Abstract
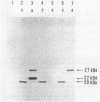
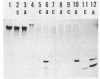
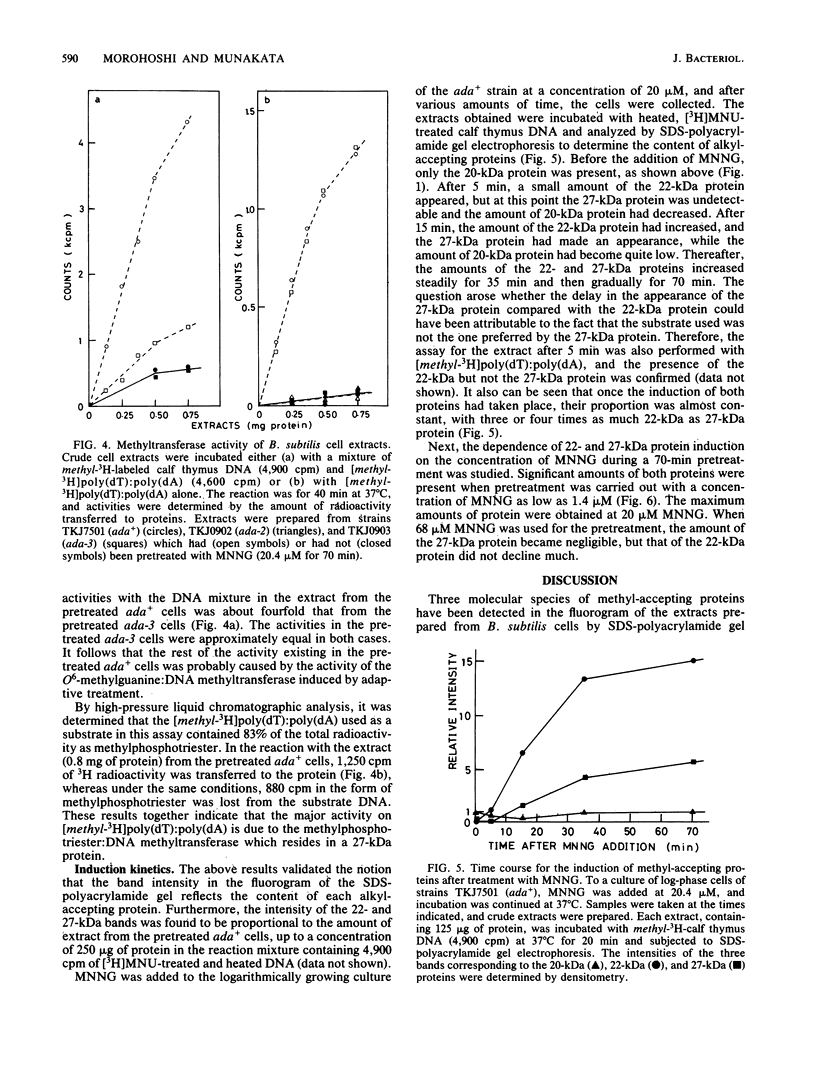
Three molecular species of methyl-accepting proteins exist in Bacillus subtilis cells, which collect methyl groups from methylated DNA. A 20-kilodalton (kDa) protein was constitutively present in the cells of the ada+ (proficient in adaptive response) strain as well as in those of six ada (deficient in adaptive response) mutant strains and was assigned to the O6-methylguanine:DNA methyltransferase. Another species of O6-methylguanine:DNA methyltransferase, which had a molecular size of 22 kDa, emerged after adaptive treatment of the ada+ but not any of the ada mutant cells. A 27-kDa methyl-accepting protein, which preferred methylated poly(dT) to methylated calf thymus DNA as a substrate, was assigned to the methylphosphotriester:DNA methyltransferase. It was produced, after adaptive treatment, in the cells of ada+, ada-3, ada-4, and ada-6 strains but not in the cells of ada-1, ada-2, or ada-5 strains. These results support and extend our proposition that ada mutants can be classified into two groups; one (the ada-4 group) is defective only in the inducible synthesis of O6-methylguanine:DNA methyltransferase (22-kDa protein), and the other (the ada-1 group) is deficient in the adaptive response in toto. The finding that inducible and constitutive methyltransferases reside in different molecular species of methyl-accepting proteins is intriguing compared with the regulatory mechanisms of the adaptive response to simple alkylating agents in other organisms.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Demple B., Jacobsson A., Olsson M., Robins P., Lindahl T. Repair of alkylated DNA in Escherichia coli. Physical properties of O6-methylguanine-DNA methyltransferase. J Biol Chem. 1982 Nov 25;257(22):13776–13780. [PubMed] [Google Scholar]
- Demple B., Sedgwick B., Robins P., Totty N., Waterfield M. D., Lindahl T. Active site and complete sequence of the suicidal methyltransferase that counters alkylation mutagenesis. Proc Natl Acad Sci U S A. 1985 May;82(9):2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden C. T., Foote R. S., Mitra S. Adaptive response of Bacillus subtilis to N-methyl-N'-nitro-N-nitrosoguanidine. J Bacteriol. 1983 Feb;153(2):756–762. doi: 10.1128/jb.153.2.756-762.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P. Isolation and characterization of Escherichia coli K-12 mutants unable to induce the adaptive response to simple alkylating agents. J Bacteriol. 1979 Sep;139(3):783–791. doi: 10.1128/jb.139.3.783-791.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Hjelmgren T., Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982 Apr 22;296(5859):770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Griffin B. Adaptive response to alkylating agents involves alteration in situ of O6-methylguanine residues in DNA. Nature. 1979 Jul 5;280(5717):76–77. doi: 10.1038/280076a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemotte P. K., Walker G. C. Induction and autoregulation of ada, a positively acting element regulating the response of Escherichia coli K-12 to methylating agents. J Bacteriol. 1985 Mar;161(3):888–895. doi: 10.1128/jb.161.3.888-895.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margison G. P., Cooper D. P., Brennand J. Cloning of the E. coli O6-methylguanine and methylphosphotriester methyltransferase gene using a functional DNA repair assay. Nucleic Acids Res. 1985 Mar 25;13(6):1939–1952. doi: 10.1093/nar/13.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. G., Edington B. V., Schendel P. F. Inducible repair of phosphotriesters in Escherichia coli. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7380–7384. doi: 10.1073/pnas.80.24.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy T. V., Lindahl T. Methyl phosphotriesters in alkylated DNA are repaired by the Ada regulatory protein of E. coli. Nucleic Acids Res. 1985 Apr 25;13(8):2683–2698. doi: 10.1093/nar/13.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S., Pal B. C., Foote R. S. O6-methylguanine-DNA methyltransferase in wild-type and ada mutants of Escherichia coli. J Bacteriol. 1982 Oct;152(1):534–537. doi: 10.1128/jb.152.1.534-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Munakata N. Bacillus subtilis mutants deficient in the adaptive response to simple alkylating agents. J Bacteriol. 1985 Mar;161(3):825–830. doi: 10.1128/jb.161.3.825-830.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi F., Munakata N. Two classes of Bacillus subtilis mutants deficient in the adaptive response to simple alkylating agents. Mol Gen Genet. 1986 Feb;202(2):200–206. doi: 10.1007/BF00331637. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Kondo H., Kawabata S., Iwanaga S., Sekiguchi M. Purification and structure of the intact Ada regulatory protein of Escherichia coli K12, O6-methylguanine-DNA methyltransferase. J Biol Chem. 1985 Jun 25;260(12):7281–7288. [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: ada protein acts as a transcriptional regulator. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M., Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980 Nov 25;255(22):10569–10571. [PubMed] [Google Scholar]
- Robins P., Cairns J. Quantitation of the adaptive response to alkylating agents. Nature. 1979 Jul 5;280(5717):74–76. doi: 10.1038/280074a0. [DOI] [PubMed] [Google Scholar]
- Schendel P. F., Robins P. E. Repair of O6-methylguanine in adapted Escherichia coli. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6017–6020. doi: 10.1073/pnas.75.12.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B. Molecular cloning of a gene which regulates the adaptive response to alkylating agents in Escherichia coli. Mol Gen Genet. 1983;191(3):466–472. doi: 10.1007/BF00425764. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Weinfeld M., Drake A. F., Saunders J. K., Paterson M. C. Stereospecific removal of methyl phosphotriesters from DNA by an Escherichia coli ada+ extract. Nucleic Acids Res. 1985 Oct 11;13(19):7067–7077. doi: 10.1093/nar/13.19.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]