Abstract
HIV is evolution gone mad and bad. The virus infects a person and rapidly diversifies to become a huge swarm of viruses, each equipped differently to resist the onslaught of diverse T cells and antibodies. We can't expect to predict details of the struggle between virus and immunity, right? Wrong—maybe we can make some predictions, say two new landmark studies with potentially huge consequences for AIDS vaccine design.
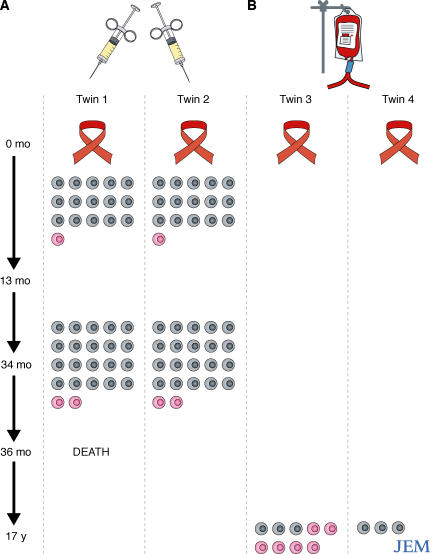
Two new studies aim to answer a seemingly simple question—what happens if two genetically identical people are infected with the same strain of HIV? In this issue, Draenert et al. (p. 529) study identical twins infected with the same strain of HIV at essentially the same time through needle sharing in injection drug use (1). The twins were diagnosed with HIV soon after infection, allowing the investigators to track initial immune responses, viral evolution, and disease outcome (Fig. 1). A second study recently published by Yang et al. in the Journal of Virology followed a different pair of HIV+ identical twins infected perinatally with blood transfusions from the same donor. However, immune responses were not monitored in detail until much later in the course of infection in this second study (2).
Figure 1.
Initial sharing of HIV-specific CTL responses is largely lost by late-stage infection. Two sets of identical twins were infected with identical stocks of HIV. The twins followed by Draenert et al. and described in this issue of JEM (1) (Twins 1 and 2) were infected by intravenous drug use and were followed during the first 3 years of infection (A). Yang et al. (2) studied twins 3 and 4, who were infected by blood transfusion, during late-stage infection (B). CTL responses shared between both twins in each pair are shown as gray cells. CTL responses mounted only by one twin in each pair are colored pink.
Strikingly, conventional markers of HIV disease progression were nearly identical within both sets of twins. The juvenile twins in the study by Yang et al. developed serious HIV complications at age 7, had similar viral loads and CD4+ T cell counts during several years of incomplete antiretroviral therapy, manifested late-stage disease at age 17, and rebounded upon initiation and adherence to a different antiretroviral regimen (2). In the Draenert et al. study, viral loads and CD4+ T cell counts were also very similar during 3 years of observation, including a precipitous CD4+ T cell decline ∼30 months after infection. This study was terminated after 3 years when one of the twins died of a drug overdose (1).
Surprisingly similar CTL and antibody responses immediately after infection
Are the similar outcomes caused by identical immune responses in both pairs of twins? Here the picture is less clear, although many details of immune responses in the pairs of twins are very similar. Both groups focused their attention primarily on CD8+ T lymphocyte (cytotoxic T lymphocyte; CTL) responses. 6 months after infection, 15 of the 16 CTL epitopes recognized by the twins in the Draenert et al. study were identical (1). By 34 months, modest differences in the CTL responses emerged, though the majority of responses were still shared between both twins. The two twins in the Yang et al. study shared three CTL responses (2). One of the twins only mounted these shared responses, whereas the other recognized six additional epitopes. Unlike the twins in the Draenert et al. study, these twins were both treated with antiretrovirals that suppressed HIV replication. It is possible that CTL responses waned in the absence of vigorous HIV replication, dampening shared responses that may have been present earlier during infection. Yang et al. took the analysis of shared responses one step further than Draenert et al., discovering that the TCRs used by CTL against the same epitope were markedly different between the twins. This finding parallels the recent observation that structurally diverse TCRs with distinct Vβ usage and high amino diversity comprise the CTL directed against an immunodominant epitope in the HIV Nef protein (3).
Draenert et al. also found essentially identical neutralizing antibody responses in both twins (1). Recent longitudinal studies of early HIV infection have shown that strain-specific neutralizing antibodies are elicited in response to the virus (4, 5). When these antibodies reach a critical threshold (a matter of weeks), a resistant virus emerges. Eventually, a neutralizing antibody response to this virus develops and a new resistant virus emerges. Apparently, the virus always stays one step ahead of the evolving neutralizing antibody response. Despite the continual diversification of antibody responses and virus as infection progresses, serum antibodies from each twin “stayed in step” with the virus from the other twin, suggesting that evolution of the viral envelope was synchronized in the two twins. This view was confirmed in general terms by sequencing of the gene that encodes the virus envelope.
A strength of the Draenert cohort (1) is its inclusion of a brother that shared a single HLA class I haplotype with his twin siblings. This brother shared needles with the twins and was infected with HIV about a year after the twins had become infected. The authors investigated a panel of CTL responses restricted by the allele HLA-B*4001, as this allele was shared among all three siblings. All mounted an immunodominant response against an identical epitope in the HIV Pol protein, but the magnitude and ontogeny of other responses restricted by HLA-B*4001 differed. This finding suggests that the magnitude and breadth of the HIV-specific CTL response restricted by a single HLA allele can be profoundly influenced by the presence or absence of other HLA alleles.
CTL divergence later in infection and the importance of escapology
A puzzle that remains unanswered is why the twins' highly concordant CTL responses early on ultimately became less uniform as the infection progressed. HIV sequence evolution may drive this diversification. In the twins studied by Draenert et al. (1), four CTL responses weakened in both brothers as the infection progressed. At the same time, mutations consistent with immune escape arose in three of the epitopes recognized by these CTL. Moreover, four CTL responses persisted in one twin and declined in the other. The reduced recognition of all four epitopes was accompanied by viral sequence changes that varied between the twins.
HIV mutation is a stochastic process and, therefore, CTL escape cannot be predicted absolutely. Certainly, some CTL responses consistently select for escape variants rapidly (6), but others do not (7). And for each epitope the pattern of escape mutations is limited but rarely completely uniform (8). Stochasticity in the timing and pattern of CTL escape may be powering a destabilizing “variability engine.” Once immune escape mutants begin to accrue, the antigenic composition of the virus changes, fundamentally altering the immune response. Recent studies show that new CTL responses emerge against escape mutants (9, 10), whereas CTL raised against wild-type epitope sequences often retain partial activity against variant epitope sequences. The adaptation of the immune response to immune escape mutants may promote further viral diversification, essentially creating a positive feedback loop that intimately tailors HIV to each host's immune response. Data from the paper by Yang et al. (2) supports this hypothesis: by 17 years after infection, all but one epitope sequence differed between the twins, even in the subset of epitopes that were recognized by both.
Despite the diversification of CD8+ T cell responses and sequence evolution after long-term infection and escape, markers of disease progression remained consistent. This could imply that CD8+ T cell responses are a relatively minor determinant of long-term HIV control and that the questions aforementioned are largely academic. Alternatively, the long-term prognosis of infection may be largely dictated by immunological and virological events that occur early during infection when immune responses and primary escape mutations are virtually identical.
Implications for vaccine design: immunogens that retaliate first
How do these findings inform HIV vaccine development? The enormous sequence heterogeneity of HIV is often cited as a chief stumbling block to the development of a vaccine. To put this diversity in context, the global sequence diversity of influenza, another variable RNA virus, during a pandemic is lower than the diversity of viruses observed within a single HIV-infected individual (11). Developing a vaccine to cope with this enormous diversity is a Herculean challenge, though the results of Draenert et al. (1) and Yang et al. (2) suggest that HIV variability may be a more manageable challenge than previously thought. Their findings show that it is the dominant sequences in each host that dictate the immune responses raised against the virus, and immune selection that consistently drives the turnover of the dominant viral population. Global HIV sequence heterogeneity (defined by sequencing the virus in a cross section of the infected population), therefore, may be little more than the aggregate effect of immune selection during millions of HIV person-years. If true, this raises the hope that the mutational history of HIV can be deciphered and used to understand the interplay of immune responses and viral sequence evolution in different populations and over time.
Importantly, the papers by Draenert et al. (1) and Yang et al. (2) suggest that there is a window in HIV infection, albeit a relatively short one, during which immune responses, viral evolution, and disease progression are relatively reproducible. The reproducibility observed in the paper by Draenert et al. raises the exciting possibility that the early events of HIV infection could, in fact, be predicted from preexisting genetic data. Such an analysis may highlight key indicators for disease progression and guide vaccine design. In particular, it may be possible to “corner” the virus with immunogens that are designed to control the virus not only in its current guise but in guises that it is predicted to adopt. Generating predictions would likely require orders of magnitude more data than is currently available, as well as an enormous amount of raw supercomputing power, but could prove well worth the effort. The stochasticity imparted by variable CTL escape and TCR utilization will likely limit the value of such models to predict the entire natural history of HIV infection. Nonetheless, these papers alter the landscape of HIV research, providing some order to a disease that is largely known for its variability.
References
- 1.Draenert, R., T.M. Allen, Y. Liu, T. Wrin, C. Chappey, C. Verrill, G. Sirera, R.L. Eldridge, M.P. Lahaie, L. Ruiz, B. Clotet, C.J. Petropoulos, B.D. Walker, and J. Martinez-Picado. Constraints on HIV-1 evolution and immunodominance revealed in monozygotic adult twins infected with the same virus. J. Exp. Med. 203:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang, O.O., J. Church, C.M. Kitchen, R. Kilpatrick, A. Ali, Y. Geng, M.S. Killian, R.L. Sabado, H. Ng, J. Suen, et al. 2005. Genetic and stochastic influences on the interaction of human immunodeficiency virus type 1 and cytotoxic T lymphocytes in identical twins. J. Virol. 79:15368–15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer-Olson, D., K.W. Brady, M.T. Bartman, K.M. O'Sullivan, B.C. Simons, J.A. Conrad, C.B. Duncan, S. Lorey, A. Siddique, R.A. Draenert, et al. 2005. Fluctuations of functionally distinct CD8+ T cell clonotypes demonstrate flexibility of the HIV-specific TCR repertoire. Blood. 10.1182/blood-2005-04-1636. [DOI] [PMC free article] [PubMed]
- 4.Richman, D.D., T. Wrin, S.J. Little, and C.J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA. 100:4144–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei, X., J.M. Decker, S. Wang, H. Hui, J.C. Kappes, X. Wu, J.F. Salazar-Gonzalez, M.G. Salazar, J.M. Kilby, M.S. Saag, et al. 2003. Antibody neutralization and escape by HIV- 1. Nature. 422:307–312. [DOI] [PubMed] [Google Scholar]
- 6.Price, D.A., P.J. Goulder, P. Klenerman, A.K. Sewell, P.J. Easterbrook, M. Troop, C.R. Bangham, and R.E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA. 94:1890–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koibuchi, T., T.M. Allen, M. Lichterfeld, S.K. Mui, K.M. O'Sullivan, A. Trocha, S.A. Kalams, R.P. Johnson, and B.D. Walker. 2005. Limited sequence evolution within persistently targeted CD8 epitopes in chronic human immunodeficiency virus type 1 infection. J. Virol. 79:8171–8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen, T.M., M. Altfeld, S.C. Geer, E.T. Kalife, C. Moore, K.M. O'sullivan, I. Desouza, M.E. Feeney, R.L. Eldridge, E.L. Maier, et al. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 79:13239–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen, T.M., X.G. Yu, E.T. Kalife, L.L. Reyor, M. Lichterfeld, M. John, M. Cheng, R.L. Allgaier, S. Mui, N. Frahm, et al. 2005. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J. Virol. 79:12952–12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feeney, M.E., Y. Tang, K. Pfafferott, K.A. Roosevelt, R. Draenert, A. Trocha, X.G. Yu, C. Verrill, T. Allen, C. Moore, et al. 2005. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J. Immunol. 174:7524–7530. [DOI] [PubMed] [Google Scholar]
- 11.Korber, B., B. Gaschen, K. Yusim, R. Thakallapally, C. Kesmir, and V. Detours. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19–42. [DOI] [PubMed] [Google Scholar]