Abstract
Somatic hypermutation of Ig genes enables B cells of the germinal center to generate high-affinity immunoglobulin variants. Key intermediates in somatic hypermutation are deoxyuridine lesions, introduced by activation-induced cytidine deaminase. These lesions can be processed further to abasic sites by uracil DNA glycosylase. Mutagenic replication of deoxyuridine, or of its abasic derivative, by translesion synthesis polymerases is hypothesized to underlie somatic hypermutation.
Rev1 is a translesion synthesis polymerase that in vitro incorporates uniquely deoxycytidine opposite deoxyuridine and abasic residues. To investigate a role of Rev1 in mammalian somatic hypermutation we have generated mice deficient for Rev1. Although Rev1−/− mice display transient growth retardation, proliferation of Rev1−/− LPS-stimulated B cells is indistinguishable from wild-type cells. In mutated Ig genes from Rev1−/− mice, C to G transversions were virtually absent in the nontranscribed (coding) strand and reduced in the transcribed strand. This defect is associated with an increase of A to T, C to A, and T to C substitutions. These results indicate that Rev1 incorporates deoxycytidine residues, most likely opposite abasic nucleotides, during somatic hypermutation. In addition, loss of Rev1 causes compensatory increase in mutagenesis by other translesion synthesis polymerases.
DNA translesion synthesis (TLS) is a backup replication pathway that, in contrast to replicative polymerases δ and ε, is capable of replicating damaged nucleotides that confer helical distortion to the DNA template. Replication of the damaged nucleotide by TLS is believed to safeguard the perpetuation of replication in the presence of unrepaired DNA damage, albeit frequently at the expense of misincorporations. The Y family of DNA polymerases in mammals is a major class of TLS polymerases comprising the polymerases η, ι, κ, and Rev1 (1). In vitro, the catalytic activity of mammalian Rev1 is limited to the highly distributive incorporation of cytosine residues opposite deoxyuridine residues and abasic nucleotides (2, 3). Analysis of TLS at site-specifically damaged DNA templates in Saccharomyces cerevisiae supports an important role of Rev1 in the bypass of abasic sites in vivo (4). In addition, Rev1-deficient chicken DT40 cells and hypomorphic mouse Rev1 mutant cells display hypersensitivity to a variety of genotoxic agents (5, 6). The infrequent mutations to deoxycytidine induced by these agents in Rev1-proficient S. cerevisiae suggests a second (noncatalytic) role for Rev1, possibly by recruiting other TLS polymerases. In agreement, TLS polymerases η, ι, κ, as well as the Rev7 TLS-associated protein, interact with a COOH-terminal domain of Rev1 (references 7–10; unpublished data).
Deoxyuridine and abasic sites are essential triggers for somatic hypermutation (SHM), a process of antibody diversification in which the variable regions of Ig heavy (IgH) and light (IgL) chain genes in proliferating B cells of the germinal center mutate at an extremely high rate (11). This is followed by clonal selection of the cells that express Ig with increased affinity toward the antigen (12). SHM is triggered by deamination to uracil of deoxycytidines within Ig genes by the activation-induced deoxycytidine deaminase (AID) (11, 13). Subsequent processing by uracil DNA glycosylase (UNG) can generate abasic sites that may be bypassed by one or more of the TLS polymerases (14). In a second phase of SHM, DNA mismatch repair may induce single-stranded gaps at sites of mispaired deoxyuridine residues, followed by filling the gaps by mutagenic TLS (11, 15). To investigate involvement of the Rev1 TLS polymerase in SHM, we have generated and analyzed Rev1-deficient mice.
RESULTS AND DISCUSSION
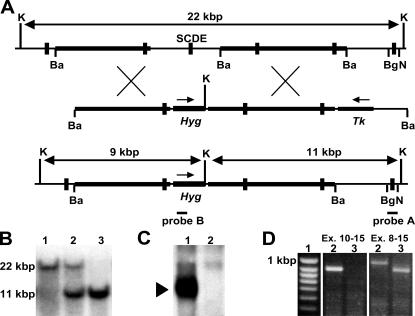
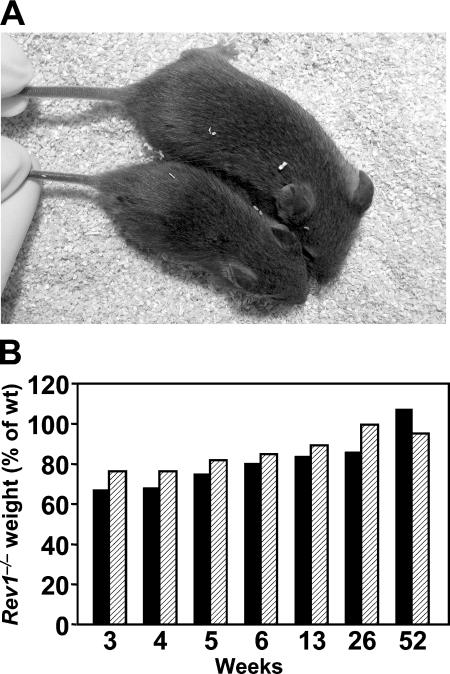
Using conventional gene targeting we deleted exon 10 of Rev1 encoding the conserved S(C/I)DE amino acid sequence essential for the catalytic activity of Y family DNA polymerases (Fig. 1, A and B). Rev1+/– chimeric mice were obtained through blastocyst injection of heterozygous embryonic stem cells and crossed to C57BL/6 and 129/OLA mice. Rev1−/− offspring from interbreeding after F1 and F2 backcrosses to both strains was obtained at 63% of the expected Mendelian ratios. Strikingly, Rev1−/− mice were not obtained beyond the F2 backcross into C57BL/6 mice, in contrast to backcrosses to 129/OLA. A similar strain dependence of the phenotype was found for mice deficient for the Rev3 TLS polymerase (16). The milder phenotypes of the 129/OLA mice are not caused by the pol ι defect of 129/OLA mice (17) due to the fact that (pol ι-proficient) Rev1−/− F1 hybrid mice of C57BL/6 and 129/OLA crossings are viable. Rev1−/− mice from all strains displayed a transiently reduced weight in the absence of gross abnormalities (Fig. 2, A and B). Together, these results are consistent with a partially strain-dependent role for Rev1 in TLS of endogenous DNA damage.
Figure 1.
Targeted disruption of Rev1. (A, top) the genomic region of Rev1 that encodes the catalytic domain of Rev1. Vertical bold sections denote exons. SCDE, exon 10 encoding the catalytic domain of Rev1. Horizontal bold sections denote regions homologous to the targeting vector. (A, middle) Targeting vector pSCDE-hygro to delete exon 10. Hyg, pPGK-hygromycine cassette; TK, pPGK-thymidine kinase cassette. Arrows indicate direction of transcription. (A, bottom) Targeted Rev1 allele. Probes A and B, DNA fragments used to analyze gene-targeting events. Ba, BamHI; Bg, BglII; K, KpnI; N, NcoI. (B) Southern blot of genomic mouse DNA digested with KpnI and hybridized with probe A. Fragment sizes indicate the following alleles: 22 kbp, wild type; 11 kbp, Rev1−/−. 1, Rev1+/+; 2, Rev1+/−; 3, Rev1−/−. (C) Western blot of MEF lysates hybridized with an α-Rev1 antiserum. Arrow indicates position of Rev1. 1, Rev1+/+; 2, Rev1−/−. (D) PCR products of Rev1 cDNA amplified from Rev1−/− kidneys. PCR products of exons 10–15 (left) and exons 8–15 (right). 1, size marker; 2, Rev1+/+; 3, Rev1−/−. In the left panel, in lane 3 no PCR product is detected as a consequence of the deletion of exon 10 in the mutant.
Figure 2.
Retarded growth of Rev1−/− mice. (A) 3-wk-old wild-type (top) and Rev1−/− (bottom) littermates illustrating the reduced body size in young Rev1−/− mice. (B) Growth characteristics of Rev1−/− mice between 3 and 52 wk after birth, expressed as the percentage of weight of wild-type mice. Males (solid bars); females (hatched bars). Each data point represents the mean of 7–10 mice.
Western blot analysis of cell lysates from Rev1−/− mouse embryonic fibroblasts (MEFs), using a COOH-terminal α-Rev1 antiserum, confirmed the absence of protein in Rev1−/− cells (Fig. 1 C). Analysis of Rev1 transcripts from mouse kidney by RT-PCR demonstrated the presence of only shortened transcripts in Rev1−/− cells (Fig. 1 D). Downstream of the deletion of exon 10 all transcripts contained translational frameshifts (unpublished data). Based on these results we conclude that both the catalytic domain and the domain that interacts with other TLS polymerases and the Rev7 protein (7–10; unpublished data) are absent from our Rev1 mutant. In agreement with a generalized defect in TLS of exogenous DNA damage, Rev1−/− MEFs are sensitive to several genotoxic agents tested, including short-wave UV light (unpublished data).
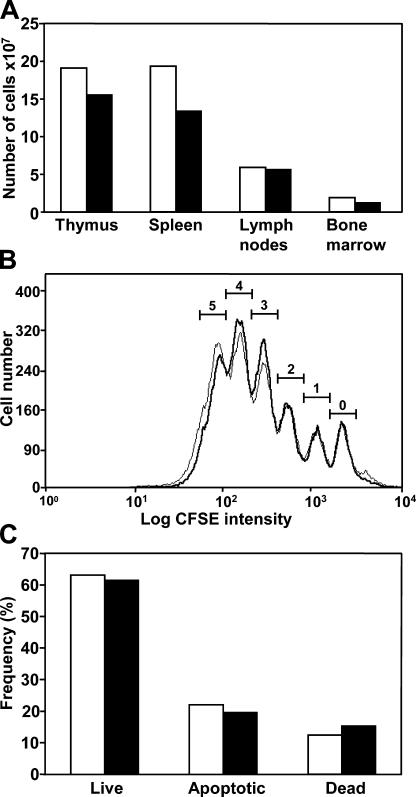
To investigate lymphopoiesis of Rev1−/− mice, we analyzed the cellularity and the composition of B and T cell subsets of primary and secondary lymphatic organs like thymus, spleen, lymph nodes, and bone marrow. Consistent with the moderately reduced size of 7-wk-old Rev1−/− mice (Fig. 2), a proportional reduction in the cellularity of lymphoid organs was observed (Fig. 3 A). The normal frequency of all tested lymphoid subsets suggests that qualitative aspects of lymphocyte development are not impaired by the absence of Rev1 (Table S1, available at http://www.jem.org/cgi/content/full/jem.20052227/DC1). To determine whether the lack of Rev1 activity has any impact on the proliferative capacity of mature B cells we determined proliferation of Rev1−/− and wild-type B cells in vitro after polyclonal stimulation with LPS. Proliferation characteristics of Rev1−/− and wild-type B cells were superimposable (Fig. 3 B), demonstrating that Rev1 deficiency does not impair proliferation of B cells. Similarly, the frequency of live, apoptotic, and dead B cells was normal in the mutant (Fig. 3 C). These results contrast with a previous report (5) showing that Rev1 deficiency causes a slower expansion rate and increased spontaneous apoptosis in the chicken DT40 B cell line.
Figure 3.
Cellularity, proliferation, and survival of Rev1−/− B cells. (A) Cellularity of primary and secondary lymphoid organs of wild-type and Rev1−/− mice. At the age of 7 wk, the cellularity of primary and secondary lymphoid organs of wild-type and Rev1−/− mice were determined. The moderate reduction in cellularity likely is related to the reduced size of Rev1−/− mice in comparison with wild-type mice, rather than to a proliferation defect. See Fig. 2 A and Fig. 4 A. (B) Proliferation of CFSE-loaded B cells from wild-type (thin line) and Rev1 mutant mice (bold line). As analyzed by flow cytometry, LPS blasts proliferate normally in the absence of Rev1. (C) Survival of LPS-activated B cells. Live, apoptotic, and dead cells were distinguished by annexinV and propidium iodine (PI) using flow cytometry. Survival and cell death are not significantly influenced by Rev1 under these conditions. Wild type (white bars); Rev1−/− (black bars).
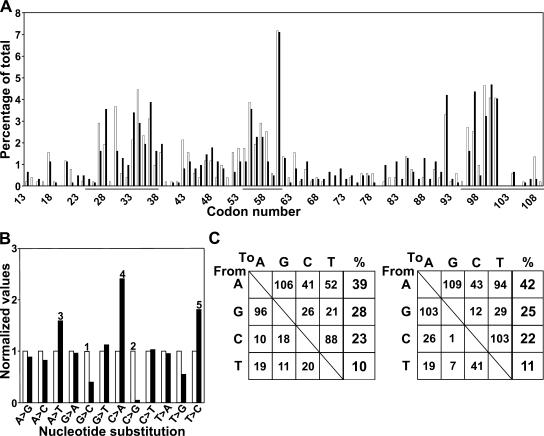
To address the role of Rev1 in mammalian SHM, single class-switched (IgM−IgD−) memory B cells expressing a Igλ1 light chain were isolated from mice derived from intercrosses of F2 backcrosses to C57BL/6. PCR amplification and direct sequencing of the rearranged Vλ1 gene segment from single cells was performed to determine the distribution, frequency, and base exchange pattern of somatic mutations as well as the frequency of memory B cells carrying a mutated Vλ1 gene (18). SHM, corrected for clonality of the mutations, occurs at a normal frequency in Rev1−/− B cells (Table I). Thus, regardless of the genotype, 40–41% of all analyzed class-switched B cells contained a mutated rearranged Vλ1 gene segment. Taking only the mutated Vλ1 sequences into account, the mutation frequency is 1.1% for wild-type and 1.3% for Rev1−/− B cells. Likewise the distribution of mutations along the Vλ1 segments, with a concentration at mutation hot spots within the complementarity-determining regions (CDR 1, 2, and 3), is similar in wild-type and Rev1−/− B cells (Fig. 4 A). In contrast, highly significant differences in mutation spectra are found (Fig. 4, B and C; Table S2, available at http://www.jem.org/cgi/content/full/jem.20052227/DC1). In the coding (nontranscribed) strand we find an almost complete absence of C to G transversions as well as a moderate decrease in G to C transversions (Fig. 4, B and C). These results indicate a catalytic role of Rev1 in incorporating dCMP during SHM in mammals, in agreement with recent data demonstrating a role of the Rev1 catalytic domain in SHM in chicken DT40 cells (19). The strand-specific mutational defect at GC basepairs mimics that of Ung −/− B cells in which AID-induced deoxyuridine residues cannot be converted into abasic sites (15, 20). This result suggests that abasic sites, generated by subsequent AID and UNG activities on deoxycytidine residues, are the substrates for Rev1 in vivo. Because mice doubly deficient for the mismatch repair gene Msh2 and for Ung have lost all transversion mutations at GC basepairs during SHM (15), the residual G to C transversions in Rev1−/− B cells are likely induced during the second, mismatch repair-dependent phase of SHM. Compensating the strand-specific defect in transversions at GC basepairs, the frequencies of A to T and C to A transversions, as well as T to C transitions, are increased significantly in mutated Vλ1 gene sequences in Rev1−/− B cells (Fig. 4, B and C). These mutations may be caused by compensatory activity of pol η (21, 22) and/or other TLS polymerases. This result indicates that the interaction between Rev1 and other TLS polymerases, including pol η, is dispensable for their activity in SHM. In addition, deletion of the BRCT domain of Rev1 that regulates TLS of short-wave UV light–induced photoproducts by other TLS polymerases, including pol η, does not affect SHM (6). This further supports the specific involvement of the catalytic activity of the Rev1 protein in transversion mutagenesis at GC basepairs.
Table I.
Mutation frequencies of hypermutated Vλ1 gene segments in Rev1−/− memory B cells
| WT | Rev1 | |
|---|---|---|
| Cells analyzed | 368 | 372 |
| Cells mutated | 41% (150) | 40% (148) |
| Number of mutations | 508 | 587 |
| Mutation frequency | 0.46% | 0.53% |
| Actual mutation frequencya | 1.1% | 1.3% |
Only mutated Vλ1 sequences are taken into account.
Figure 4.
Somatic hypermutation of hypermutated Vλ1 genes from memory B cells of wild-type and Rev1−/− mice. (A) Distribution of point mutations in Rev1-deficient B cells. No significant differences are found between both genotypes. Wild-type mice (white bars); Rev1−/− mice (black bars). Mutations concentrate in CDR 1, 2, and 3 (underlined). Numbering of the codons is according to Kabat and Wu (22). (B) Nucleotide substitution profile (normalized to wild-type controls) of the hypermutated Vλ1 segments of the Igλ1 gene derived from Rev1−/− memory B cells. Highly significant differences are found between both genotypes. Wild-type mice (white bars); Rev1−/− mice (black bars). Although the frequency of C to G and, to a lesser extent, G to C transversions are decreased, A to T and C to A transversions as well as T to C transitions are increased. Significant changes (Chi square test) are marked by 1, 2, 3, 4, and 5. The p-values are 0.006, 2 × 10−5, 0.005, 0.02, and 0.03, respectively. (C) Nucleotide substitution profile in hypermutated Vλ1 sequences from Rev1-proficient and -deficient B cells. The relative contribution of substitutions at all four basepairs is unaltered indicating no overall defect in SHM. (Left) Wild type, n = 508 mutations. (Right) Rev1−/−, n = 587 mutations.
In conclusion, we have demonstrated an important role of mammalian Rev1 in determining the mutation spectrum of hypermutating Ig genes, in a strand-biased fashion. This likely is achieved by the direct incorporation of deoxycytidine opposite abasic sites, generated at cytidines via AID-mediated deamination and UNG activity, respectively.
MATERIALS AND METHODS
Generation of Rev1-deficient mice.
Replacement of exon 10 of Rev1 in the 129/OLA-derived embryonic stem cell line E14 with a pPGK-hygromycine cassette (Fig. 1 A) was performed according to established procedures. Gene targeting was analyzed by Southern blotting of KpnI-digested DNA and hybridization with probes A (Fig. 1 B) and B. Rev1+/− cells were used to generate C57BL/6-129/OLA chimeric mice that were crossed to 129/OLA and C57BL/6 mice. Tail DNA of progeny was genotyped in a multiplex PCR reaction (35 cycles at 94°C for 60 s, 53°C for 60 s, and 72°C for 90 s), using primers SCDEKA1 (5′-ATTGTGAGTCTCTAGCGTTTG-3′) and SCDEKA2 (5′-GCTGGAATTGAAATTCTAGG-3′) amplifying the wild-type allele, and primers SCDEKA1 and PGKPR1 (5′-GCTTCCATTGCTCAGCGGTG-3′) amplifying the mutant allele. All required permits were obtained (government permit number VVM/BD 00.235 D06 for generating the transgenic animals and University Animal Ethical Committee permit numbers DEC 01058 and DEC 03139).
Characterization of Rev1 expression.
Rev1 cDNA was generated from kidney-derived RNA as described (6). Primers p13 (5′-GCAGATATACCAGATTC-3′) and p14 (5′-GAGCTGATGGACGACTT-3′) amplify a PCR fragment containing exons 8–15. Primers pSCDE (5′-GTCAGCTGCGATGAAGCACTG-3′) and p14 amplify a product encompassing exons 10–15 (Fig. 1 D). PCR products were cloned and sequenced. Rev1 protein in lysates from immortalized MEFs was analyzed by immunoblotting as described (6).
Flow cytometry.
Single-cell suspensions from thymus, spleen, lymph nodes, and bone marrow were stained with specific antibodies conjugated to FITC, PE, or APC or biotin; biotinylated monoclonal antibodies were revealed with Streptavidin-PerCP or Streptavidin-APC. Antibodies were purchased from BD Biosciences unless mentioned otherwise: CD8b-FITC (53–5.8), CD19-FITC (1D3), IgD-FITC (11-26c.2a), IgM-Fluos (331.12, self conjugated), CD3-PE (17A2), CD8-PE (53–6.7), CD19-PE (1D3), Vλ-PE (LS136, self conjugated), CD4-APC (RM4-5), CD19-APC (1D3), cKit-APC (CD117, clone 2B8), Thy1-APC (CD90, clone 53–2.1), biotinylated TCRγδ (GL3), biotinylated IgM (II/41), and streptavidin-APC. Analysis was performed on a FACScalibur.
Proliferation of LPS-stimulated B cells.
Isolated splenocytes were labeled in serum-free IMDM medium for 10 min at 37°C with 5 μM carboxy fluorescein diacetate succinimidyl ester (CFSE; Molecular Probes). Next, the cells were cultured for 3 d in the presence of 25 μg/ml LPS and 7 μg/ml dextran sulfate. LPS blasts were stained with a PE-conjugated anti-CD19, and subsequently with APC-conjugated annexin V and propidium iodine, and analyzed on a FACScalibur (Becton Dickinson).
Analysis of somatic hypermutation.
Single live IgD− and IgM− (FITC−) and CD19+ (APC+) and Vλ1+ (PE+) B cells were isolated from spleens of 3–5-mo-old mice. Analysis of SHM in the rearranged Vλ1 gene segment by single-cell PCR was performed as described (18).
Mutation analysis.
Multiple, nonimmunized, wild-type (n = 7) and Rev1−/− (n = 5) mice were analyzed, minimizing the incidence of clonally related sequences (Table S2). Data represent mutations between codons 13 and 97 of Vλ1 (23) after exclusion of putatively clonally related mutants.
Online supplemental material.
Table S1 is a composition of B and T cell subsets of primary and secondary lymphatic organs of wild-type and Rev1−/− mice. No qualitative differences are found between the genotypes. Table S2 shows a complete mutational spectrum at the Vλ1 gene segment of wild-type and Rev1−/− mice. All mutated Rev1-deficient and proficient clones used for analysis are shown. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20052227/DC1.
Supplemental Material
Acknowledgments
We wish to thank Angela H.W. de Kleynen, Leon H. Mullenders, and Johan W.A. Verspuy for their contributions to this work and Peter Krijger for discussions.
Financial support was obtained from the Leiden University Medical Center and the Dutch Cancer Foundation (grant RUL 2001-2517 to N. de Wind) and ZonMW (VIDI program 917.56.328 to H. Jacobs). P. Langerak is supported by a startup grant from the Netherlands Cancer Institute (2.129 SFN to H. Jacobs).
The authors have no conflicting financial interests.
J.G. Jansen, P. Langerak, H. Jacobs, and N. de Wind contributed equally to this work.
References
- 1.Prakash, S., R.E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317–353. [DOI] [PubMed] [Google Scholar]
- 2.Masuda, Y., M. Takahashi, S. Fukuda, M. Sumii, and K. Kamiya. 2002. Mechanisms of dCMP transferase reactions catalyzed by mouse Rev1 protein. J. Biol. Chem. 277:3040–3046. [DOI] [PubMed] [Google Scholar]
- 3.Zhang, Y., X. Wu, O. Rechkoblit, N.E. Geacintov, J.S. Taylor, and Z. Wang. 2002. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 30:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbs, P.E.M., J. McDonald, R. Woodgate, and C.W. Lawrence. 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 169:575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson, L.J., and J.E. Sale. 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 22:1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen, J.G., A. Tsaalbi-Shtylik, P. Langerak, F. Calleja, C.M. Meijers, H. Jacobs, and N. de Wind. 2005. The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis. Nucleic Acids Res. 33:356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, C., P.L. Fischhaber, M.J. Luk-Paszyc, Y. Masuda, J. Zhou, K. Kamiya, C. Kisker, and E.C. Friedberg. 2003. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 22:6621–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohashi, E., Y. Murakumo, N. Kanjo, J. Akagi, C. Masutani, F. Hanaoka, and H. Ohmori. 2004. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 9:523–531. [DOI] [PubMed] [Google Scholar]
- 9.Tissier, A., P. Kannouche, M.P. Reck, A.R. Lehmann, R.P. Fuchs, and A. Cordonnier. 2004. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst.). 3:1503–1514. [DOI] [PubMed] [Google Scholar]
- 10.Acharya, N., L. Haracska, R.E. Johnson, I. Unk, S. Prakash, and L. Prakash. 2005. Complex formation of yeast rev1 and rev7 proteins: a novel role for the polymerase-associated domain. Mol. Cell. Biol. 25:9734–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuberger, M.S., R.S. Harris, J. Di Noia, and S.K. Petersen-Mahrt. 2003. Immunity through DNA deamination. Trends Biochem. Sci. 28:305–312. [DOI] [PubMed] [Google Scholar]
- 12.Rajewsky, K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. [DOI] [PubMed] [Google Scholar]
- 13.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 14.Diaz, M., and C. Lawrence. 2005. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 26:215–220. [DOI] [PubMed] [Google Scholar]
- 15.Rada, C., J.M. Di Noia, and M.S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171. [DOI] [PubMed] [Google Scholar]
- 16.Van Sloun, P.P., I. Varlet, E. Sonneveld, J.J. Boei, R.J. Romeijn, J.C. Eeken, and N. de Wind. 2002. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol. Cell. Biol. 22:2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald, J.P., E.G. Frank, B.S. Plosky, I.B. Rogozin, C. Masutani, F. Hanaoka, R. Woodgate, and P.J. Gearhart. 2003. 129-derived strains of mice are deficient in DNA polymerase iota and have normal immunoglobulin hypermutation. J. Exp. Med. 198:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs, H., Y. Fukita, G.T. van der Horst, J. de Boer, G. Weeda, J. Essers, N. de Wind, B.P. Engelward, L. Samson, S. Verbeek, et al. 1998.Hypermutation of immunoglobulin genes in memory B cells of DNA repair-deficient mice. J. Exp. Med. 187:1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross, A.L., and J.E. Sale. 2005. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol. Immunol. In press. [DOI] [PubMed]
- 20.Rada, C., G.T. Williams, H. Nilsen, D.E. Barnes, T. Lindahl, and M.S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 21.Delbos, F., A. de Smet, A. Faili, S. Aoufouchi, J.C. Weill, and C.A. Reynaud. 2005. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 201:1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martomo, S.A., W.W. Yang, R.P. Wersto, T. Ohkumo, Y. Kondo, M. Yokoi, C. Masutani, F. Hanaoka, and P.J. Gearhart. 2005. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl. Acad. Sci. USA. 102:8656–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabat, E.A., and T.T. Wu. 1971. Attempts to locate complementarity-determining residues in the variable positions of light and heavy chains. Ann. N. Y. Acad. Sci. 190:382–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.