurine (2-AP), a PKR inhibitor in macrophages (15), suppressed IFN-α–induced APOBEC3G mRNA to basal levels (Fig. 1 A) with a subsequent reduction in protein expression (Fig. 1 D), suggesting that APOBEC3G expression may be downstream of PKR activation. Collectively, these data indicate that IFN-α achieves APOBEC3G augmentation by promoting gene expression rather than inhibiting proteasome-dependent degradation and that there is a connecting link between PKR and APOBEC3G.
Figure 1.
IFN-α induces APOBEC3G expression. (A) Macrophages were not treated (control) or treated with 1–100 ng/ml IFN-α for 18 h in the absence or presence of 1 mM 2-AP, and RNA was analyzed for APOBEC3G by RT-PCR (30 cycles). (B) Macrophages were treated or not with 10 ng/ml IFN-α for the indicated times, and RNA was subjected to RT-PCR to amplify APOBEC3G and PKR mRNA. Representative experiment, n = 3. (C) Macrophages were treated with IFN-α for 2–16 h, and APOBEC3G expression was assessed by real-time PCR as compared with untreated cells cultured in parallel. Data (mean ± SD of triplicates) are presented as fold change from 2-h control macrophages. (D) Cell lysates were obtained after macrophage treatment with 10 ng/ml IFN-α and/or 1 mM 2-AP and assessed for APOBEC3G protein (∼42 kD) by Western blot.
IFN-α reverses HIV suppression of APOBEC3G
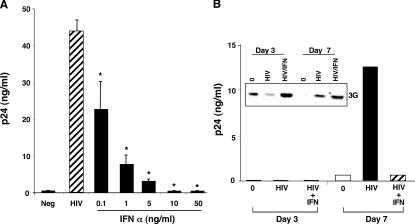
Because IFN-α impedes HIV infection in macrophages (13), induced APOBEC3G may represent a previously unrecognized mechanism underlying this antiviral activity. Associated with the dose-dependent enhancement of APOBEC3G (Fig. 1), IFN-α blocked HIV infection (Fig. 2 A). Although HIV typically suppressed APOBEC3G protein levels (Fig. 2 B, inset, day 3), consistent with the fact that Vif promotes APOBEC3G degradation, the addition of IFN-α reversed the loss of APOBEC3G protein in the infected cultures, coincident with reduced viral replication (Fig. 2 B). Whether the emergence of low APOBEC3G expression around 7 d after infection reflects diminished proteasomal degradation, endogenous IFN-α release (16), and/or other sequelae, increased APOBEC3G in IFN-α–treated infected macrophages remained consistent with restricted HIV expansion (Fig. 2 B). In virus-free cells, APOBEC3G levels characteristically diminish with time in culture. The influence of IFN-α on HIV infection in macrophages appears to occur early in the viral life cycle. For example, a single treatment may prevent productive HIV infection for up to 7 d, compatible with targeting of an event(s) in the virus life cycle before massive replication. An early increase of APOBEC3G by IFN-α likely promotes enhanced APOBEC3G packaging into the initial viral particles, which in turn renders these virions defective in establishing new rounds of infection.
Figure 2.
IFN-α increases APOBEC3G coincident with anti-HIV activity. (A) Dose-dependent inhibition of HIV infection by a single addition of 0–50 ng/ml IFN-α immediately after macrophage (48-well plates) cultures were exposed to HIV for 2 h and washed to remove free virus. Supernatant p24 levels were measured on day 5. n = 3, mean ± SD; *, P < 0.001. (B) Macrophages (six-well plates) were infected with HIV, and 10 ng/ml IFN-α was added after infection and added back every 2–3 d. p24 levels were measured in the supernatants. Inset: APOBEC3G was detected in lysates of macrophages infected with HIV in the presence or absence of IFN-α 3 and 7 d after infection by Western blot (n = 2).
IFN uniquely regulates APOBEC3G
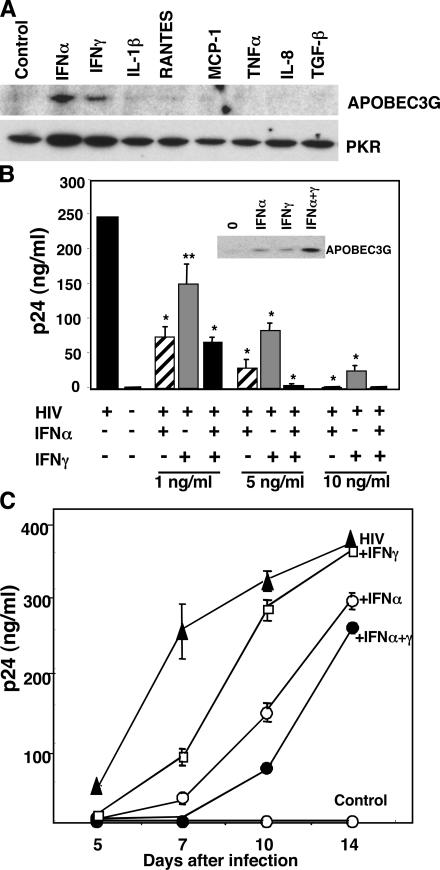
To establish whether IFN-α was a unique regulator of this antiviral molecule in macrophages, we analyzed a panel of cytokines for their ability to alter the expression profile of APOBEC3G protein. As shown in Fig. 3 A, of the cytokines evaluated, only IFN-α and IFN-γ were found to detectably enhance this retroviral restrictor, whereas the remaining inflammatory mediators (TNF-α, IL-1β, IL-8, MCP-1, RANTES, and TGF-β) tested did not promote APOBEC3G production nor augment PKR. Although only a limited number of proteins were analyzed and other potential inducers cannot be excluded, since its recent discovery, there has yet to be any report of APOBEC3G induction by other endogenous factors. Moreover, because IFN-α was reportedly ineffective in enhancing APOBEC3G expression in a T cell line (17) and did not increase frequency of G to A hypermutations in hepatitis B virus stably infected HepG2 cell lines (18), this may represent a key macrophage anti-viral pathway.
Figure 3.
APOBEC3G is inducible by IFN-α and IFN-γ. (A) Macrophages were treated with the indicated cytokines at 100 ng/ml or TGFβ at 10 ng/ml for 20 h. Induction of APOBEC3G in macrophage lysates was examined by Western blot using anti-APOBEC3G and anti-PKR antibodies. (B) Macrophages were infected with HIV (104 TCID50), and IFN-α, IFN-γ, or combined IFNs (1–10 ng/ml) were added. Infected macrophage supernatants were assayed by p24 ELISA. Data shown are mean ± SD; **, P < 0.05; *, P < 0.01. Inset: APOBEC3G protein was assayed by Western blot 3 d after single or dual IFN treatment (5 ng/ml). (C) Kinetics of HIV infection (p24) in macrophages treated only once with IFN at the onset of infection (n = 3, mean ± SD).
IFN-α and IFN-γ collaboratively increase APOBEC3G and inhibit HIV
Because both IFN-α and IFN-γ increase APOBEC3G and inhibit HIV infection in macrophages, and macrophages remain a crucial target and reservoir of HIV in the blood even after HAART (19, 20) as well as in the tissues (21), we sought to determine the relative efficiency of the IFNs on their innate cellular defenses. Although IFN-α is clinically effective against HIV and multiple additional viruses, its toxicity represents a clinical concern (22). By comparison, IFN-γ, functioning as an immune stimulant, has both anti–HIV-1 and PKR activation potential, but to a lesser extent than IFN-α (13). To test the possibility that IFN-γ together with IFN-α might enable use of reduced levels of IFN-α, while achieving a comparable antiviral effect, we compared IFN-α and IFN-γ concentrations required for inhibition of HIV. IFN-α was typically severalfold more potent than IFN-γ in the in vitro infection assay. Costimulation with IFN-α and IFN-γ at suboptimal concentrations in a single application after viral inoculation resulted in complete or near complete antiviral activity, as compared with treatment with IFN-α or IFN-γ alone (Fig. 3 B, day 7 p24 shown). This dual treatment further bolstered APOBEC3G (Fig. 3 B, inset) expression that correlated with enhanced viral resistance. Nonetheless, as shown in Fig. 3 C, a single treatment with IFN-α, IFN-γ, or with IFN-α plus IFN-γ did not inhibit viral replication indefinitely, and reexposure to the IFNs was required to sustain APOBEC3G and maximum viral suppression beyond 1 wk. Whether administered individually or coordinately, these data provide the first indication that APOBEC3G-mediated interference with the production of a functional provirus can be regulated by a host cytokine as a new point of attack in the viral life cycle.
Preferential regulation of APOBEC3 family genes by IFN-α
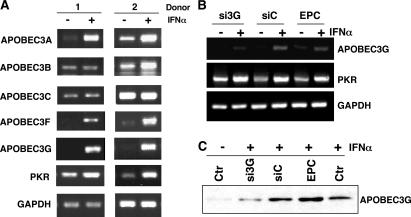
APOBEC3 represents a family of deaminase proteins, some of which reportedly restrict HIV infection (APOBEC3G and APOBEC3F) or are influenced by IFN (APOBEC3A; references 23–26), and we examined whether IFN-α could trigger the expression of additional superfamily genes. In macro-phages from multiple donors, RT-PCR showed that IFN-α not only up-regulated APOBEC3G, but also APOBEC3A and APOBEC3F with a similar profile. Family members APOBEC3B and APOBEC3C were not consistently augmented by IFN-α (Fig. 4 A, two representative donors shown). Consequently, IFN-α appears to preferentially regulate members of the APOBEC3 cluster that reportedly have antiviral activity. In this regard, our analysis of the 5′ region of APOBEC3 family genes (National Center for Biotechnology Information, National Institutes of Health [NIH]) revealed differential expression of IFN-stimulated response elements (ISRE). We identified the existence of an ISRE sequence −47 to −59 from the APOBEC3G translation start codon (GAAAGTGAAAC), and an identical ISRE sequence is also found in APOBEC3F, reflecting their coordinate regulation. A potential promoter region in APOBEC3A, containing an ISRE (−1,787 to −1,800), is located further from the start codon relative to APOBEC3G and is consistent with IFN-α inducibility. Thus, our data support several APOBEC3 nucleic acid–editing enzymes (APOBEC3A, APOBEC3F, and APOBEC3G) as ISRE-competent IFN-α–inducible targets. Among them, both APOBEC3F and APOBEC3G can reportedly restrain Vif-dependent infection mechanisms (1, 24, 25). Although we observed that APOBEC3A is also coordinately expressed after macrophage exposure to IFN-α, to date, no antiviral activity has been ascribed to this family member.
Figure 4.
IFN-α augments induction of APOBEC3 family members. (A) RNA was isolated from macrophages (two donors shown) untreated or treated with 10 ng/ml IFN-α for 4 h and evaluated by RT-PCR with primers corresponding to each of the indicated APOBEC3 genes and PKR (30 cycles). GAPDH was used as a parallel control. (B) Monocytes were transfected with APOBEC3G siRNA, cultured for 6 d, stimulated with IFN-α for 4 h, and APOBEC3G and PKR expression was monitored by RT-PCR in comparison with the GAPDH control. (C) Western blot for APOBEC3G protein 6 d after monocyte transfection with siRNA or control RNA, or electroporation only control (EPC), and treatment with or without IFN-α for 20 h. Representative experiment, n = 3.
Knockdown of APOBEC3G by small interfering RNA (siRNA) blunts inhibition of HIV by IFN-α
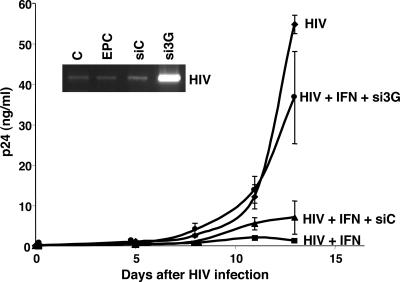
IFN-α executes its antiviral activity through multiple mechanisms, including PKR and RNase L and/or RNA deaminases (13). As our evidence implicates APOBEC3 family induction as one of the potential contributing mechanisms, we considered that removal of APOBEC3G would result in at least a partial block of IFN-α antiviral activity. Using siRNA synthesized based on the sequence of APOBEC3G, we show that transfection of siRNA into macrophages decreased APOBEC3G RNA and protein induction by IFN-α, in contrast to control siRNA (Fig. 4, B and C), without any effect on IFN-α–induced PKR. When the cells were depleted of APOBEC3G by siRNA and then treated with a low dose of IFN-α (1 ng/ml), IFN-α could no longer completely inhibit HIV in the absence of APOBEC3G (Fig. 5). By comparison, the same dose of IFN-α effectively inhibited HIV infection in both untransfected and negative control siRNA-transfected macrophages (Fig. 5), confirming that APOBEC3G is a key downstream anti-HIV mechanism induced by IFN-α. Perhaps not surprisingly, at a higher dose of 10 ng/ml, IFN-α inhibition of HIV infection in macrophages was not substantially altered by siRNA transfection of APOBEC3G (not depicted), likely a result of the multiple antiviral mechanisms triggered by IFN-α. Furthermore, by nested PCR, which detects early viral de novo DNA synthesis (Fig. 5, inset), it was apparent that depletion of APOBEC3G by siRNA resulted in substantially augmented reverse transcription with elevated levels of HIV proviral DNA (day 2 after exposure to HIVBaL shown) once this obstacle was removed.
Figure 5.
APOBEC3G siRNA blocks IFN-α anti-HIV activity. Monocytes were transfected with APOBEC3G siRNA, cultured for 6 d, infected with HIV, and treated or not with 1 ng/ml IFN-α. P24 levels were monitored in the supernatants of untreated and treated macrophage cultures at the indicated time points after infection with HIV. Inset: Nested PCR to evaluate initial rounds of HIV replication (day 2 after infection) in cells transfected with APOBEC3G-specific siRNA or negative control siRNA, electroporation only control (EPC), or left untreated (C), and then infected with HIV. Increased proviral DNA was evident in cells depleted of APOBEC3G. Representative experiment, n = 3.
In what appears to be a unique and powerful pathway, IFN-α increases APOBEC3G expression as a component of its complex repertoire of antiviral mechanisms. This selective induction of APOBEC3G by IFN-α provides a potential mechanism to overcome HIV Vif sequestration of APOBEC3G, thus allowing enhanced APOBEC3G packaging into the budding virions to dampen further infection potential. The net effect is that IFN-α tilts the balance between APOBEC3G and Vif in favor of APOBEC3G as opposed to unimpaired Vif-driven APOBEC3G degradation, which enables HIV replication in subsequent host cells. Demonstration of IFN-α–inducible intracellular APOBEC3G suggests an important strategic approach to regulation of this newly recognized intracellular defense against the deadly virus. Beyond the role of APOBEC3G in retroviral infections, recent evidence documents that APOBEC3G also blocks hepatitis B virus replication, pointing to a broader antiviral profile (18). Moreover, the evidence that multiple APOBEC3 family proteins are inducible by IFN-α indicates the potential for additional elements in the regulatory control of the viral life cycle. With the recent recognition of several new host cell–associated proteins controlled by and/or required for HIV infection, such as p21 and annexin II (27, 28), as well as those that represent potent opposition to HIV infection (TRIM5α, APOBEC3G, and lentivirus susceptibility-1; references 1 and 29), new classes of inhibitors may emerge as future antiviral candidates.
MATERIALS AND METHODS
Isolation and culture of human blood monocytes.
Human peripheral blood mononuclear cells, obtained by leukapheresis of normal volunteers at the Department of Transfusion Medicine (NIH), were diluted in endotoxin-free PBS without Ca2+ and Mg2+ (BioWhittaker) and separated by density centrifugation on lymphocyte sedimentation medium (Organon Teknika Corp.) at 400 g for 30 min. The monocytes in the mononuclear cell layer were enriched by elutriation (27, 28) and plated at 1.5 × 106/ml per well in 48-well plates or 6 × 106/2 ml per well in 6-well plates (Corning) in DMEM (BioWhittaker) with antibiotics and glutamine for 3 h, followed by the addition of 10% FCS. Adherent monocytes were cultured for 7–10 d to enable differentiation into monocyte-derived macrophages.
Infection of macrophages with M-tropic HIV-1.
Unless indicated otherwise, adherent macrophages were incubated with HIV-1BaL(HIV) at 1–2 × 103/TCID50/ml (Advanced Biotechnologies, Inc.) in DMEM containing 10% FCS for 2 h at 37°C. After infection, cells were washed with PBS three times and cultured in DMEM containing 10% FCS for up to 14 d (27, 28). IFN-α and IFN-γ (National Cancer Institute-Frederick Cancer Research and Development Center [NCI-FCRDC]) were added once at the indicated concentrations or every 2–3 d. To measure viral infectivity, supernatants (200 μl) were collected at the indicated intervals and replaced with fresh medium. Infection was measured by supernatant p24 using HIV-1 p24 ELISA kits (PerkinElmer) with data presented as mean ± SD and by nested PCR of extracted DNA as described previously (27, 28). PCR product from the second amplification was visualized by ethidium bromide staining after agarose gel electrophoresis.
RNA isolation and analysis.
6 × 106 macrophages cultured in six-well plates were treated with 1–100 ng/ml IFN-α and/or 1mM 2-AP (Biomol) for the times indicated. Total RNA was extracted using RNeasy Total RNA kit (QIAGEN). For RT-PCR, GeneAmp RNA PCR kit (Perkin Elmer, Branchburg, NJ) was used according to protocol. A total of 0.15–0.3 μgRNA from each sample was used for first-strand cDNA synthesis. The cDNA was then divided and used for PCR amplification of APOBEC3G and other APOBEC3 family genes. PKR and GAPDH were used as controls. All PCRs were performed by 30-cycle amplification, except that 25 cycles were used for GAPDH (94°C for 4 min, followed by 30 cycles at 94°C for 30 s, 55°C for 3 s, 72°C for 30 s, and finally extension at 72°C for 10 min). Primers were as follows: APOBEC3A: 5′-TTCTTTGCAGTTGGACCCGG-3′ (forward), 5′-CTCATCTAGTCCATCCCAGG-3′ (reverse); APOBEC3B: 5′-TGGTCGGAGCTACACTTGGC-3′ (forward), 5′-CAGACAGGAATTCGGCCAGC-3′ (reverse); APOBEC3C: 5′-CTTCCTCTCTTGGTTCTGCG-3′ (forward), 5′-CCATGATCTCCACAGCGACC-3′ (reverse); APOBEC3F: 5′-TACGCAAAGCCTATGGTCGG-3′ (forward), 5′-GCTCCAAGATGTGTACCAGG-3′ (reverse); APOBEC3G: 5′-TTACCTGCTTCACCTCCTGG-3′ (forward), 5′-TCATCTAGTCCATCCCAGGG-3′ (reverse); PKR: 5′-GCCTTTTCATCCAAATGGAATTC-3′ (forward), 5′-GAAATCTGTTCTGGGCTCATG-3′ (reverse); and GAPDH: 5′-CCTTGGAGAAGGCTGGGG-3′ (forward), 5′-CAAAGTTGTCATGGATGACC-3′ (reverse).
For real-time PCR, cDNA was prepared using the GeneAmp RNA PCR Core kit (Applied Biosystems), and cDNA corresponding to 7.5 ng RNA was used for each reaction in triplicate. The primers for APOBEC3G and GAPDH and the FAM dye-labeled probes were from Applied Biosystems. PCR reactions (15 s at 95°C for melting and 1 min at 60°C for annealing/extending; a total of 40 cycles) were conducted with the Real Time PCR System 7500 (Applied Biosystems). Gene expression was determined using the relative quantification: ΔΔCT = (CT Target − CT GAPDH)Test − (CT Target − CT GAPDH)Control. CT is the fractional cycle number that reaches a fixed threshold, CTest is the test of interest, and CControl is the reference control (RNA from 2-h untreated cells). ΔCT is the difference between gene expression in treated cells and reference control cells. The fold increase was calculated using 2-ΔΔCT (30).
Western blot for APOBEC3G protein.
Macrophages were treated with or without IFN-α, IFN-γ, and/or 2-AP for the times indicated, and cells were lysed with ice-cold buffer containing 10 mM Hepes, pH 7.9, 10 mM KCl, and 0.1 mM EDTA, and 0.1 mM EGTA, 0.5 mM PMSF, 1 mM DTT, and 0.5% NP-40. 15–75 μg protein was used for electrophoresis (10% polyacrylamide gel) and blotted onto nitrocellulose membranes. After blocking with 5% milk in Tris-buffered saline with 0.05% Triton X-100 (TBS-T), membranes were probed with monoclonal anti-APOBEC3G at 1:1,000 (Immunodiagnostics) or polyclonal anti-PKR at 1:1,000 (Cell Signaling) at 4°C overnight. Membranes were washed with TBS-T three times for 10 min, followed by secondary antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Inc.), and detected by chemiluminescence (Pierce Chemical Co.). A similar protocol was followed for detection of APOBEC3G in cells treated with 100 ng/ml IFN-γ, TNF-α, IL-1β, IL-8, MCP-1, or RANTES (NCI-FCRDC) or 10 ng/ml TGF-β (R&D Systems) for 20 h.
siRNA inhibition of APOBEC3G.
Monocytes (30 × 106 cells) were treated with 36 μl siRNA (20 nM) or siRNA (control; QIAGEN) in 300 μl Dendritic Nucleofector Solution (Amaxa Biosystems) or siRNA buffer only at room temperature. APOBEC3G siRNA was synthesized based on the sequence r(CGGUCAAGAUGGACCAGCA)dTdT/r(UGCUGGUCCAUCUUGACCG)dAdG. 100-μl aliquots were added to a cuvette for electropora-tion using the U-02 program (27, 28). After electroporation, the cells from three replicate cuvettes were pooled and resuspended in 3 ml RPMI before 0.2 ml (2 × 106) was added into a 24-well plate containing 1 ml RPMI preincubated in a humidified 37°C/5% CO2 incubator for 30 min and incubated for 3 h. The medium was changed to complete DMEM (1.5 ml/well), and the cells were cultured for 6 d before infection with HIV and/or treatment with 0.1–10 ng/ml IFN-α.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute of Dental and Craniofacial Research.
The authors have no conflicting financial interests.
References
- 1. Sheehy, A.M., N.C. Gaddis, J.D. Choi, and M.H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 418:646–650. [DOI] [PubMed] [Google Scholar]
- 2. Zhang, H., B. Yang, R.J. Pomerantz, C. Zhang, S.C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 424:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 424:99–103. [DOI] [PubMed] [Google Scholar]
- 4. Harris, R.S., K.N. Bishop, A.M. Sheehy, H.M. Craig, S.K. Petersen-Mahrt, I.N. Watt, M.S. Neuberger, and M.H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell. 113:803–809. [DOI] [PubMed] [Google Scholar]
- 5. Newman, E.N., R.K. Holmes, H.M. Craig, K.C. Klein, J.R. Lingappa, M.H. Malim, and A.M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170. [DOI] [PubMed] [Google Scholar]
- 6. Chiu, Y.L., V.B. Soros, J.F. Kreisberg, K. Stopak, W. Yonemoto, and W.C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 435:108–114. [DOI] [PubMed] [Google Scholar]
- 7. Marin, M., K.M. Rose, S.L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398–1403. [DOI] [PubMed] [Google Scholar]
- 8. Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X.F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 302:1056–1060. [DOI] [PubMed] [Google Scholar]
- 9. Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N.R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 114:21–31. [DOI] [PubMed] [Google Scholar]
- 10.Meylan, P.R., J.C. Guatelli, J.R. Munis, D.D. Richman, and R.S. Kornbluth. 1993. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology. 193:138–148. [DOI] [PubMed] [Google Scholar]
- 11.Baca-Regen, L., N. Heinzinger, M. Stevenson, and H.E. Gendelman. 1994. Alpha interferon-induced antiretroviral activities: restriction of viral nucleic acid synthesis and progeny virion production in human immunodeficiency virus type 1-infected monocytes. J. Virol. 68:7559–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wahl, S.M., T. Greenwell-Wild, G. Peng, G. Ma, J.M. Orenstein, and N. Vazquez. 2003. Viral and host cofactors facilitate HIV-1 replication in macrophages. J. Leukoc. Biol. 74:726–735. [DOI] [PubMed] [Google Scholar]
- 13.Samuel, C.E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams, B.R. 2001. Signal integration via PKR. Sci. STKE. 89:re 2. [DOI] [PubMed] [Google Scholar]
- 15.Gusella, G.L., T. Musso, S.E. Rottschafer, K. Pulkki, and L. Varesio. 1995. Potential requirement of a functional double-stranded RNA-dependent protein kinase (PKR) for the tumoricidal activation of macrophages by lipopolysaccharide or IFN-alpha beta, but not IFN-gamma. J. Immunol. 154:345–354. [PubMed] [Google Scholar]
- 16.Szebeni, J., C. Dieffenbach, S.M. Wahl, C.N. Venkateshan, A. Yeh, M. Popovic, S. Gartner, L.M. Wahl, M. Peterfy, R.M. Friedman, et al. 1991. Induction of alpha interferon by human immunodeficiency virus type 1 in human monocyte-macrophage cultures. J. Virol. 65:6362–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose, K.M., M. Marin, S.L. Kozak, and D. Kabat. 2004. Transcriptional regulation of APOBEC3G, a cytidine deaminase that hypermutates human immunodeficiency virus. J. Biol. Chem. 279:41744–41749. [DOI] [PubMed] [Google Scholar]
- 18.Noguchi, C., H. Ishino, M. Tsuge, Y. Fujimoto, M. Imamura, S. Takahashi, and K. Chayama. 2005. G to A hypermutation of hepatitis B virus. Hepatology. 41:626–633. [DOI] [PubMed] [Google Scholar]
- 19.Lambotte, O., Y. Taoufik, M.G. de Goer, C. Wallon, C. Goujard, and J.F. Delfraissy. 2000. Detection of infectious HIV in circulating monocytes from patients on prolonged highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 23:114–119. [DOI] [PubMed] [Google Scholar]
- 20.Garbuglia, A.R., M. Zaccarelli, S. Calcaterra, G. Cappiello, R. Marini, and A. Benedetto. 2001. Dynamics of viral load in plasma and HIV DNA in lymphocytes during highly active antiretroviral therapy (HAART): high viral burden in macrophages after 1 year of treatment. J. Chemother. 13:188–194. [DOI] [PubMed] [Google Scholar]
- 21.Orenstein, J.M., C. Fox, and S.M. Wahl. 1997. Macrophages as a source of HIV during opportunistic infections. Science. 276:1857–1861. [DOI] [PubMed] [Google Scholar]
- 22.Lane, H.C., V. Davey, J.A. Kovacs, J. Feinberg, J.A. Metcalf, B. Herpin, R. Walker, L. Deyton, R.T. Davey Jr., J. Falloon, et al. 1990. Interferon-alpha in patients with asymptomatic human immunodeficiency virus (HIV) infection. A randomized, placebo-controlled trial. Ann. Intern. Med. 112:805–811. [DOI] [PubMed] [Google Scholar]
- 23.Zheng, Y.H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B.M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liddament, M.T., W.L. Brown, A.J. Schumacher, and R.S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385–1391. [DOI] [PubMed] [Google Scholar]
- 25.Wiegand, H.L., B.P. Doehle, H.P. Bogerd, and B.R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor, M.W., W.M. Grosse, J.E. Schaley, C. Sanda, X. Wu, S.C. Chien, F. Smith, T.G. Wu, M. Stephens, M.W. Ferris, et al. 2004. Global effect of PEG-IFN-alpha and ribavirin on gene expression in PBMC in vitro. J. Interferon Cytokine Res. 24:107–118. [DOI] [PubMed] [Google Scholar]
- 27.Ma, G., T. Greenwell-Wild, K. Lei, W. Jin, J. Swisher, N. Hardegen, C.T. Wild, and S.M. Wahl. 2004. Secretory leukocyte protease inhibitor binds to annexin II, a cofactor for macrophage HIV-1 infection. J. Exp. Med. 200:1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez, N., T. Greenwell-Wild, N.J. Marinos, W.D. Swaim, S. Nares, D.E. Ott, U. Schubert, P. Henklein, J.M. Orenstein, M.B. Sporn, and S.M. Wahl. 2005. Human immunodeficiency virus type 1-induced macrophage gene expression includes the p21 gene, a target for viral regulation. J. Virol. 79:4479–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stremlau, M., C.M. Owens, M.J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 427:848–853. [DOI] [PubMed] [Google Scholar]
- 30.Livak, K.J., and T.D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. [DOI] [PubMed] [Google Scholar]