Abstract
Killer cell lectin-like receptor G1 (KLRG1) is an inhibitory receptor expressed on subsets of natural killer (NK) cells and T cells, for which no endogenous ligands are known. Here, we show that KLRG1 binds three of the classical cadherins (E-, N-, and R-), which are ubiquitously expressed in vertebrates and mediate cell–cell adhesion by homotypic or heterotypic interactions. By expression cloning using the mouse KLRG1 tetramer as a probe, we identified human E-cadherin as a xenogeneic ligand. We also identified a syngeneic interaction between mouse KLRG1 and mouse E-cadherin. Furthermore, we show that KLRG1 binds N- and R-cadherins. Finally, we demonstrate that E-cadherin binding of KLRG1 prevents the lysis of E-cadherin–expressing targets by KLRG1+ NK cells. These results suggest that KLRG1 ligation by E-, N-, or R-cadherins may regulate the cytotoxicity of killer cells to prevent damage to tissues expressing the cadherins.
NK cells play a pivotal role in innate immunity by their direct cytolytic activity and production of regulatory cytokines (1). NK cell recognition of targets involves two types of receptors, activating receptors and inhibitory receptors (2). Integration of opposing signals from these two types of receptors determines the activation of NK cells. These receptors are classified into two structurally defined groups, Ig-like receptors and C-type lectin-like receptors. The former group includes killer cell Ig-like receptors (KIRs) (3, 4), whereas the latter group includes CD94/NKG2 (KLRD/KLRC), rodent Ly49 (KLRA), NKG2D (KLRK), NKR-P1 (KLRB), and KLRG1. Many of the receptors, such as KIR, CD94/NKG2, NKG2D, and Ly49, have been shown to bind either MHC class I molecules or molecules structurally related to MHC class I (2, 4, 5). However, the MHC class I–specific receptors do not appear to account for all NK cell specificities, some of which may be conveyed by orphan receptors expressed on the NK cells (6).
KLRG1 is an orphan C-type lectin-like receptor that was originally identified as the mast cell function–associated antigen (MAFA) expressed on the rat basophilic leukemia cell line RBL-2H3 (7). Antibody-mediated ligation of KLRG1 inhibits release of inflammatory mediators from RBL-2H3 cells induced by cross-linking of FcɛRI. In contrast, in mouse and human, KLRG1 is expressed on subsets of NK cells and T cells (8–12). In normal mice, ∼30% of resting NK cells express KLRG1, and viral infections increase the percentage of KLRG1-expressing NK cells (13). In humans, ∼60% of NK cells from healthy adult donors express KLRG1 (14). Although T cell expression of KLRG1 in normal mice is restricted to small subpopulations of effector–memory type T cells (11, 12, 15), expression of KLRG1 is up-regulated in mouse CD8+ T cells by infection with pathogens (12, 16). In humans, KLRG1 is expressed on ∼40% of CD8+ T cells and ∼20% of CD4+ T cells from healthy adult donors (14). Furthermore, over 90% of CD8+ T cells specific to CMV or EBV express KLRG1 during the latent stages of these chronic infections (17). Consistent with the inhibitory activity in RBL-2H3 cells (7), KLRG1 has an immune receptor tyrosine-based inhibitory motif (ITIM) in its cytoplasmic domain, and mAb-mediated cross-linking of KLRG1 inhibits NK cell function (13). Although rat KLRG1 has been shown to bind saccharides (7), endogenous ligands for KLRG1 have not been identified.
To identify endogenous KLRG1 ligands, we generated a KLRG1 tetramer and a KLRG1 reporter cell line, which allowed us to identify cell lines expressing KLRG1 ligands. By expression cloning using the KLRG1 tetramer as a probe, we identify human E-cadherin as a xenogeneic ligand. We also show that mouse KLRG1 binds three members of the mouse classical cadherin family and KLRG1 binding by its ligand inhibits NK cell cytotoxicity.
RESULTS AND DISCUSSION
Expression of putative KLRG1 ligands on mouse and human cell lines
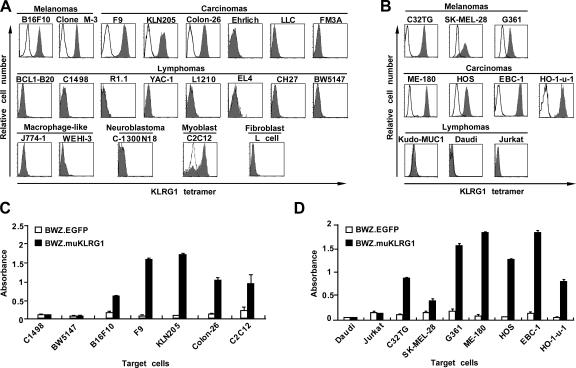
To identify endogenous ligands for KLRG1, we generated a fluorescently labeled tetramer of the entire extracellular domain of mouse KLRG1. The KLRG1 tetramer was tested for binding to a panel of mouse and human cell lines. Of the mouse cell lines, the KLRG1 tetramer bound a myoblast cell line, two melanoma lines, and three carcinoma cell lines (Fig. 1 A). The binding was inhibited when the KLRG1 tetramer was preincubated with an excess of the anti–mouse KLRG1 mAb 2F1 (unpublished data). The mouse KLRG1 tetramer also bound human melanoma and carcinoma cell lines (Fig. 1 B), suggesting that the KLRG1 ligands are highly conserved across species. These results were confirmed using the KLRG1 reporter cell line BWZ.muKLRG1, which enabled us to visualize binding of the mouse KLRG1 ectodomain to a putative ligand by converting the signal into β-galactosidase expression. All of the mouse cell lines that bound the KLRG1 tetramer induced the expression of β-galactosidase in the mouse KLRG1 reporter cells, whereas cell lines that did not bind the KLRG1 tetramer did not stimulate the KLRG1 reporter cells (Fig. 1 C). Similar results were obtained for the human cell lines (Fig. 1 D). These results suggest that putative KLRG1 ligands are expressed ubiquitously in melanoma and carcinoma cell lines.
Figure 1.
Expression of a putative KLRG1 ligand on mouse and human cell lines. (A and B) A panel of mouse (A) and human (B) cell lines were stained with mouse KLRG1 tetramer (filled histograms) or PE-streptavidin alone (solid lines) and analyzed by FACS. LLC, Lewis lung cancer. (C and D) The mouse KLRG1 reporter cell line BWZ.muKLRG1(black bars) or the control line BWZ.EGFP (white bars) were cocultured with the indicated mouse (C) or human (D) cell lines and then assayed for β-galactosidase activity. Representative data (means ± SD of triplicate samples) from one of three independent experiments are shown.
Biochemical characterization of putative KLRG1 ligands
We next characterized the biochemical nature of the putative KLRG1 ligand. The binding of the KLRG1 tetramer to the mouse and human cell lines was sensitive to pretreatment of the cells with trypsin-EDTA (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20051986/DC1), suggesting that the putative KLRG1 ligand on these cells is a membrane protein. We also precipitated the putative KLRG1 ligand expressed on the human lung carcinoma line EBC-1, which was stained most intensely with the mouse KLRG1 tetramer among the cell lines tested. Mouse KLRG1 specifically precipitated a protein with an apparent mass on SDS-PAGE of 115 kD under nonreducing conditions and 120 kD under reducing conditions (Fig. 2 A). These results suggest that the putative KLRG1 ligand expressed on EBC-1 cells is a protein with an apparent mass of 120 kD and has a single or a small number of intramolecular disulfide bond(s).
Figure 2.
Expression cloning of a KLRG1 ligand expressed on EBC-1 cells. (A) Biochemical characterization of the putative KLRG1 ligand. The human EBC-1 cell line was surface labeled with 125I, and cell lysates were precipitated with streptavidin-beads loaded with (+) or without (−) biotinylated mouse KLRG1. The precipitates were separated by electrophoresis on an SDS-PAGE gel under reducing (R) or nonreducing (NR) conditions, and 125I-labeled proteins were visualized by phosphorimaging. Arrowheads indicate the bands for the putative KLRG1 ligand. (B) BW5147 cells were transduced with a retroviral cDNA library prepared from EBC-1 cells, and the KLRG1 tetramer+ population was enriched. The enriched KLRG1 tetramer+ population (right) or untransduced control BW5147 cells were stained with the mouse KLRG1 tetramer (filled histograms) or PE-streptavidin alone (solid lines). (C) The KLRG1 tetramer+ population and the control BW5147 cells were tested for stimulation of the KLRG1 reporter cells. Representative data (mean ± SD of triplicate samples) from one of three independent experiments are shown. (D) The KLRG1 tetramer+ population (right) or control BW5147 cells (left) were stained with an anti–human E-cadherin mAb and PE-anti–mouse IgG (filled histograms) or PE-anti–mouse IgG alone (solid lines).
Binding of KLRG1 to E-cadherin
To identify the ligand expressed on EBC-1, we constructed a cDNA library from EBC-1 and transduced mouse BW5147 thymoma cells, which do not bind the KLRG1 tetramer. The cells were stained with the mouse KLRG1 tetramer to enrich the KLRG1 tetramer-positive population by FACS. After three rounds of enrichment, we obtained a population that was uniformly stained with the KLRG1 tetramer (Fig. 2 B), and this population also stimulated the mouse KLRG1 reporter cells (Fig. 2 C). PCR recovery of the integrated cDNA gave an ∼5-kb DNA fragment, which encoded human E-cadherin, a Ca2+-dependent adhesion molecule that plays pivotal roles in tissue morphogenesis and in the formation of cellular junctions (18, 19). Consistent with these data, a mAb to human E-cadherin stained the KLRG1 tetramer-positive population (Fig. 2 D). The biochemical nature of E-cadherin, such as sensitivity to trypsin-EDTA treatment, the apparent migration on SDS-PAGE, and the presence of a single intramolecular disulfide bond, is also consistent with that of the putative KLRG1 ligand expressed on EBC-1 cells. To confirm a syngeneic interaction between KLRG1 and E-cadherin, mouse E-cadherin was expressed on BW5147 cells (Fig. 3 A). BW5147 cells expressing mouse E-cadherin bound the mouse KLRG1 tetramer and stimulated the mouse KLRG1 reporter cells (Fig. 3, A and B). In the complementary experiment, a dodecameric form of a mouse E-cadherin–Fc fusion protein was used to stain BW5147 cells transduced with mouse KLRG1 using a bicistronic expression vector carrying cDNA for expression of an EGFP marker. The E-cadherin–Fc fusion protein stained the EGFP+ KLRG1+ cells, and the staining was blocked by a mAb against KLRG1 (Fig. 3, C and D). Conversely, the E-cadherin–Fc fusion protein did not stain the EGFP-negative population of KLRG1 transductants or control transductants. Collectively, these results indicate that KLRG1 binds E-cadherin.
Figure 3.
Binding of mouse KLRG1 to mouse E-cadherin. (A) BW5147 cells transduced with mouse E-cadherin (right) and control BW5147 cells (left) were analyzed for E-cadherin expression and binding of the KLRG1 tetramer. For E-cadherin expression (top), the cells were stained with mAb to mouse E-cadherin (ECCD-2) and a secondary antibody (filled histograms) or the secondary antibody alone (solid lines). For binding of the mouse KLRG1 tetramer (bottom), the cells were stained with the KLRG1 tetramer (filled histograms) or PE-streptavidin alone (solid lines). (B) BW5147 cells transduced with mouse E-cadherin and control BW5147 cells were tested for stimulation of the KLRG1 reporter cells. Representative data (mean ± SD of triplicate samples) from one of three independent experiments are shown. (C) BW5147 cells transduced with the KLRG1-IRES-EGFP vector (right) or the control EGFP vector (left) were stained with the anti-KLRG1 antibody and PE-anti–mouse IgG. (D) BW5147 cells transduced with the KLRG1-IRES-EGFP vector (bottom) or the control EGFP vector (top) were stained with E-cadherin–Fc fusion protein and PE-anti–human IgG Fc. Where indicated, the cells were treated with the anti-KLRG1 mAb or an isotype-matched control mAb, before staining with the E-cadherin–Fc fusion protein.
Binding of KLRG1 to three members of classical cadherins
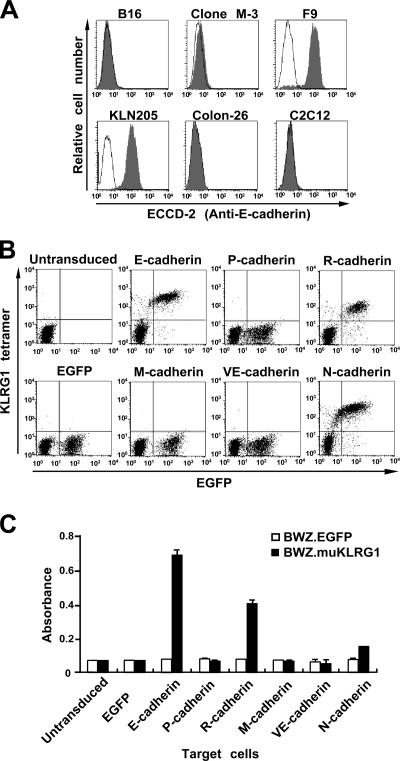
Because the KLRG1 tetramer stained melanoma cells, which do not usually express E-cadherin (20), we examined E-cadherin expression in the mouse cell lines that bound the KLRG1 tetramer. We found that two melanomas, B16 and clone M-3, the Colon-26 carcinoma, and the C2C12 myoblast cell line expressed little or no E-cadherin (Fig. 4 A). By RT-PCR analysis, we found that these cell lines express other members of the classical cadherin family, such as N- or M-cadherin (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20051986/DC1). These observations led us to test whether KLRG1 binds other cadherins. We independently transduced BW5147 cells with each of the five mammalian classical cadherins (E-, N-, P-, R-, or M-) or one of the type II cadherins (VE-) using the bicistronic expression vector, which also expresses the EGFP marker, and tested those cells for KLRG1 tetramer binding. The expression of cadherins was confirmed by flow cytometry or Western blotting (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20051986/DC1). The KLRG1 tetramer stained cells transduced with E-, N-, or R-cadherins, but not those transduced with P-, M-, or VE-cadherins (Fig. 4 B). These results were confirmed by the KLRG1 reporter assays (Fig. 4 C). In RT-PCR analysis, all the mouse cell lines that bound the KLRG1 tetramer expressed at least one of the E-, N-, or R-cadherins (Fig. S2). These data indicate that KLRG1 recognizes three members (E-, N-, and R-) of the classical cadherin family.
Figure 4.
Binding of KLRG1 to three members of the classical cadherin family. (A) E-cadherin expression on mouse cell lines that bound the KLRG1 tetramer. The indicated cell lines were stained with a mAb to mouse E-cadherin (ECCD-2) and FITC-anti–mouse IgG (filled histograms) or FITC-anti–mouse IgG alone (solid lines). (B and C) BW5147 cells were retrovirally transduced with vectors for bicistronic expression of EGFP and the mouse cadherins indicated. The cells were stained with the KLRG1 tetramer (B) or tested for stimulation of the mouse KLRG1 reporter cells (C). In C, representative data (mean ± SD of triplicate samples) from one of three independent experiments are shown.
Classical cadherins primarily mediate cell–cell adhesion by homotypic interactions (18, 19). E-cadherin also participates in adhesion of intestinal intraepithelial lymphocytes to epithelial cells by heterotypic interactions with the αEβ7 integrin (21). Thus, classical cadherins function both as ligands and as receptors in mediating cell–cell adhesion. Our results reveal a novel function of the cadherins as ligands for an inhibitory receptor expressed on immune cells.
Binding of KLRG1 by E-cadherin inhibits NK cell cytotoxicity
Because KLRG1 has an ITIM consensus sequence in its cytoplasmic region and mAb-mediated cross-linking of KLRG1 inhibits NK cell functions (13), we investigated the functional consequence of the KLRG1 receptor binding to its ligand. We expressed KLRG1 in the mouse NK cell line NK03 (Fig. 5 A), which does not normally express KLRG1, and examined its capacity to kill F9 cells, which express E-, N-, and R-cadherins. The KLRG1-expressing NK cells killed F9 cells less efficiently than the uninfected control NK cells, and the anti–mouse KLRG1 mAb restored the cytotoxicity of the KLRG1 expressing NK cells to levels similar to that of the control NK cells (Fig. 5 B). We also tested killing of the E-cadherin–expressing BW5147 cells by the same effectors. The KLRG1-expressing NK cells again showed a reduced capacity to kill E-cadherin–expressing targets, and the reduction was abolished by addition of an anti-KLRG1 mAb (Fig. 5 C). These results indicate that E-cadherin binding to KLRG1 on NK cells inhibits NK cell cytotoxicity.
Figure 5.
Ligation of KLRG1 on NK cells by E-cadherin inhibits NK cell cytotoxicity. (A) Expression of KLRG1 on the mouse NK cell line NK03 (left) or the cell line transduced (right) with KLRG1. The cells were stained with the mAb to KLRG1 and a secondary antibody (filled histograms) or the secondary antibody alone (solid lines). (B) Cytotoxicity of the control (left) or KLRG1 transduced (right) mouse NK cell line NK03 against mouse F9 cells was assessed in a 4-h 51Cr-release assay. The assays were conducted in the absence (◯) or the presence (•) of the anti-KLRG1 mAb, or in the presence of an isotype-matched control mAb (▵). (C) Cytotoxicity of the control (left) or KLRG1 transduced (right) mouse NK cells against E-cadherin expressing (bottom) or control (top) BW5147 targets, both of which lack Fcγ receptors. The assays were conducted in the absence (◯) or the presence (•) of the anti-KLRG1 mAb, or in the presence of an isotype-matched control mAb (▵). Representative data (mean ± SD of triplicate samples) from one of three independent experiments are shown.
As E-, N-, and R-cadherins are ubiquitously expressed among solid tissues, these tissues may be protected from attack by killer cells expressing KLRG1. In this context, it is of note that the activation history of individual NK and T cells appears to regulate KLRG1 expression on the cells. NK cell expression of KLRG1 is up-regulated by viral infection in animals (13) and, in contrast, is down-regulated in MHC class I–deficient animals (22), in which NK cells are hyporesponsive (23). Expression of KLRG1 on CD8+ T cells, especially those showing specificity to viral antigens, is also induced by viral infection (8, 12, 16, 17). Moreover, KLRG1 is expressed on effector–memory CD8+ T cells (15), which have experienced antigen stimulation and immediately respond to antigenic stimuli. A higher threshold of activation for these “immune experienced” NK and T cells expressing KLRG1 would be imposed when their targets express E-, N-, or R-cadherins. This would prevent excessive damage to vital tissues expressing the cadherins and would also contribute to terminating the immune response.
Normally, classical cadherins are localized at tight junctions in epithelia and similar structures in endothelia, both of which form the physical barriers. Migration of leukocytes through these barriers is regulated by chemokines, whose action is modulated by the inhibitory receptor Ly49A in NK cells (24). In this context, KLRG1 may regulate transendothelial migration of NK and T cells that express KLRG1. Similar localization at the tight junctions has been observed for the poliovirus receptor (CD166). CD166 recognition by DNAM-1, an activating receptor expressed on NK cells, as well as other leukocytes, is involved in the transendothelial migration of monocytes (25). Further studies using KLRG1-deficient animals or in vivo blocking of KLRG1 function using mAbs against KLRG1 will provide insight into the physiological function of KLRG1.
The malignancy of epithelial tumors is often associated with down-regulation of E-cadherin, which makes tumor cells invasive and metastatic (26, 27). Because our results predict that tumor cells lacking E-cadherin expression would be more prone to killing by KLRG1+ NK cells, it is tempting to hypothesize that NK cells expressing KLRG1 are instrumental in surveillance of malignant tumors. This idea is analogous to the advocated role of inhibitory MHC class I receptors in surveillance of tumor cells that have lost or down-regulated MHC class I expression in the “missing self hypothesis” (28). Although KLRG1 expression on NK cells is inducible by viral infections (13), there is also a compartment of NK cells that constitutively express KLRG1, which dominates ∼30% of mouse resting NK cells and ∼50% of human peripheral blood NK cells (8, 9, 14, 22, 29). The KLRG1+ subsets of resting NK cells may be involved in surveillance of malignant tumors that have lost their “identity” by losing or down-regulating the expression of E-, N-, or R-cadherins, although this hypothesis needs validation by in vivo studies.
Finally, our results uncover a novel role of the classical cadherins, which are ubiquitously expressed among solid tissues and have a fundamental role in maintaining morphology of the solid tissues in vertebrates, in which these proteins are used to regulate the cytotoxicity of cells of the innate and acquired immune systems.
MATERIALS AND METHODS
Cell lines, reagents, and antibodies.
The BWZ.36 cell line was provided by N. Shastri (University of California Berkeley, Berkeley, CA). The retrovirus vectors, pMXs and pMXs-IRES-GFP, and the retrovirus packaging cell line Plat-E were provided by T. Kitamura (The University of Tokyo) and were used for retroviral transduction throughout this study.
Culture supernatant from hybridoma cells obtained from M. Takeichi (Institute of Physical and Chemical Research CDB, Hyogo, Japan) was used to provide the rat anti–E-cadherin (ECCD-2) mAb. The other mAbs used were a mouse monoclonal anti–human E-cadherin (67A4; Santa Cruz Biotechnology, Inc.) and a hamster monoclonal anti–mouse KLRG1 (2F1; eBioscience).
For flow cytometry, PE-anti–mouse IgG F(ab′)2 or FITC-anti–mouse IgG F(ab′)2 fragments were used as the secondary reagents. Data were acquired with a FACSCalibur system and were analyzed with CellQuest software (BD Biosciences).
Preparation of biotinylated KLRG1 protein and its tetramer.
A cDNA encoding the ectodomain of mouse KLRG1 amplified by RT-PCR from C57BL/6J spleen cells was cloned into pET3Nbio (30). Expression, refolding, biotinylation, and tetramerization of the KLRG1 ectodomain were performed as described previously (30). The tetramer was used at 2.5–5 μg/ml for staining.
KLRG1 reporter assays.
The construct for the KLRG1 chimeric receptor, muKLRG1rep-pMXs-IG, included the extracellular and transmembrane regions of the mouse KLRG1 and the cytoplasmic region of the mouse CD3ζ chain. The muKLRG1rep-pMXs-IG vector and the empty pMXs-IRES-GFP vector were used for transduction of the BWZ.36 cell line to establish the reporter BWZ.muKLRG1 and the control BWZ.EGFP, respectively. The cell lines were cultured with the indicated target cells for 16 h, and β-galactosidase activity was determined by colorimetric assay using chlorophenol red-β-d-galactopyranoside (Wako) as a substrate.
Precipitation and detection of a putative KLRG1 ligand.
KLRG1 ligands were precipitated from lysates of EBC-1 cells that had been surface labeled with 125I using streptavidin-agarose beads loaded with the biotinylated mouse KLRG1 protein. The precipitates were analyzed on a 10% SDS-PAGE gel, and the gel was subjected to phosphorimaging with a BAS-2500 system (Fuji Photo Film).
Expression cloning.
A cDNA library containing 1.4 × 106 independent clones was constructed using the pMXs vector and used for transduction of BW5147 cells. The cells positive for the KLRG1 tetramer were enriched by three rounds of sorting with a FACSVantage SE. Genomic DNA extracted from the enriched population was used for the recovery of the cDNA inserts by PCR amplification with primers specific for the pMXs vector. The amplified cDNA fragments were cloned into the pBluescript II SK(+) vector (Stratagene) and sequenced.
Transduction with cadherins or KLRG1.
cDNAs for mouse E- and N-cadherins were provided by M. Takeichi (Institute of Physical and Chemical Research CDB, Hyogo, Japan). cDNA for mouse P-cadherin was obtained from the Institute of Physical and Chemical Research BioResource Center (Ibaraki, Japan). cDNAs for R-, VE-, and M-cadherin were amplified by RT-PCR from the brain of a C57BL/6J mouse and from C2C12 cells, respectively. These cDNAs were independently subcloned into a pMXs vector or a pMXs-IRES-EGFP vector and used for retroviral transduction. A BW5147 cell line expressing E-cadherin was established from BW5147 cells transduced with the pMXs vector with E-cadherin cDNA by limiting dilution.
Preparation of E-cadherin–Fc fusion protein.
Preparation of the E-cadherin–Fc fusion protein will be described elsewhere (unpublished data). In brief, cDNA fragments encoding the extracellular region of mouse E-cadherin and the Fc segment of human IgG1 fused with an IgA tail piece (a gift from H. Arase, Osaka University, Osaka, Japan) were cloned into a pRc/CMV1 vector (Invitrogen). 293T cells were transfected with a 5:1 mixture of the E-cadherin–Fc expression vector and an expression vector for mouse furin (pCMVmFur; a gift from K. Nakayama, Kyoto University, Kyoto, Japan), and the culture supernatants containing 0.2 μg/ml of E-cadherin–Fc protein were used to stain cells for 30 min at 37°C, followed by staining with PE-goat anti–human IgG Fc antibody preabsorbed with hamster serum. Where indicated, cells were incubated with 10 μg/ml of hamster anti–mouse KLRG1 mAb (2F1) or control hamster antibody, before staining with E-cadherin–Fc fusion protein.
Cytotoxicity assays.
The mouse NK cell line NK03, which was established from spleen NK cells of C57BL/6J mice in our laboratory (unpublished data), was transduced with mouse KLRG1 cDNA subcloned into the pMXs-IRES-EGFP vector. The cells with high levels of EGFP expression were sorted using a FACSVantage SE. The cytotoxicity of NK cells against various target cells was examined using a standard 4 h 51Cr-release assay, as described (30). Where indicated, 5 μg/ml of the indicated mAb was included in the assays. F9 cells and BW5147 cells lack expression of Fcγ receptors, excluding the possibility of reverse antibody-dependent cell cytotoxicity.
Online supplemental material.
Table S1 lists the cell lines used to identify the KLRG1 ligands. Fig. S1 describes the sensitivity of the putative KLRG1 ligand(s) to trypsin-EDTA treatment. Fig. S2 describes RT-PCR analysis for expression of cadherins in mouse cell lines. Fig. S3 describes expression of cadherins on BW5147 cells transduced with various cadherins. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20051986/DC1.
Supplemental Material
Acknowledgments
We thank N. Shastri, M. Takeichi, T. Kitamura, H. Karasuyama, H. Arase, K. Nakayama, and K. Nakamura for materials, and K. Maenaka, A. Shibuya, K. Iizuka, and A. Ochiai for discussion of our work.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and a grant from the Mochida Memorial Foundation for Medical and Pharmaceutical Research.
The authors have no conflicting financial interests.
References
- 1.Biron, C.A., K.B. Nguyen, G.C. Pien, L.P. Cousens, and T.P. Salazar-Mather. 1999. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 17:189–220. [DOI] [PubMed] [Google Scholar]
- 2.Lanier, L.L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274. [DOI] [PubMed] [Google Scholar]
- 3.Long, E.O. 1999. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 17:875–904. [DOI] [PubMed] [Google Scholar]
- 4.Moretta, L., and A. Moretta. 2004. Killer immunoglobulin-like receptors. Curr. Opin. Immunol. 16:626–633. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama, W.M. 1998. Natural killer cell receptors. Curr. Opin. Immunol. 10:298–305. [DOI] [PubMed] [Google Scholar]
- 6.Kumar, V., and M.E. McNerney. 2005. A new self: MHC-class-I-independent natural-killer-cell self-tolerance. Nat. Rev. Immunol. 5:363–374. [DOI] [PubMed] [Google Scholar]
- 7.Abramson, J., R. Xu, and I. Pecht. 2002. An unusual inhibitory receptor–the mast cell function-associated antigen (MAFA). Mol. Immunol. 38:1307–1313. [DOI] [PubMed] [Google Scholar]
- 8.Blaser, C., M. Kaufmann, and H. Pircher. 1998. Virus-activated CD8 T cells and lymphokine-activated NK cells express the mast cell function-associated antigen, an inhibitory C-type lectin. J. Immunol. 161:6451–6454. [PubMed] [Google Scholar]
- 9.Hanke, T., L. Corral, R.E. Vance, and D.H. Raulet. 1998. 2F1 antigen, the mouse homolog of the rat “mast cell function-associated antigen”, is a lectin-like type II transmembrane receptor expressed by natural killer cells. Eur. J. Immunol. 28:4409–4417. [DOI] [PubMed] [Google Scholar]
- 10.Butcher, S., K.L. Arney, and G.P. Cook. 1998. MAFA-L, an ITIM-containing receptor encoded by the human NK cell gene complex and expressed by basophils and NK cells. Eur. J. Immunol. 28:3755–3762. [DOI] [PubMed] [Google Scholar]
- 11.Beyersdorf, N.B., X. Ding, K. Karp, and T. Hanke. 2001. Expression of inhibitory “killer cell lectin-like receptor G1” identifies unique subpopulations of effector and memory CD8 T cells. Eur. J. Immunol. 31:3443–3452. [DOI] [PubMed] [Google Scholar]
- 12.Voehringer, D., C. Blaser, P. Brawand, D.H. Raulet, T. Hanke, and H. Pircher. 2001. Viral infections induce abundant numbers of senescent CD8 T cells. J. Immunol. 167:4838–4843. [DOI] [PubMed] [Google Scholar]
- 13.Robbins, S.H., K.B. Nguyen, N. Takahashi, T. Mikayama, C.A. Biron, and L. Brossay. 2002. Cutting edge: inhibitory functions of the killer cell lectin-like receptor G1 molecule during the activation of mouse NK cells. J. Immunol. 168:2585–2589. [DOI] [PubMed] [Google Scholar]
- 14.Voehringer, D., M. Koschella, and H. Pircher. 2002. Lack of proliferative capacity of human effector and memory T cells expressing killer cell lectinlike receptor G1 (KLRG1). Blood. 100:3698–3702. [DOI] [PubMed] [Google Scholar]
- 15.Kaech, S.M., J.T. Tan, E.J. Wherry, B.T. Konieczny, C.D. Surh, and R. Ahmed. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198. [DOI] [PubMed] [Google Scholar]
- 16.McMahon, C.W., A.J. Zajac, A.M. Jamieson, L. Corral, G.E. Hammer, R. Ahmed, and D.H. Raulet. 2002. Viral and bacterial infections induce expression of multiple NK cell receptors in responding CD8+ T cells. J. Immunol. 169:1444–1452. [DOI] [PubMed] [Google Scholar]
- 17.Ibegbu, C.C., Y.X. Xu, W. Harris, D. Maggio, J.D. Miller, and A.P. Kourtis. 2005. Expression of killer cell lectin-like receptor G1 on antigen-specific human CD8+ T lymphocytes during active, latent, and resolved infection and its relation with CD57. J. Immunol. 174:6088–6094. [DOI] [PubMed] [Google Scholar]
- 18.Takeichi, M. 1991. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 251:1451–1455. [DOI] [PubMed] [Google Scholar]
- 19.Vleminckx, K., and R. Kemler. 1999. Cadherins and tissue formation: integrating adhesion and signaling. Bioessays. 21:211–220. [DOI] [PubMed] [Google Scholar]
- 20.Tang, A., M.S. Eller, M. Hara, M. Yaar, S. Hirohashi, and B.A. Gilchrest. 1994. E-cadherin is the major mediator of human melanocyte adhesion to keratinocytes in vitro. J. Cell Sci. 107(Pt 4):983–992. [DOI] [PubMed] [Google Scholar]
- 21.Cepek, K.L., S.K. Shaw, C.M. Parker, G.J. Russell, J.S. Morrow, D.L. Rimm, and M.B. Brenner. 1994. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the αEβ7 integrin. Nature. 372:190–193. [DOI] [PubMed] [Google Scholar]
- 22.Corral, L., T. Hanke, R.E. Vance, D. Cado, and D.H. Raulet. 2000. NK cell expression of the killer cell lectin-like receptor G1 (KLRG1), the mouse homolog of MAFA, is modulated by MHC class I molecules. Eur. J. Immunol. 30:920–930. [DOI] [PubMed] [Google Scholar]
- 23.Raulet, D.H., R.E. Vance, and C.W. McMahon. 2001. Regulation of the natural killer cell receptor repertoire. Annu. Rev. Immunol. 19:291–330. [DOI] [PubMed] [Google Scholar]
- 24.Inngjerdingen, M., B. Rolstad, and J.C. Ryan. 2003. Activating and inhibitory Ly49 receptors modulate NK cell chemotaxis to CXC chemokine ligand (CXCL) 10 and CXCL12. J. Immunol. 171:2889–2895. [DOI] [PubMed] [Google Scholar]
- 25.Reymond, N., A.M. Imbert, E. Devilard, S. Fabre, C. Chabannon, L. Xerri, C. Farnarier, C. Cantoni, C. Bottino, A. Moretta, et al. 2004. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J. Exp. Med. 199:1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeichi, M. 1993. Cadherins in cancer: implications for invasion and metastasis. Curr. Opin. Cell Biol. 5:806–811. [DOI] [PubMed] [Google Scholar]
- 27.Birchmeier, W., K.M. Weidner, J. Hulsken, and J. Behrens. 1993. Molecular mechanisms leading to cell junction (cadherin) deficiency in invasive carcinomas. Semin. Cancer Biol. 4:231–239. [PubMed] [Google Scholar]
- 28.Karre, K., H.G. Ljunggren, G. Piontek, and R. Kiessling. 1986. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defense strategy. Nature. 319:675–678. [DOI] [PubMed] [Google Scholar]
- 29.Robbins, S.H., M.S. Tessmer, T. Mikayama, and L. Brossay. 2004. Expansion and contraction of the NK cell compartment in response to murine cytomegalovirus infection. J. Immunol. 173:259–266. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto, N., M. Mitsuki, K. Tajima, W.M. Yokoyama, and K. Yamamoto. 2001. The functional binding site for the C-type lectin-like natural killer cell receptor Ly49A spans three domains of its major histocompatibility complex class I ligand. J. Exp. Med. 193:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.