Abstract
HIV-1 infects immature dendritic cells (iDCs), but infection is inefficient compared with activated CD4+ T cells and only involves a small subset of iDCs. We analyzed whether this could be attributed to specific cellular restrictions during the viral life cycle. To study env-independent restriction to HIV-1 infection, we used a single-round infection assay with HIV-1 pseudotyped with vesicular stomatitis virus G protein (HIV-VSVG). Small interfering RNA–mediated depletion of APOBEC3G/3F (A3G/3F), but not TRIM5α, enhanced HIV-1 infection of iDCs, indicating that A3G/3F controls the sensitivity of iDCs to HIV-1 infection. Furthermore, sequences of HIV reverse transcripts revealed G-to-A hypermutation of HIV genomes during iDC infection, demonstrating A3G/3F cytidine deaminase activity in iDCs. When we separated the fraction of iDCs that was susceptible to HIV, we found the cells to be deficient in A3G messenger RNA and protein. We also noted that during DC maturation, which further reduces susceptibility to infection, A3G levels increased. These findings highlight a role for A3G/3F in explaining the resistance of most DCs to HIV-1 infection, as well as the susceptibility of a fraction of iDCs. An increase in the A3G/3F-mediated intrinsic resistance of iDCs could result in a block of HIV infection at its mucosal point of entry.
The spread of HIV-1 over time has occurred mainly through sexual transmission. This transmission necessitates that minute amounts of virus traverse mucosal surfaces to reach replication-competent sites in lymphoid tissue. Model systems have indicated that one of the key events during this transmission is the transfer of HIV infection from DCs to CD4+ T cells (for review see references 1–3).
HIV infects DCs in vitro only when relatively high viral inocula are used (for review see reference 1) (4–6). Presently, the mechanism responsible for the poor replication of HIV in immature DCs (iDCs), the type of DCs that likely encounters HIV in the mucosal tissues during sexual transmission, has not been elucidated.
In the absence of viral replication, DCs also can capture and transfer viral material to CD4+ T cells via an infectious synapse, which results in vigorous infection (7–10). The contribution of this “capture–transfer” pathway remains unclear in vivo.
Langerhans cells/DCs replicate at low levels, preferentially HIV-1 R5 compared with HIV-1 X4 (5, 11, 12). This preferential replication of R5 strains versus X4 strains appears to be caused in part by levels of coreceptors on the surface of iDCs and is also partly due to a block during viral fusion with iDCs (11, 12). However, infection of iDCs is still inefficient, irrespective of the viral env, when compared with activated CD4+ T cells (for review see reference 1).
In this study, we asked whether iDCs expressed an env-independent intrinsic resistance to HIV-1 infection that could operate after viral entry. The idea of intrinsic resistance derives from the discovery of cellular restriction factors to HIV-1 infection (for review see reference 13). Two families of restriction factors in human cells have been recently identified: the TRIM family and the APOBEC family (14, 15). Specifically, one member of each family can operate, at least in some circumstances, after viral entry and before viral integration into the nucleus (e.g., TRIM5α [T5α] and APOBEC3G [A3G]) (15, 16). Human T5α restricts HIV-1 only weakly (15), whereas A3G effectively restricts HIV-1 after entry in primary cells such as resting CD4+ T cells (16). In fact, A3G is a potent host antiretroviral factor that can restrict HIV-1 infection through at least two mechanisms. First, A3G is a DNA deaminase that is incorporated into virions during viral production and subsequently triggers massive G-to-A hypermutation in the nascent retroviral DNA (17, 18). This mode of action is counteracted by Vif that prevents incorporation of A3G into virions (19). Second, cellular A3G can function as a post-entry restriction factor for HIV during reverse transcription in resting CD4+ T cells (16), where it resides in a low molecular mass (LMM) active form. This second mechanism does not seem to be counteracted by Vif, because incoming viral particles contain little Vif. The potential contribution of these cellular restriction factors to the low levels of HIV-1 infection in iDCs has not been tested. In this paper, we demonstrate an env-independent restriction to HIV-1 infection in human iDCs using single-round infection assays with viral particles pseudotyped with vesicular stomatitis virus G (VSVG). We will demonstrate a role for A3G, and to a lesser extent for APOBEC3F (A3F), in restriction of HIV-1 infection in DCs.
RESULTS
HIV-1 replication is restricted in iDCs
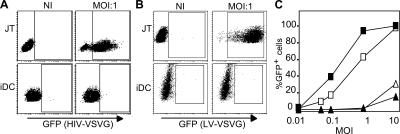
We generated iDCs after 6 d of culture of monocytes in GM-CSF and IL-4, as previously described, and verified that the iDCs expressed the expected markers (i.e., CD14− and MHC class II+) (9, 20). To overcome the entry block that has been described in iDCs, we used single-round infection assays with HIV-1 Δ env and pseudotyped with VSVG (HIV-VSVG; Fig. 1 A). Similar results were obtained using a lentiviral vector (LV) that is deleted in env, vif, vpr, vpu, and nef (LV-VSVG), indicating that the low susceptibility of iDCs to HIV infection was independent of these virulence genes (Fig. 1 B). iDCs were at least 50–100 times less susceptible to HIV-VSVG infection than Jurkat CD4+ T cells (JTs; Fig. 1, A–C). HIV-VSVG infection was equally efficient in activated peripheral blood lymphocytes and in JTs (unpublished data).
Figure 1.
HIV-1 infection is restricted in iDCs. (A) Infection of iDCs and JTs with full-length HIV-1 vector encoding GFP and pseudotyped with VSVG (HIV-VSVG; MOI = 1). GFP expression was followed by FACS after 3 d of infection. One representative experiment out of two is shown. (B) iDCs and JTs were infected with a multiply deleted LV encoding GFP pseudotyped with VSVG (LV-VSVG; MOI = 1). One representative experiment out of two is shown. Boxes in A and B indicate that GFP+ cells are in that gate. (C) Infection of iDCs or Jurkat cells with increasing amounts of LV-VSVG (open symbols) or HIV-VSVG (closed symbols; MOI = 0.01–10). One representative experiment out of two is shown. ▪ and □, GFP expression in JTs; ▴ and ▵, GFP expression in iDCs.
A3G/3F, but not T5α, are active restriction factors in iDCs
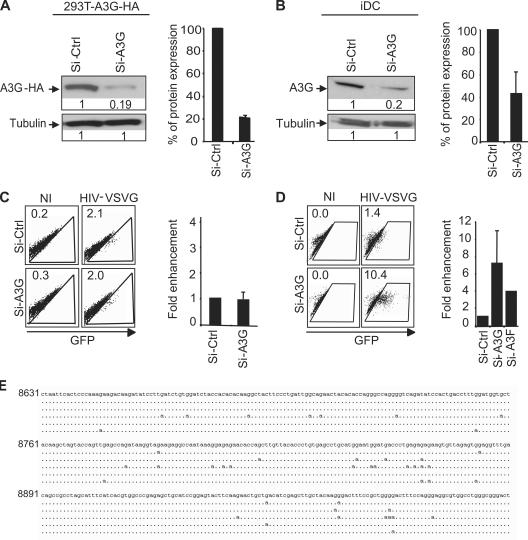
To explore whether a cellular restriction factor operates in iDCs, we used RNA interference to target the cellular restriction factors, members of the APOBEC family (A3G and A3F), as well as T5α. Monocyte-derived DCs (mDCs) were transfected with small interfering RNA (siRNA) sequences specific for the target gene using lipofectamine reagent. The extent of down-regulation was monitored by Western blotting followed by densitometry analysis. Double transfection of a mix of three siRNA targeting A3G induced an efficient knockdown in 293T cells stably expressing A3G-HA (293T-A3G-HA cells; 80% down-regulation; Fig. 2 A) or in iDC (60–80% down-regulation; Fig. 2 B), which is an efficiency comparable to that reported for RNA interference of A3G in resting CD4 T cells (16). In 293T-A3G-HA cells, siRNA-mediated interference of A3G did not increase HIV-1 infection (Fig. 2 C), but HIV infection was enhanced >7-fold (range = 2–22-fold, depending on the donor) in A3G knockdowned iDCs in comparison with control cells treated with equal amounts of siRNA targeting an irrelevant sequence (Fig. 2 D). Based on these results, we conclude that HIV-1 infection in iDCs is restricted at least in part by A3G.
Figure 2.
A3G restricts HIV-1 infection in iDC 293T. A3G-HA cells (A) or iDCs (B) were transfected with siRNA against A3G (Si-A3G) or siRNA control (Si-Ctrl). After two rounds of transfection, cells were analyzed by Western blot (A and B, left). The percentage of protein expression on Western blot was quantified by density of specific bands and normalized with Si-Ctrl (A and B, right). 293T-A3G-HA cells (C) or iDCs (D) transfected by Si-A3G and Si-Ctrl were infected with HIV-VSVG. GFP expression was followed by FACS (numbers are the percentage of GFP+ cells; C and D, left), and the fold enhancement of GFP expression was calculated by normalization with nontreated (NT) cells (C and D, right). A pool of five independent experiments is shown for A3G (±SEM), and two independent experiments are shown for A3F (±SEM). NI, noninfected. (E) A3G/3F induce G-to-A hypermutation of HIV genomes during iDC infection. Sequences represent individual HIV-1 reverse transcripts obtained after an 8-h infection of iDCs and are representative of the 7 hypermutated sequences (17%) that were obtained among 40 reverse transcripts. The standard HIV NL4.3 numbering is used.
A second APOBEC family member, A3F, also possesses antiviral function, though it is 10–100-fold less than potent than A3G (21, 22). We targeted A3F with a combination of two siRNA in iDCs, which induced an efficient knockdown of this protein in 293T cells transiently expressing A3F-HA (unpublished data). A3F knockdown in iDCs increased HIV infection approximately threefold (i.e., less efficiently than A3G knockdown [approximately sevenfold]; Fig. 2 D). Double knockdown experiments (A3G+A3F) were not feasible, possibly because of the large amounts of siRNA used to knockdown both genes (unpublished data).
To substantiate the involvement of A3G/3F proteins in the early block observed in DCs, we analyzed the sequences of viral reverse transcripts 8 h after infection (Fig. 2 E). A substantial fraction of the incoming viral DNA molecules (∼17%) harbored G-to-A mutations, the hallmark of an A3G/3F action.
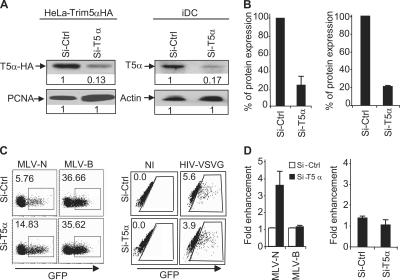
Because T5α is another factor that can restrict HIV-1 infection in several cell types (15, 23), we used RNA interference to knock down human T5α in HeLa stably overexpressing T5α-HA (HeLa-T5α-HA; ∼80% down-regulation of T5α; Fig. 3, A and B, left) and in iDCs (∼80% down-regulation of T5α; Fig. 3, A and B, right). Although RNA interference of T5α in HeLa-T5α-HA relieved the block imposed to murine leukemia virus–N in human cells as expected (Fig. 3, C and D, left), interference of endogenous human T5α in iDCs did not increase HIV-VSVG infection (Fig. 3, C and D, right). Thus, human T5α is not a potent cellular restriction factor to HIV infection in iDCs.
Figure 3.
Human T5α does not restrict HIV-1 infection in iDCs. HeLa-T5α-HA cells (A and B, left) or iDCs (A and B, right) were transfected with siRNA against T5α (Si-T5α) or siRNA control (Si-Ctrl). After two rounds of transfection, the cells were lysed and analyzed by Western blot (A). The percentage of protein expression on Western blot was quantified by the density of specific bands and normalized with Si-Ctrl (B). HeLa-T5α-HA (C and D, left) or iDC (C and D, right) knockdowns for T5α were infected with MLV-GFP or HIV-VSVG. GFP expression was followed by FACS (C), and the fold enhancement of GFP expression was calculated by normalization with cells treated with Si-Ctrl. Boxes indicate the percentage of GFP+ cells. (D). Results are a pool of three independent experiments (±SD). MLV, murine leukemia virus.
A3G messenger RNA (mRNA) is absent in permissive iDCs
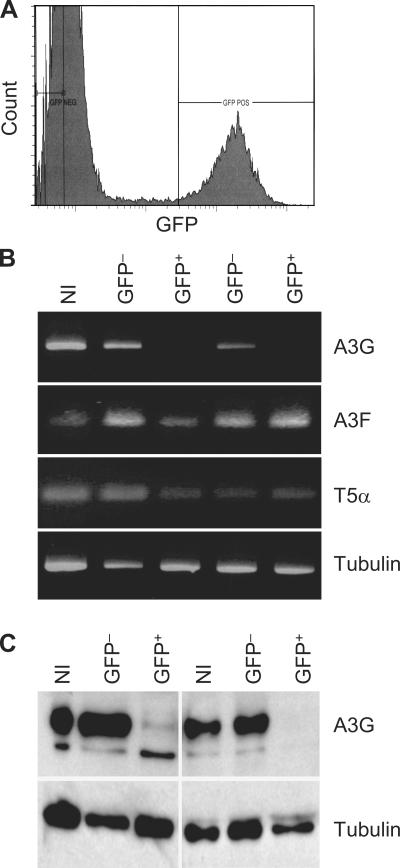
In spite of the inefficiency of infection of iDCs, a small fraction of the cells do support productive infection (24). To pursue the basis for the permissiveness of a subset of iDCs, we infected iDCs with HIV-VSVG, and 5 d later, GFP+ and GFP− cells were sorted by FACS (Fig. 4 A). We performed RT-PCR for A3G/A3F and T5α on GFP+ and GFP− cells. Strikingly, A3G mRNA was severely decreased in GFP+ permissive iDCs in comparison with GFP− iDCs, whereas A3F and T5α mRNA were equally present in both fractions of iDCs (Fig. 4 B). Tubulin was used as a loading control, indicating that this result was not explained by differences in cell counts between the GFP+ and GFP− populations. We also performed a Western blot analysis. A3G protein levels were decreased in the GFP+ iDCs in comparison with GFP− iDCs (Fig. 4 C). Because HIV Vif does not affect A3G mRNA in infected cells (19), this result suggests that the susceptibility of a fraction of iDCs to HIV-1 is caused by the absence of A3G mRNA before infection of these cells.
Figure 4.
HIV-VSVG infects iDCs that express low levels of A3G. (A) iDCs were infected with HIV-VSVG and sorted after 5 d as GFP+ (infected) and GFP− (noninfected) fractions. (B) RNA was extracted and subjected to RT-PCR for A3G (30 cycles), A3F (35 cycles), T5α (35 cycles), and tubulin. (C) Proteins extracted from the same cells were used in Western blot for A3G and tubulin. The figure shows two different cell preparations. NI, uninfected control cells.
DC maturation positively modulates A3G expression levels
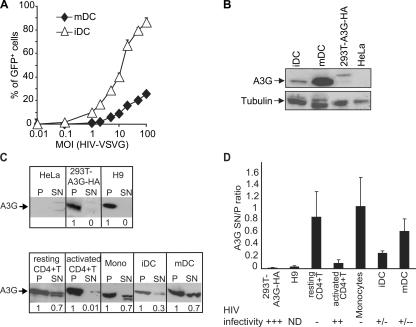
To analyze whether DC maturation modulates A3G activity, we activated iDCs with maturing agents such as LPS. LPS triggered the up-regulation of CD83, HLA-DR, CD80, and CD86 by FACS (unpublished data). In contrast to the activation of resting CD4+ T cells, which increases HIV-1 infection, LPS-induced maturation of DCs further decreased infection of HIV-VSVG relative to iDCs ∼5–10-fold (Fig. 5 A). We next quantified A3G expression by Western blot analysis in iDCs and mDCs. mDCs expressed three to fivefold more A3G than iDCs (Fig. 5 B). A3F antibodies did not function for Western blot analysis, thus precluding this type of analysis for A3F in mDCs (unpublished data). RNA interference of A3G in mDCs was inefficient, perhaps because of the low capacity of mDCs for internalization, which prevented us from directly testing the capacity of A3G to control HIV-1 infection in mDCs. Nevertheless, these results suggest that DC maturation further restricts HIV-1 infection through an increase in A3G expression.
Figure 5.
DC maturation increases A3G levels and further restricts HIV-1 infection. (A) Infection of iDCs and mDCs with HIV-VSVG. DCs were infected with increasing amounts of HIV-VSVG. GFP expression was followed by FACS after 3 d of infection. One representative experiment out of two is shown. (B) Analysis of A3G expression in iDCs, mDCs, 293T-A3G-HA cells, and HeLa cells by Western blot. (C) DCs contain A3G in the SN fraction (A3G SN). Control cells (top) and primary cells (bottom) were lysed and centrifuged at high speed. Equal volumes of P and SN fractions were analyzed by Western blotting. Numbers represent the amount of A3G in SN and P; A3G expression in SN was quantified, with a value of 1 assigned to the P. One representative experiment out of three is shown. (D) A3G SN correlates with restriction to HIV infection in DCs. Single-round infection experiments were performed with a VSVG-pseudotyped HIV-GFP virus. GFP+ cells were evaluated by FACS 3 d after infection. Infectivity of the different cell types is represented: high (+++), >75%; moderate (++), 40–50%; low, (+/−) <10–15%; very low (+/−−), <5%; none (−); and not determined (ND). One representative experiment out of three is shown (±SD).
To evaluate whether A3G was present in an LMM active form in DCs as previously observed in resting CD4+ T cells (16), we designed an assay where we separated cytosolic proteins from a pellet (P) fraction (containing heavy membranes, endosomes, and nucleus) by high speed centrifugation. We reasoned that because A3G LMM was most likely in a cytosolic unbound form, we could extract cytosolic A3G in a supernatant (SN) fraction (A3G SN) and leave other higher molecular forms of A3G in the P fraction. Using this assay, we found that A3G was exclusively in the P in cells highly sensitive to HIV infection, such as 293T and H9 CD4+ T cells, and no A3G SN was detected (Fig. 5 C). We next analyzed A3G distribution in primary cells and identified a strong proportion of A3G SN in cells restrictive to HIV infection, such as monocytes and resting CD4+ T cells, but not in activated CD4+ T cells, where A3G SN levels were low (Fig. 5 C). These controls are consistent with the fact that our simplified assay can determine the proportion of A3G LMM described in (16), which only identified A3G SN in cells restrictive to HIV infection (similar to the A3G LMM) and does not provide specific information on the high molecular mass forms of A3G. When we subjected iDCs and mDCs to the same analysis, A3G SN was present in iDCs but was strongly up-regulated in mDCs (Fig. 5 C). In all these cells, the proportion of A3G SN correlated with the HIV restriction phenotype (Fig. 5 D); i.e., mDCs showed a high level of A3G SN, in accordance with their low susceptibility to HIV, whereas iDCs had an intermediate level of A3G SN and an intermediate resistance to HIV infection (Fig. 5, A and D).
DISCUSSION
We have investigated two features in the infection of iDCs, the cells that are implicated in transmission of HIV to T cells: first, the poor susceptibility of iDCs relative to activated T cells, and second, the permissiveness of a small subset of iDCs to replication of HIV-1. Although the resistance of DCs to HIV-1 has been known (for review see references 1, 2), the underlying mechanisms have remained unclear. Envelope-dependent blocks during entry explain the fact that CXCR4low CCR5+ iDCs are poorly infectable by HIV-1 X4 and allow entry of R5 virus (5, 25). We and others also have described a block during viral fusion for HIV-1 X4 with iDCs (9, 11) independent of CXCR4 levels (12). However, even when HIV is pseudotyped with VSVG, iDCs are less infectable than JTs or activated PBLs, indicating that a cellular restriction also operates post-entry in iDCs.
We therefore considered the two families of restriction factors (TRIM/APOBEC) that have been identified in human cells (for review see reference 13). Among their family members, T5α and A3G are known to block HIV-1 after viral entry in select cells. T5α did not operate as a strong restriction factor in iDCs. This is not surprising given the fact that human T5α restricts HIV-1 only weakly in human cells (15). On the other hand, human A3G does function as a potent post-entry cellular restriction factor for HIV-1 in resting CD4+ T cells (16). A3G acts through cytidine deaminase activity (17, 18), as well as through less well-understood nonenzymatic mechanisms (26).
Three separate lines of evidence indicated that A3G could restrict HIV-1 infection in iDC. First, RNA interference of A3G in iDCs considerably increased HIV-1 infection in these cells. Interestingly A3F down-regulation also enhanced HIV infection in iDCs, though to a lesser extent than A3G down-regulation, suggesting that both A3G/3F may contribute to restricting HIV infection in iDCs. To confirm the involvement of A3G/3F in the early block of HIV infection in DCs, we analyzed the sequences of viral reverse transcripts. A substantial fraction of the incoming viral DNA molecules (17%) harbored G-to-A mutations, the hallmark of an active cytidine deaminase like A3G/3F, and these hypermutations harbored the signatures of both A3F and A3G action. Based on the known respective frequencies of their different signatures (21), it can be deduced that both enzymes contribute comparably to the total mutations found in reverse transcripts. It appears, however, that A3G is probably the main contributor to the restriction, because its RNA interference–mediated ablation leads to the strongest relief of the block in iDCs.
Second, iDC that were infectable with HIV-VSVG did not express A3G mRNA or A3G protein. Therefore, our result indicates that low A3G mRNA in a subset of iDCs possibly increases permissivity to HIV-1. Interestingly, A3F was expressed in both infected and uninfected iDC populations, which correlates with its less important contribution to the restriction of HIV infection in iDCs, as determined by RNA interference studies.
Third, we tried to modulate A3G expression and function in DCs by inducing DC maturation. Unlike resting CD4+ T cells that become permissive upon activation (16), DCs are more restrictive after maturation/activation. Interestingly A3G levels were up-regulated in mDCs, further indicating a link between A3G and HIV-1 infection in iDCs.
The recent description of A3G in an enzymatically active LMM form in resting CD4+ T cells and monocytes (16, 27) suggests that A3G possibly exists in that active LMM form in DCs as well. We recapitulated a simplified version of the assay in which we separated A3G SN that behaved similarly to A3G LMM. Our results demonstrated that the presence of A3G SN correlated with restriction. Monocytes and resting CD4+ T cells had the highest quantity of SN A3G and were not infectable. 293T and activated CD4+ T cells had low amounts of A3G SN and were completely permissive. Interestingly, mDCs had more A3G SN than iDCs. Collectively, our results indicate that A3G restriction explains, at least in part, the post-entry block observed in iDCs. Attempts to modulate A3G/A3F function in DCs could offer the potential to counteract HIV infection before its dissemination during the early events of HIV-1 transmission.
MATERIALS AND METHODS
Virus stocks.
These were produced by transient transfection of 293T cells. Infection assays were performed as previously described (9, 20). Details about the viral constructs used are in Supplemental materials and methods, available at http://www.jem.org/cgi/content/full/jem.20061519/DC1.
Preparation of primary mDCs.
Monocyte isolation from buffy coats of healthy donors (obtained according to guidelines of the ethical committees of the University of Geneva and the Rockefeller University) and subsequent generation of iDCs was performed as previously described (20). DC maturation was induced by 48 h of LPS stimulation at 20 ng/ml (Escherichia coli, strain 055:B5; Difco).
Cell culture reagents.
HeLa-T5α-HA cells and 293T-A3G cells (a gift of D. Trono, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) were grown and maintained in DMEM with 10% FCS, 100 μg/ml penicillin and streptomycin, and 2 mM glutamine.
RNA interference in iDCs.
mDCs were transfected with siRNA sequences specific for the target gene using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations, with modifications available in Supplemental materials and methods.
Sequencing of viral DNA.
iDCs were infected with DNase-treated HIV-VSVG-GFP virions at a multiplicity of infection (MOI) of 10. Viral reverse transcripts were extracted with the DNeasy kit (QIAGEN) 8 h after infection and individually amplified by standard PCR using O.nef1 and O.U5.1 primers (see Supplemental materials and methods). They were then cloned with a cloning kit (TOPO-TA; Invitrogen) and sequenced. Finally, Clone Manager software (version 8; Scientific & Educational Software) was used for the alignments.
RT-PCR from iDC-infected cells.
iDCs were infected with HIV-VSVG expressing GFP (2 ng p24 gag/105 cells) for 5 d. Cells were sorted as GFP+ (infected) or GFP− (noninfected). Total RNA was extracted with TRIzol (Invitrogen). RT-PCR was performed with 0.5 μg RNA using SuperScript one-step RT-PCR (Invitrogen). PCR for A3G and T5α was performed by 30 cycles of amplification at 94°C for 15 s, 55°C for 30 s, and 72°C for 1 min, or at 94°C for 15 s, 48°C for 30 s, and 72°C for 1 min, respectively, with gene-specific primers (see Supplemental materials and methods).
A3G subcellular localization.
Cells (see Supplemental materials and methods) were lysed with ice-cold lysis buffer (125 mM NaCl, 50 mM Hepes, pH 7.4, 0.2% NP40, 1 mM dithiothreitol, 0.1 mM PMSF, EDTA-free protease inhibitor cocktail; Sigma-Aldrich) for 30 min and centrifuged at 35,000 rpm (MC M150GX; Sorvall) for 1.5 h. SN and P were separated, and P was resuspended in a volume equal to that of SN and sonicated for 20 s (Sonic B12; Branson). Equal volumes of P and SN were loaded on gel and analyzed by standard Western blotting. Protein expression was quantified by densitometry analysis of specific bands. Ratios of A3G SN were calculated as a ratio of A3G SN/P, where the A3G P fraction had an arbitrary value of 1 based on densitometry analysis.
Online supplemental material.
Supplemental materials and methods contains information about virus stocks, RNA interference in iDCs, siRNA and primer sequences, antibodies, and A3G subcellular localization. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20061519/DC1.
Supplemental Material
Acknowledgments
We thank J. Stoye, D. Littman, D. Trono, and J. Sodroski for reagents and F. Leuba for excellent technical help. We thank M. Stevenson for helpful discussions regarding RNA interference in primary cells.
This work was supported by the Geneva Cancer League, the Leenaards Foundation, and the Swiss National Science Foundation (V. Piguet), and National Institutes of Health grant AI40045 (to R.M. Steinman). This work was also supported by the Human Science Frontier Program (V. Piguet and R.M. Steinman).
The authors have no conflicting financial interests.
Abbreviations used: A3F and A3G, APOBEC3F and APOBEC3G, respectively; iDC, immature DC; JT, Jurkat CD4+ T cell; LMM, low molecular mass; LV, lentiviral vector; mDC, monocyte-derived DC; MOI, multiplicity of infection; mRNA, messenger RNA; P, pellet; siRNA, small interfering RNA; SN, supernatant; T5α, Trim5α; VSVG, vesicular stomatitis virus G.
References
- 1.Piguet, V., and A. Blauvelt. 2002. Essential roles for dendritic cells in the pathogenesis and potential treatment of HIV disease. J. Invest. Dermatol. 119:365–369. [DOI] [PubMed] [Google Scholar]
- 2.Steinman, R.M., A. Granelli-Piperno, M. Pope, C. Trumpfheller, R. Ignatius, G. Arrode, P. Racz, and K. Tenner-Racz. 2003. The interaction of immunodeficiency viruses with dendritic cells. Curr. Top. Microbiol. Immunol. 276:1–30. [DOI] [PubMed] [Google Scholar]
- 3.Margolis, L., and R. Shattock. 2006. Selective transmission of CCR5-utilizing HIV-1: the ‘gatekeeper’ problem resolved? Nat. Rev. Microbiol. 4:312–317. [DOI] [PubMed] [Google Scholar]
- 4.Pope, M., S. Gezelter, N. Gallo, L. Hoffman, and R.M. Steinman. 1995. Low levels of HIV-1 infection in cutaneous dendritic cells promote extensive viral replication upon binding to memory CD4+ T cells. J. Exp. Med. 182:2045–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granelli-Piperno, A., E. Delgado, V. Finkel, W. Paxton, and R.M. Steinman. 1998. Immature dendritic cells selectively replicate macrophagetropic (M-tropic) human immunodeficiency virus type 1, while mature cells efficiently transmit both M- and T-tropic virus to T cells. J. Virol. 72:2733–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawamura, T., S.S. Cohen, D.L. Borris, E.A. Aquilino, S. Glushakova, L.B. Margolis, J.M. Orenstein, R.E. Offord, A.R. Neurath, and A. Blauvelt. 2000. Candidate microbicides block HIV-1 infection of human immature Langerhans cells within epithelial tissue explants. J. Exp. Med. 192:1491–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald, D., L. Wu, S.M. Bohks, V.N. Kewalramani, D. Unutmaz, and T.J. Hope. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 300:1295–1297. [DOI] [PubMed] [Google Scholar]
- 8.Turville, S.G., J.J. Santos, I. Frank, P.U. Cameron, J. Wilkinson, M. Miranda-Saksena, J. Dable, H. Stossel, N. Romani, M. Piatak Jr., et al. 2004. Immunodeficiency virus uptake, turnover, and 2-phase transfer in human dendritic cells. Blood. 103:2170–2179. [DOI] [PubMed] [Google Scholar]
- 9.Arrighi, J.F., M. Pion, E. Garcia, J.M. Escola, Y. van Kooyk, T.B. Geijtenbeek, and V. Piguet. 2004. DC-SIGN–mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J. Exp. Med. 200:1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moris, A., A. Pajot, F. Blanchet, F. Guivel-Benhassine, M. Salcedo, and O. Schwartz. 2006. Dendritic cells and HIV-specific CD4+ T cells: HIV antigen presentation, T cell activation, viral transfer. Blood. 108:1643–1651. [DOI] [PubMed] [Google Scholar]
- 11.Cavrois, M., J. Neidleman, J. Kreisberg, D. Fenard, C. Callebaut, and W.C. Greene. 2006. Human Immunodeficiency Virus fusion to dendritic cells declines as cells mature. J. Virol. 80:1992–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pion, M., J.F. Arrighi, J. Jiang, C.A. Lundquist, O. Hartley, C. Aiken, and V. Piguet. 2006. Analysis of HIV-1-X4 Fusion with Immature Dendritic Cells Identifies a Specific Restriction that Is Independent of CXCR4 Levels. J. Invest. Dermatol. 10.1038/sj.jid.5700518. [DOI] [PubMed]
- 13.Bieniasz, P.D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109–1115. [DOI] [PubMed] [Google Scholar]
- 14.Sheehy, A.M., N.C. Gaddis, J.D. Choi, and M.H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 418:646–650. [DOI] [PubMed] [Google Scholar]
- 15.Stremlau, M., C.M. Owens, M.J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 427:848–853. [DOI] [PubMed] [Google Scholar]
- 16.Chiu, Y.L., V.B. Soros, J.F. Kreisberg, K. Stopak, W. Yonemoto, and W.C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 435:108–114. [DOI] [PubMed] [Google Scholar]
- 17.Harris, R.S., K.N. Bishop, A.M. Sheehy, H.M. Craig, S.K. Petersen-Mahrt, I.N. Watt, M.S. Neuberger, and M.H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell. 113:803–809. [DOI] [PubMed] [Google Scholar]
- 18.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 424:99–103. [DOI] [PubMed] [Google Scholar]
- 19.Stopak, K., C. de Noronha, W. Yonemoto, and W.C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 12:591–601. [DOI] [PubMed] [Google Scholar]
- 20.Garcia, E., M. Pion, A. Pelchen-Matthews, L. Collinson, J.F. Arrighi, G. Blot, F. Leuba, J.M. Escola, N. Demaurex, M. Marsh, and V. Piguet. 2005. HIV-1 trafficking to the Dendritic Cell-T-Cell infectious synapse uses a pathway of tetraspanin sorting to the immunological synapse. Traffic. 6:488–501. [DOI] [PubMed] [Google Scholar]
- 21.Simon, V., V. Zennou, D. Murray, Y. Huang, D.D. Ho, and P.D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop, K.N., R.K. Holmes, and M.H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450–8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayah, D.M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 430:569–573. [DOI] [PubMed] [Google Scholar]
- 24.Granelli-Piperno, A., A. Golebiowska, C. Trumpfheller, F.P. Siegal, and R.M. Steinman. 2004. HIV-1-infected monocyte-derived dendritic cells do not undergo maturation but can elicit IL-10 production and T cell regulation. Proc. Natl. Acad. Sci. USA. 101:7669–7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaitseva, M., A. Blauvelt, S. Lee, C.K. Lapham, V. Klaus-Kovtun, H. Mostowski, J. Manischewitz, and H. Golding. 1997. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat. Med. 3:1369–1375. [DOI] [PubMed] [Google Scholar]
- 26.Newman, E.N., R.K. Holmes, H.M. Craig, K.C. Klein, J.R. Lingappa, M.H. Malim, and A.M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170. [DOI] [PubMed] [Google Scholar]
- 27.Kreisberg, J.F., W. Yonemoto, and W.C. Greene. 2006. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J. Exp. Med. 203:865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.