Abstract
The polycomb group (PcG) protein Bmi1 plays an essential role in the self-renewal of hematopoietic and neural stem cells. Derepression of the Ink4a/Arf gene locus has been largely attributed to Bmi1-deficient phenotypes in the nervous system. However, its role in hematopoietic stem cell (HSC) self-renewal remained undetermined. In this study, we show that derepressed p16Ink4a and p19Arf in Bmi1-deficient mice were tightly associated with a loss of self-renewing HSCs. The deletion of both Ink4a and Arf genes substantially restored the self-renewal capacity of Bmi1−/− HSCs. Thus, Bmi1 regulates HSCs by acting as a critical failsafe against the p16Ink4a- and p19Arf-dependent premature loss of HSCs. We further identified a novel role for Bmi1 in the organization of a functional bone marrow (BM) microenvironment. The BM microenvironment in Bmi1−/− mice appeared severely defective in supporting hematopoiesis. The deletion of both Ink4a and Arf genes did not considerably restore the impaired BM microenvironment, leading to a sustained postnatal HSC depletion in Bmi1−/−Ink4a-Arf−/− mice. Our findings unveil a differential role of derepressed Ink4a and Arf on HSCs and their BM microenvironment in Bmi1-deficient mice. Collectively, Bmi1 regulates self-renewing HSCs in both cell-autonomous and nonautonomous manners.
Polycomb group (PcG) genes are involved in cellular memory by maintaining gene silencing through chromatin modifications (1, 2). Recent studies have implicated the role of PcG genes in stem cell self-renewal, a process in which cellular memory is precisely maintained through cell division (2, 3). Among PcG genes, Bmi1 plays a central role in the inheritance of the stemness of hematopoietic and neural stem cells (3–8), and its forced expression promotes hematopoietic stem cell (HSC) self-renewal (8). These findings highlight the importance of epigenetic regulation in stem cell self-renewal.
One of the major Bmi1 targets is the Ink4a/Arf locus (9). This locus encodes a cyclin-dependent kinase inhibitor, p16Ink4a, and a tumor suppressor, p19Arf. p16Ink4a inhibits the binding of cyclin D to Cdk4/6 and keeps retinoblastoma protein (Rb) hypophosphorylated. Hypophosphorylated Rb represses E2F-dependent transcription by sequestrating E2F, ultimately leading to cell cycle arrest or senescence. p19Arf inhibits MDM2 and ARF-BP1, which mediate the ubiquitin-dependent degradation of p53, leading to the accumulation of p53 protein. This results in activation of the p53 target genes involved in cell cycle arrest, apoptosis, or senescence (10). In Bmi1-deficient mice, the expression of Ink4a and Arf is markedly increased in hematopoietic cells (7, 8), and the enforced expression of Ink4a and Arf in HSCs resulted in cell cycle arrest and p53-dependent apoptosis, respectively (7). Conversely, Bmi1;Ink4a/Arf compound mutant mice (hereafter referred to as Bmi1 −/− Ink4a-Arf −/− mice) exhibited a substantial recovery of hematopoietic cells, as indicated by restored lymphocyte counts (9, 11), as well as of the self-renewal capacity of neural stem cells (11, 12). However, the real impact of derepressed Ink4a and Arf in self-renewing HSCs has not yet been determined using a genetic approach.
To address this question, we performed a detailed analysis of HSCs in Bmi1 −/− Ink4a-Arf −/− mice and identified a critical role for Bmi1-dependent repression of the p16Ink4a–Rb and p19Arf–p53 pathways in the maintenance of self-renewing HSCs. We further demonstrated evidence of the involvement of Bmi1 in the regulation of HSCs and their BM microenvironment in a way that is not associated with the Ink4a and Arf locus.
RESULTS AND DISCUSSION
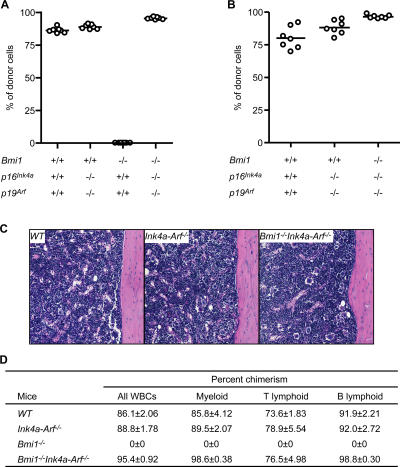
To clarify the contribution of derepressed Ink4a and Arf to the self-renewal defect of Bmi1 −/− HSCs, we evaluated the competitive repopulation capacity of Bmi1 −/− Ink4a-Arf −/− HSCs. Total BM cells from 4-wk-old wild-type, Ink4a-Arf −/−, Bmi1 −/−, and Bmi1 −/− Ink4a-Arf −/− mice (C57BL/6-Ly5.2) were infused into lethally irradiated recipients (C57BL/6-Ly5.1) along with the same number of competitor BM cells from C57BL/6-Ly5.1 mice. Bmi1 −/− Ink4a-Arf −/− BM cells exhibited a mostly normal long-term repopulating activity of the recipient BM in both primary and secondary transplantations, whereas Bmi1 −/− BM cells did not contribute to long-term repopulation at all (Fig. 1, A and B). Bmi1 −/− Ink4a-Arf −/− BM cells fully repopulated recipients' BM in cellularity (Fig. 1 C) and also manifested a full differentiation capacity along myeloid and lymphoid lineages (Fig. 1 D). As evident in Fig. S1 (available at http://www.jem.org/cgi/content/full/jem.20052477/DC1), the frequencies of Bmi1 −/− and Bmi1 −/− Ink4a-Arf −/− CD34−c-Kit+Sca-1+lineage marker− (KSL) cells, which are highly enriched for long-term repopulating HSCs (13), were comparable with that of the wild type. Bmi1 −/− mice displayed a HSC frequency no less than that of the wild type, and Bmi1 −/− Ink4a-Arf −/− mice exhibited almost the same HSC frequency as that of the wild type. Thus, the number of HSCs infused was comparable among recipients in the competitive repopulation assay. Even with 10 times more donor cells, Bmi1 −/− BM cells did not contribute to the repopulation at all, highlighting a severe defect of Bmi1 −/− HSC function (Fig. S2). All of these data clearly demonstrate that the defective self-renewal capacity of Bmi1 −/− HSCs could be substantially rescued by the deletion of Ink4a and Arf, thus defining the Ink4a/Arf locus as a critical Bmi1 target for the maintenance of HSC self-renewal.
Figure 1.
Substantial recovery of the defective repopulation capacity of Bmi1−/− HSCs by the deletion of Ink4a and Arf. (A) Competitive lymphohematopoietic repopulating assay. 1 × 106 pooled test BM cells from 4-wk-old mice (B6-Ly5.2) were mixed with 1 × 106 competitor BM cells from 12-wk-old wild-type mice (B6-Ly5.1) and injected into lethally irradiated recipient mice (B6-Ly5.1; n = 7). The percent chimerism of donor cells in the recipient peripheral blood cells 12 wk after transplantation is presented. (B) Secondary transplantation analysis. 2 × 106 pooled BM cells from primary recipients were injected into lethally irradiated secondary recipient mice (B6-Ly5.1; n = 7). The percent chimerism of donor cells 12 wk after transplantation is presented. (A and B) The mean values are indicated as horizontal bars. (C) Hematoxylin and eosin staining of sections of decalcified femur from primary recipients that had transplanted donor cells of the indicated genotype 12 wk before. Donor cell chimerism in the recipient peripheral blood cells was around 90%, as depicted in A and D, and the absolute BM cell numbers were comparable (4.2 × 107 WT cells, 4.75 × 107 Ink4a-Arf −/− cells, and 4.15 × 107 Bmi1 −/− Ink4a-Arf −/− cells for one pair of femur and tibia). (D) The percent chimerism of donor cells in each lineage 12 wk after primary transplantation is presented as the mean ± SD. WBC, white blood cell.
The deletion of Arf alone scarcely restored the self-renewal defect of Bmi1 −/− HSCs, and the chimerism of Bmi1 −/− Arf −/− hematopoietic cells in peripheral blood gradually decreased with time (unpublished data). This presents a striking contrast to the major role of Arf derepression in Bmi1 −/− phenotypes in neural stem cell self-renewal and cerebellar granule neuron progenitor proliferation (11).
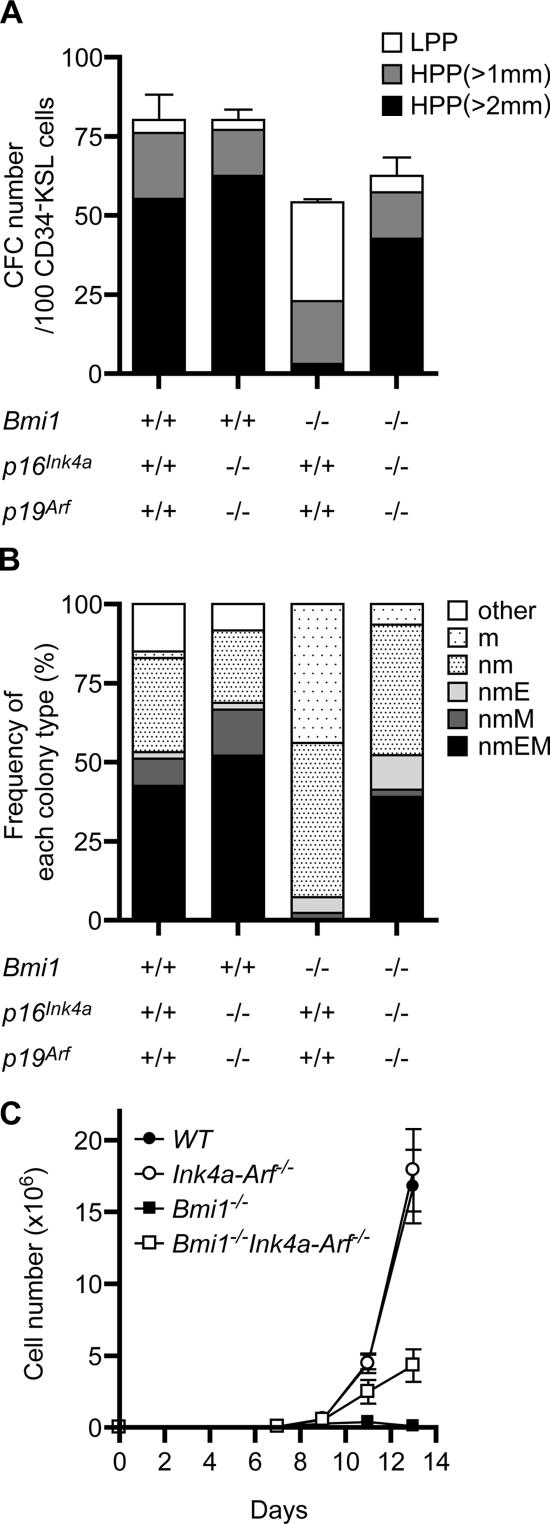
To further evaluate the proliferative and differentiation capacity of Bmi1 −/− Ink4a-Arf −/− HSCs, we purified the CD34−KSL HSC fraction, and an in vitro single-cell culture was performed for 14 d in the presence of stem cell factor (SCF), IL-3, thrombopoietin (TPO), and erythropoietin (EPO). Although Bmi1 −/− HSCs contained 3.3-fold fewer high proliferative potential (HPP) colony-forming cells (CFCs) than the wild type, Bmi1 −/− Ink4a-Arf −/− HSCs contained a comparable number of HPP-CFCs with the wild type (Fig. 2 A). We have previously demonstrated that CFU-neutrophil/macrophage/erythroblast/megakaryocyte (nmEM), which retains multilineage differentiation capacity, is a major subpopulation among CD34−KSL HSCs but not among CD34+KSL multipotential progenitor cells and that its frequency is well correlated with that of functional HSCs (14). Of note, the morphological analysis of HPP colonies revealed that Bmi1 −/− CD34−KSL cells present a drastic reduction in their frequency of CFU-nmEM, whereas Bmi1 −/− Ink4a-Arf −/− HSCs show a substantial recovery in their frequency of CFU-nmEM compared with the wild type (Fig. 2 B). In an in vitro culture of pooled CD34−KSL HSCs, however, Bmi1 −/− Ink4a-Arf −/− HSCs exhibited a considerable but only partial recovery of proliferation (Fig. 2 C). In vitro culture systems are a kind of stringent condition in which numerous signaling entities are missing that are supportive for HSCs and are present in the in vivo microenvironment. Thus, these findings suggest that the deletion of Ink4a and Arf does not completely restore the defective proliferative and differentiation capacity of Bmi1 −/− HSCs.
Figure 2.
The deletion of Ink4a and Arf largely restores the proliferative and differentiation capacity of Bmi1−/− HSCs in vitro. (A) Single HSC growth assay. 96 individual CD34−KSL HSCs were sorted clonally into 96-well microtiter plates in the presence of SCF, IL-3, TPO, and EPO. The numbers of high (HPP) and low proliferative potential (LPP) CFCs were retrospectively evaluated by counting colonies on day 14 (HPP- and LPP-CFCs, colony diameters of >1 and <1 mm, respectively). The results are shown as the mean ± SD (error bars) of triplicate cultures. (B) Frequency of each colony type. Colonies derived from HPP-CFCs were recovered and morphologically identified as neutrophils (n), macrophages (m), erythroblasts (E), or megakaryocytes (M). (C) Growth of CD34−KSL HSCs in vitro. 50 freshly isolated CD34−KSL cells were cultured in the presence of SCF, IL-3, and TPO. The results are shown as the mean ± SD of triplicate cultures.
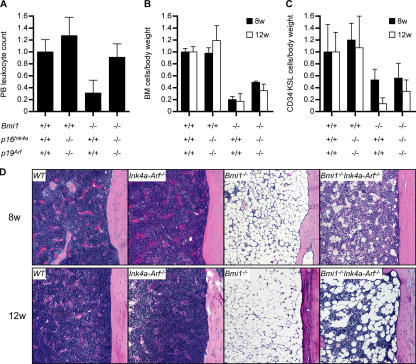
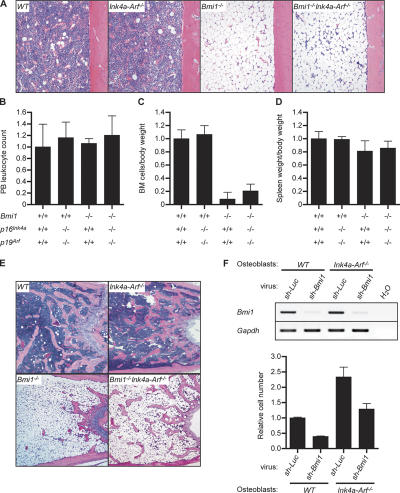
We next examined hematopoiesis in Bmi1 −/− Ink4a-Arf −/− mice in detail. The peripheral leukocyte count of Bmi1 −/− Ink4a-Arf −/− mice recovered to the same level as that of the wild type (Fig. 3 A). However, the recovery of BM cellularity as well as the number of CD34−KSL HSCs was incomplete in Bmi1 −/− Ink4a-Arf −/− mice, and, unexpectedly, their numbers progressively decreased over time (Fig. 3, B and C). Histological analysis of femurs showed a severely hypoplastic fatty marrow in Bmi1 −/− mice as previously described (4). This phenotype was not completely rescued by the deletion of Ink4a and Arf genes and severely progressed even in Bmi1 −/− Ink4a-Arf −/− mice (Fig. 3 D). Given that the BM repopulation capacity of Bmi1 −/− Ink4a-Arf −/− HSCs is mostly normal (Fig. 1, A and B), these data indicate defects of the BM microenvironment in the absence of Bmi1. This possibility was confirmed by transplanting wild-type BM cells into irradiated Bmi1 −/− mice. Although the peripheral blood leukocyte count and the spleen weight in Bmi1 −/− recipients recovered to the wild-type level after transplantation, the histological analysis of recipients' femurs and their BM cell counts demonstrated that the Bmi1 −/− BM microenvironment is defective in supporting the BM repopulation by wild-type HSCs (Fig. 4, A–D). Surprisingly, the deletion of both Ink4a and Arf did not considerably restore the impaired capacity of the Bmi1 −/− BM microenvironment to support hematopoiesis by wild-type HSCs (Fig. 4, A and C).
Figure 3.
Incomplete recovery of defective hematopoiesis in Bmi1−/−Ink4a-Arf−/− mice. (A) Peripheral blood leukocyte count in 8-wk-old mice (n ≥ 4). (B) BM mononuclear cell count per body weight (n ≥ 3). (C) Quantification of the number of CD34−KSL cells per body weight (n ≥ 3). All data were normalized relative to the wild type and are shown as the mean ± SD (error bars). (D) Hematoxylin and eosin staining of sections of decalcified femur from 8- and 12-wk-old mice.
Figure 4.
Impaired BM microenvironment in Bmi1−/− mice. (A–D) Wild-type, Ink4a-Arf −/−, Bmi1 −/−, and Bmi1 −/− Ink4a-Arf −/− recipient mice were transplanted with 2 × 106 wild-type BM cells. At 4 wk after transplantation, recipient mice were analyzed on their BM cellularity (femur, A), peripheral blood leukocyte count (B), BM cell number per body weight (C), and spleen weight per body weight (D). Donor cell chimerism in recipient peripheral blood mononuclear cells was 80.1 ± 4.2, 78.7 ± 2.4, 98.8 ± 0.38, and 82.5 ± 10.1% with wild-type, Ink4a-Arf −/−, Bmi1 −/−, and Bmi1 −/− Ink4a-Arf −/− recipients, respectively (n ≥ 3). (E) Hematoxylin and eosin staining of sections of decalcified distal femur from 8-wk-old mice. (F) Analyses of Bmi1 knockdown osteoblasts. Primary cultured wild-type and Ink4a-Arf −/− osteoblasts were infected with lentiviruses expressing shRNA against either luciferase (Luc; control) or Bmi1. The infection efficiency was almost 100% in all knockdown experiments. The knockdown efficiencies were evaluated by detecting Bmi1 mRNA expression by RT-PCR analysis (top), and their growth was monitored at day 5 of culture (bottom). The results are shown as the mean ± SD (error bars) of triplicate cultures.
The regulation of self-renewal and differentiation of HSCs requires a specific BM microenvironment. In BM, a subpopulation of osteoblasts has been implicated as an important component of the HSC niche, indicating that the bone surface is the major HSC niche (15–17). The size of the osteoblastic niche is largely dependent on the amount of trabecular bone (15, 16). In Bmi1 −/− BM, development of the trabecular bone was severely impaired, particularly in the metaphyseal area (Fig. 4 E). This indicates a profound reduction in the osteoblastic niche and raises the possibility of an insufficient production of osteoblasts.
To further characterize the role of Bmi1 in osteoblasts as niche cells, we analyzed primary cultured Bmi1 −/− osteoblasts. Bmi1 −/− osteoblasts showed a normal level of alkaline phosphatase activity, which is one of the representative osteoblastic differentiation markers (Fig. S3 A, available at http://www.jem.org/cgi/content/full/jem.20052477/DC1). RT-PCR analysis of the osteoblast-specific marker genes (Osteopontin, Osteocalcin, Runx2, Ostetix, and Col1a1) as well as known HSC niche factor genes (N-cadherin, Angiopoietin-1, -2, Jagged-1, and SCF) was unable to discern any gross difference between the wild-type and Bmi1 −/− osteoblasts, although p16Ink4a and p19Arf were also derepressed in Bmi1 −/− osteoblasts (Fig. S3 B). We then took advantage of the Bmi1 knockdown technique. Osteoblasts were infected with a lentivirus expressing short hairpin RNA (shRNA) against Bmi1, which efficiently inhibited the transcription of Bmi1 (Fig. 4 F). Consistent with in vivo trabecular bone formation, Bmi1 knockdown led to a reduced osteoblast proliferation (Fig. 4 F). Nonetheless, Bmi1 knockdown osteoblasts similarly supported the survival and multilineage differentiation capacity of CD34−KSL HSCs during a 5-d ex vivo culture (Fig. S3 C). Collectively, these findings suggest that Bmi1 controls the BM microenvironment, at least in part, by regulating osteoblast niche size. In contrast with the case of HSCs, however, the deletion of both Ink4a and Arf again did not substantially restore the impaired development of the trabecular bone (Fig. 4 E) or the impaired proliferation of Bmi1 knockdown osteoblasts (Fig. 4 F), confirming that the Ink4a and Arf genes are not the major targets for Bmi1 in the maintenance of the BM microenvironment, as demonstrated in Fig. 4 (A and C). The BM microenvironment consists of not only osteoblasts but also stromal cells, endothelial cells (18), and so on. It would be intriguing to ask whether Bmi1 also functions in the other components of the BM microenvironment.
Our findings in this study clearly demonstrate that the derepression of Ink4a and Arf genes is responsible for defective HSC self-renewal. However, we have previously reported that Bmi1 −/− HSCs undergo the first cell division in a fashion similar to that of the wild type and showed no apoptosis in a single HSC culture. In addition, cell cycle analysis of BM primitive hematopoietic cells (KSL and Lin− cells) did not detect any difference between the wild-type and Bmi1 −/− mice (8). These findings indicate that the derepression of Ink4a and Arf genes in Bmi1 −/− mice do not grossly affect the cell cycle or survival of HSCs.
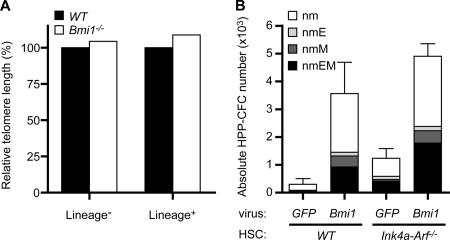
It has been well recognized that the activated p16Ink4a–Rb and p19Arf–p53 pathways are profoundly associated with cellular senescence (19). Cellular senescence is a program activated by normal cells in response to various types of stress. These include telomere attrition, DNA damage, oxidative stress, oncogenic stress, and others. Senescence of HSCs is supposed to be induced by telomere-dependent and -independent pathways (20, 21). We first measured the telomere length of wild-type and Bmi1 −/− lineage marker− immature cells and lineage marker+ differentiated cells by fluorescence in situ hybridization. The loss of Bmi1 did not alter the telomere length at all (Fig. 5 A). In the absence of Bmi1, the derepression of Ink4a and Arf genes causes the premature senescence of mouse embryonic fibroblasts (9). Bmi1 knockdown osteoblasts indeed exhibited a higher senescence-associated (SA) β-galactosidase activity, which was canceled in the absence of Ink4a and Arf genes (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20052477/DC1), suggesting that Bmi1 controls the cellular senescence of osteoblasts by regulating the expression of Ink4a and Arf genes. We then analyzed freshly isolated Bmi1 −/− CD34−KSL HSCs in terms of the SA–β-galactosidase activity and SA gene expression profiles, but all appeared negative (unpublished data). It is possible that the senescent HSCs do not express specific combinations of marker antigens for HSC identification any more. Thus, the possibility that derepressed Ink4a and Arf genes facilitate the premature senescence of HSCs remains to be determined.
Figure 5.
Additional targets for Bmi1 exist other than Ink4a and Arf genes in the maintenance of HSCs. (A) The relative telomere length of the BM lineage− and lineage+ cells measured by flow fluorescence in situ hybridization. (B) CD34−KSL cells were transduced with either GFP control or Bmi1 retroviruses and were cultured in the presence of SCF and TPO. At day 10 of culture, colony assays were performed to evaluate the content of HPP-CFCs in culture. GFP+ colonies derived from HPP-CFCs were examined as to their colony types with morphological analysis. The results are shown as the mean ± SD (error bars) of triplicate cultures. Neutrophils, n; macrophages, m; erythroblasts, E; megakaryocytes, M.
In contrast to the strong impact of derepressed p16Ink4a and p19Arf on HSC self-renewal, the loss of p16Ink4a and p19Arf has been reported to have a limited role in this process (22). In our analyses, freshly isolated Ink4a-Arf −/−HSCs did not show any advantages in competitive BM repopulation assays either. However, Ink4a-Arf −/− HSCs retained their self-renewal capacity better than the wild type during long-term ex vivo culture (unpublished data). These findings suggest that a tight repression of Ink4a and Arf genes by Bmi1 accounts for a positive effect of forced Bmi1 expression on HSC self-renewal and multipotential progenitor expansion (8). To confirm this, we transduced Ink4a-Arf −/− HSCs with a Bmi1 retrovirus, cultured for 10 d in the presence of SCF and TPO, and subjected the cells to colony assays. Unexpectedly, the overexpression of Bmi1 in Ink4a-Arf −/− HSCs again induced a similar mode of multipotential progenitor expansion to that in wild-type HSCs (Fig. 5 B). These data, together with the incomplete recovery in the proliferative capacity of Bmi1 −/− Ink4a-Arf −/− HSCs in vitro, indicate that additional targets for Bmi1 exist other than Ink4a and Arf genes, which are implicated in the regulation of HSC self-renewal and multipotential progenitor expansion, although they are largely dispensable in vivo.
Together, all of these observations implicate Bmi1 in both the cell-autonomous and nonautonomous regulation of the HSC system. Similar to BM hematopoiesis, an incomplete recovery of lymphocyte numbers in Bmi1 −/− Ink4a-Arf −/− mice could be ascribed to certain defects in the Bmi1 −/− microenvironment of the spleen and thymus (9, 11). Our findings further unveiled the differential impact of derepressed Ink4a and Arf on HSCs and their BM microenvironment in Bmi1-deficient mice, thus defining Ink4a and Arf as the major targets for Bmi1 in the maintenance of HSC self-renewal but not of the BM microenvironment.
Finally, Bmi1 has been demonstrated to be essential for the maintenance of leukemic stem cells in a mouse model of acute myelogenous leukemia induced by the Hoxa9-Meis1 fusion gene (5). It has also been demonstrated that the Rb and p53-dependent cellular senescence plays a critical role to oppose neoplastic transformation triggered by the activation of oncogenic pathways (19). It will be important to investigate whether the up-regulation of Bmi1 contributes to repression of the oncogene-induced senescence pathway in the leukemic transformation and maintenance of the self-renewal capacity of leukemic stem cells.
MATERIALS AND METHODS
Mice.
Bmi1 +/− mice and Ink4a-Arf −/− mice (provided by R.A. DePinho, Harvard Medical School, Boston, MA) that had been backcrossed at least eight times onto a C57BL/6 (B6-Ly5.2) background were used. Mice congenic for the Ly5 locus (B6 Ly5.1) were bred and maintained at the Animal Research Center of the Institute of Medical Science (University of Tokyo). All experiments using mice received approval from the Tokyo University Administrative Panel for Animal Care.
Competitive repopulation assay.
Hematopoietic cells from B6-Ly5.2 mice were mixed with BM competitor cells (B6-Ly5.1) and were transplanted into B6-Ly5.1 mice irradiated at a dose of 9.5 Gy. Donor cell chimerism in the recipient peripheral blood cells was evaluated as previously described (8). The ability of the Bmi1 −/− microenvironment to support hematopoiesis was evaluated by transplanting 2 × 106 wild-type BM cells (B6-Ly5.1) into 4-wk-old mutant mice (B6-Ly5.2) sublethally irradiated (Bmi1 −/− and Bmi1 −/− Ink4a-Arf −/− mice, 4.5 Gy; others, 6.5 Gy).
Purification of mouse HSCs and single-cell colony assay.
Mouse HSCs (CD34−KSL cells) were purified from BM cells of 8-wk-old mice on a flow cytometry system (FACSVantage; Becton Dickinson) as previously described (8). Single CD34−KSL cells were sorted clonally into 96-well plates containing 200 μl SF-O3 (Sanko Junyaku) supplemented with 5 × 10−5 M 2-β-mercaptoethanol, 2 mM l-glutamine, 10% FBS, 20 ng/ml of mouse SCF, 20 ng/ml of mouse IL-3, 50 ng/ml of human TPO, and 1 unit/ml of human EPO (PeproTech).
Primary BM-derived osteoblast culture and Bmi1 knockdown.
Femurs and tibiae were cut into small pieces after BM cells were fully flushed out. Then, bone fragments were cultured in α-MEM supplemented with 2 mM l-glutamine, 10% FCS, and 5 × 10−5 M 2-β-mercaptoethanol. Suspension cells were removed by replacing the medium. Osteoblastic phenotypes were evaluated by the expression of alkaline phosphatase. A lentivirus vector (CS-H1-shRNA-EF-1α-EGFP) expressing shRNA against mouse Bmi1 (target sequence TAAAGGATTACTACACGCTAATG) and Luciferase was prepared, and the viruses were produced as previously described (23).
RT-PCR.
Semiquantitative RT-PCR was performed using normalized cDNA with quantitative PCR using TaqMan rodent GAPDH control reagent (PerkinElmer) as previously described (8).
Quantification of telomere length.
Telomere length was quantified on a flow cytometer (FACSCalibur; BD Biosciences) using flow fluorescence in situ hybridization with a Telomere PNA Kit/FITC for flow cytometry (DakoCytomation).
Transduction of CD34−KSL cells.
The retrovirus vector pGCDNsam-ires-EGFP (provided by M. Onodera, University of Tsukuba, Ibaraki, Japan), the production and concentration of recombinant retrovirus, and the transduction of CD34−KSL cells have been described previously (8). After transduction, the cells were further incubated for 9 d in S-Clone SF-O3 supplemented with 5 × 10−5 M 2-β-mercaptoethanol, 2 mM l-glutamine, 1% FBS, 50 ng/ml SCF, and 50 ng/ml TPO and subjected to in vitro colony assay using a methylcellulose medium (StemCell Technologies Inc.) supplemented with 20 ng/ml of mouse SCF, 20 ng/ml of mouse IL-3, 50 ng/ml of human TPO, and 1 unit/ml of human EPO. GFP+ colony numbers were counted on day 10. Colonies derived from HPP-CFCs (colony diameter of >1 mm) were recovered and morphologically examined. The transduction efficiency was >80% as judged from the GFP expression.
Online supplemental material.
Fig. S1 provides data for flow cytometric profiles and frequencies of HSCs in mutant mice. Fig. S2 provides data for the competitive BM repopulating assay using 10 times more test cells than the competitor cells. Fig. S3 provides data for differentiation and the HSC-supporting capacity of osteoblasts in the absence of Bmi1. Fig. S4 provides data for the senescence of Bmi1 knockdown osteoblasts. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20052477/DC1.
Supplemental Material
Acknowledgments
We thank Dr. M. Onodera for providing pGCDNsam-ires-EGFP, Dr. R.A. DePinho for Ink4a +/− mice, and H. Tsukui and Y. Yamazaki for excellent technical assistance.
This work was supported, in part, by grants from the Ministry of Education, Culture, Sport, Science and Technology of Japan, the Core Research for Evolutional Science and Technology of Japan Science and Technology Corporation, and the Mochida Memorial Foundation for Medical and Pharmaceutical Research.
The authors have no conflicting financial interests.
References
- 1.Lund, A.H., and M. van Lohuizen. 2004. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16:239–246. [DOI] [PubMed] [Google Scholar]
- 2.Valk-Lingbeek, M.E., S.W.M. Bruggeman, and M. van Lohuizen. 2004. Stem cells and cancer: the polycomb connection. Cell. 118:409–418. [DOI] [PubMed] [Google Scholar]
- 3.Iwama, A., H. Oguro, M. Negishi, Y. Kato, and H. Nakauchi. 2005. Epigenetic regulation of hematopoietic stem cell self-renewal by polycomb group genes. Int. J. Hematol. 81:294–300. [DOI] [PubMed] [Google Scholar]
- 4.van der Lugt, N.M., J. Domen, K. Linders, M. van Roon, E. Robanus-Maandag, H. te Riele, M. van der Valk, J. Deschamps, M. Sofroniew, M. van Lohuizen, and A. Berns. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8:757–769. [DOI] [PubMed] [Google Scholar]
- 5.Lessard, J., and G. Sauvageau. 2003. Bmi-1 determines proliferative capacity of normal and leukemic stem cells. Nature. 423:255–260. [DOI] [PubMed] [Google Scholar]
- 6.Molofsky, A.V., R. Pardal, T. Iwashita, I.K. Park, M.F. Clarke, and S.J. Morrison. 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 425:962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park, I.K., D. Qian, M. Kiel, M.W. Becker, M. Pihalja, I.L. Weissman, S.J. Morrison, and M.F. Clarke. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 423:302–305. [DOI] [PubMed] [Google Scholar]
- 8.Iwama, A., H. Oguro, M. Negishi, Y. Kato, Y. Morita, H. Tsukui, H. Ema, T. Kamijo, Y. Katoh-Fukui, H. Koseki, et al. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product, Bmi-1. Immunity. 21:843–851. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs, J.J.L., K. Kieboom, S. Marino, R.A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi1 regulates proliferation and senescence through the ink4a locus. Nature. 397:164–168. [DOI] [PubMed] [Google Scholar]
- 10.Sharpless, N.E., and R.A. DePinho. 1999. The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev. 9:22–30. [DOI] [PubMed] [Google Scholar]
- 11.Bruggeman, S.W.M., M.E. Valk-Lingbeek, P.P. van der Stoop, J.J.L. Jacobs, K. Kieboom, E. Tanger, D. Hulsman, C. Leung, Y. Arsenijevic, S. Marino, and M. van Lohuizen. 2005. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 19:1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molofsky, A.V., S. He, M. Bydon, S.J. Morrison, and R. Pardal. 2005. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 19:1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osawa, M., K. Hanada, H. Hamada, and H. Nakauchi. 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cells. Science. 273:242–245. [DOI] [PubMed] [Google Scholar]
- 14.Takano, H., H. Ema, K. Sudo, and H. Nakauchi. 2004. Asymmetric division and lineage commitment at the level of hematopoietic stem cells: inference from differentiation in daughter cell and granddaughter cell pairs. J. Exp. Med. 199:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvi, L.M., G.B. Adams, K.W. Weibrecht, J.M. Weber, D.P. Olson, M.C. Knight, R.P. Martin, E. Schipani, P. Divieti, F.R. Bringhurst, et al. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 425:841–846. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, J., C. Niu, L. Ye, H. Huang, X. He, W.G. Tong, J. Ross, J. Haug, T. Johnson, J.Q. Feng, et al. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 425:836–841. [DOI] [PubMed] [Google Scholar]
- 17.Arai, F., A. Hirao, M. Ohmura, H. Sato, S. Matsuoka, K. Takubo, K. Ito, G.Y. Koh, and T. Suda. 2004. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 118:149–161. [DOI] [PubMed] [Google Scholar]
- 18.Kiel, M.J., O.H. Yilmaz, T. Iwashita, O.H. Yilmaz, C. Terhorst, and S.J. Morrison. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121. [DOI] [PubMed] [Google Scholar]
- 19.Campisi, J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 120:513–522. [DOI] [PubMed] [Google Scholar]
- 20.Allsopp, R.C., G.B. Morin, R. DePinho, C.B. Harley, and I.L. Weissman. 2003. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 102:517–520. [DOI] [PubMed] [Google Scholar]
- 21.Allsopp, R.C., G.B. Morin, J.W. Horner, R. DePinho, C.B. Harley, and I.L. Weissman. 2003. Effect of TERT over-expression on the long-term transplantation capacity of hematopoietic stem cells. Nat. Med. 9:369–371. [DOI] [PubMed] [Google Scholar]
- 22.Stepanova, L., and B.P. Sorrentino. 2005. A limited role for p16Ink4a and p19Arf in the loss of hematopoietic stem cells during proliferative stress. Blood. 106:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama, K., K. Wada, H. Miyoshi, K. Ohashi, M. Tachibana, R. Furuki, H. Mizuguchi, T. Hayakawa, A. Nakajima, T. Kadowaki, et al. 2004. RNA interfering approach for clarifying the PPARgamma pathway using lentiviral vector expressing short hairpin RNA. FEBS Lett. 560:178–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.