Abstract
Suppressor of cytokine signaling (SOCS)3 is a major negative feedback regulator of signal transducer and activator of transcription (STAT)3-activating cytokines. Transgenic mouse studies indicate that high levels of SOCS3 in T cells result in type 2 T helper cell (Th2) skewing and lead to hypersensitivity to allergic diseases. To define the physiological roles of SOCS3 in T cells, we generated T cell–specific SOCS3 conditional knockout mice. We found that the mice lacking SOCS3 in T cells showed reduced immune responses not only to ovalbumin-induced airway hyperresponsiveness but also to Leishmania major infection. In vitro, SOCS3-deficient CD4+ T cells produced more transforming growth factor (TGF)-β1 and interleukin (IL)-10, but less IL-4 than control T cells, suggesting preferential Th3-like differentiation. We found that STAT3 positively regulates TGF-β1 promoter activity depending on the potential STAT3 binding sites. Furthermore, chromatin immunoprecipitation assay revealed that more STAT3 was recruited to the TGF-β1 promoter in SOCS3-deficient T cells than in control T cells. The activated STAT3 enhanced TGF-β1 and IL-10 expression in T cells, whereas the dominant-negative form of STAT3 suppressed these. From these findings, we propose that SOCS3 regulates the production of the immunoregulatory cytokines TGF-β1 and IL-10 through modulating STAT3 activation.
Cytokines play essential roles in the control of immune systems; they not only act as growth factors but also regulate the differentiation, maintenance, and activation of naive, effector, and memory state of immune cells. Their cytoplasmic signal transduction pathways are well defined. Upon binding of cytokines to their receptors and subsequent receptor dimerization, receptor-associated JAKs become activated and phosphorylate tyrosine residues in the cytoplasmic domains of receptors, which serve as the binding sites for Src homology 2 (SH2) domain of STAT molecules. After phosphorylation of STATs by JAKs, STATs dimerize and translocate into the nucleus to induce transcription of cytokine-responsive genes (1, 2).
The cytokine milieu and their intracellular signaling molecules are also involved in naive CD4+ Th differentiation. It is well established that IL-12/STAT4 and IL-4/STAT6 are necessary for Th1 and Th2 differentiation, respectively. In addition, IFN-γ–STAT1 pathway is also necessary for Th1 differentiation (3, 4). The molecular mechanism for generating Th3 regulatory cells, which is a unique Th cell subset that primarily secretes TGF-β1, is poorly understood. TGF-β1 secreted from Th3 cells provides help for IgA induction and has suppressive properties for both Th1 and Th2 cells (5, 6). Because TGF-β1 KO mice exhibited severe multiorgan inflammations (7, 8), TGF-β1 has been thought to be an important immune regulatory cytokine. TGF-β1 is also suggested to be involved in the regulatory function of CD4+ CD25+ regulatory T cells (9, 10), though the molecular mechanism of TGF-β1 induction in such regulatory-type T cells remains to be elucidated. Because production of TGF-β1 is greatly enhanced by IL-4 and IL-10 in Th cells, while suppressed by IFN-γ (11), cytokine signals may play critical roles in the induction and regulation of TGF-β1 production.
In the physiologic condition as well as in pathological conditions, functions of cytokines are strictly controlled. Cytokine signaling pathways are negatively regulated by the family of proteins called suppressors of cytokine signaling (SOCSs), which are characterized by the presence of an SH2 domain and a COOH terminal conserved domain termed the SOCS-box. Several reports have indicated that SOCS proteins are necessary for regulation of normal immune responses (12). Among them, SOCS3, which associates with the tyrosine kinase Lck, calcineurin, and CD28, has been shown to inhibit IL-2 production during T cell activation (13–16). During Th differentiation, SOCS3 is selectively expressed in Th2 cells, whereas SOCS1 expression is higher in Th1 than in Th2 cells (17, 18). In the analysis of Lck promoter-driven SOCS3-transgenic mice, the high expression of SOCS3 in Th cells led to skewing to Th2-type differentiation. This is probably because SOCS3 binds to IL-12Rβ2 and inhibits IL-12–mediated STAT4 activation, thereby blocking Th1 development (18, 19). Importantly, SOCS3 levels were high in T cells from allergic disease patients (18). These observations implied that SOCS3 might be crucial for Th cell differentiation and activation. However, as most of these conclusions have been drawn by overexpression studies or in pathological conditions such as asthma and atopy, analysis of SOCS3-deficient mice has been necessary to clarify the physiological function of SOCS3 in T cells more precisely. Because mice lacking SOCS3 die during embryogenesis as the result of a placental defect by an enhanced activation of the leukemia inhibitory factor (LIF) signaling pathway (20, 21), we generated T cell–specific SOCS3-deficient (conditional KO [cKO]) mice by a conditional gene targeting approach using Cre-loxP system (22). We showed that not only Th2-type responses in OVA-induced asthma model but also immune responses against Leishmania major infection were reduced in cKO mice. In vitro analysis of T cells demonstrated that SOCS3-deficient CD4+ T cells produced more TGF-β1 and IL-10, but less IL-4 than WT T cells, suggesting a preferential Th3-like differentiation. We found that STAT3 bound to the TGF-β1 promoter and elevated the promoter activity, and SOCS3 deletion enhanced STAT3 recruitment to the promoter. It has been shown that STAT3 also binds to the IL-10 promoter and elevates IL-10 gene expression (23). In conclusion, we propose that STAT3 and SOCS3 reciprocally regulate Th cell function and differentiation by controlling the induction of the immunosuppressive cytokine, TGF-β1, and IL-10.
RESULTS
Generation of T cell–specific SOCS3-deficient mice
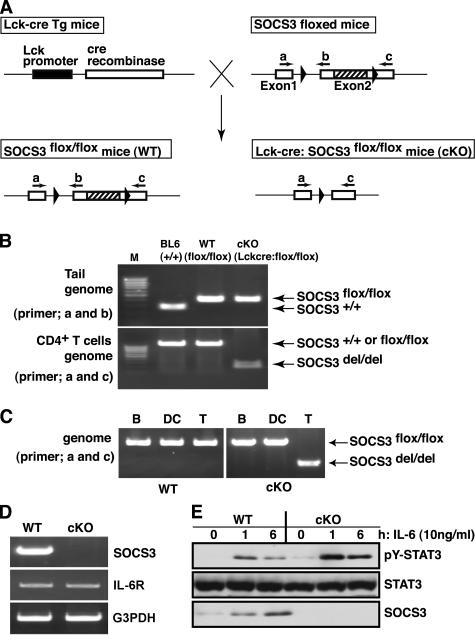
To delete the SOCS3 gene in a T cell–specific manner, proximal p56Lck promoter-cre transgenic mice were crossed with SOCS3-flox/flox mice (22) (Fig. 1 A). Resulting SOCS3-flox/flox: Lck-cre Tg mice are designated as cKO mice. SOCS3 WT alleles, floxed alleles, and the Lck-cre transgene were determined by PCR using genomic DNA from tails and CD4+ T cells. A 380-bp product corresponding to the nondeleted floxed alleles was amplified by primer set of a and b from tail DNA of SOCS3-cKO and WT-flox/flox mice (C57BL/6 showed a 280-bp fragment because of the lack of floxed alleles). An approximately 1.2-kbp fragment was amplified from DNA isolated from CD4+ T cells of WT-flox/flox mice by the primer set of a and c, whereas a 250-bp fragment corresponding to the deleted allele was amplified from SOCS3-cKO mice CD4+ T cells (Fig. 1 B). Deletion of the SOCS3-flox gene was specific to T cells, and no deletion was observed in B cells and DCs in cKO mice (Fig. 1 C). Next, to confirm the deletion of mRNA, we performed RT-PCR analysis in IL-6–stimulated splenic CD4+ T cells from SOCS3-cKO mice. Although SOCS3 was induced after IL-6 stimulation in WT CD4+ T cells and the expression of IL-6 receptor was at an almost equal level, SOCS3 mRNA was undetectable in CD4+ T cells from SOCS3-cKO mice (Fig. 1 D). Western blotting analysis using antibody specific for SOCS3 also confirmed the absence of SOCS3 protein in CD4+ T cells from SOCS3-cKO mice (Fig. 1 E). Thus, we concluded that cre-mediated deletion of SOCS3 occurred efficiently and specifically in T cells in SOCS3-cKO mice.
Figure 1.
Generation of T cell–specific SOCS3-deficient mice. (A) Schema of SOCS3 floxed and deleted loci. Exon 2 was flanked by two LoxP sites (arrowheads). (B) PCR genotyping of floxed alleles using the primer set of a and b against tail genome and deleted alleles using the primer set of a and c against genomic DNA of CD4+ T cells from indicated mice. (C) PCR detection of undeleted and deleted floxed alleles using primer set a and c against genomic DNA from B cells, DCs, and CD4+ T cells. (D) RT-PCR analysis for mRNA expression of SOCS3, IL-6R, and glyceraldehydes-3-phoshate dehydrogenase (G3PDH) in IL-6–stimulated MACS purified splenic CD4+ T cells from cKO and WT mice. (E) Western blotting analysis for SOCS3 and phosphorylated STAT3 in splenic CD4+ T cells.
We examined IL-6–mediated STAT3 activation in SOCS3-deficient T cells. As shown in Fig. 1 E, IL-6–induced STAT3 activation was enhanced and prolonged in SOCS3-deficient CD4+ T cells. This confirmed a negative regulatory function of SOCS3 for the gp130–STAT3 pathway. As in macrophages (22), SOCS3 deficiency in T cells did not much affect IL-10–induced STAT3 activation (unpublished data).
Next, we examined development of T cells in SOCS3 cKO mice. Total mononuclear cell numbers of lymphoid organs such as thymus, spleen, and lymph nodes in SOCS3-cKO mice were almost the same as those in WT mice. Flow cytometric analysis revealed that the ratio of CD4+ or CD8+ SP cells was not altered in SOCS3 cKO mice, although the numbers of CD4− CD8− DN cells were slightly higher in SOCS3-cKO mice (unpublished data). T cell numbers, the CD4/CD8 ratio, and other T cell markers (TCRβ, CD25, CD69, CD62L) were not altered in the spleen and lymph node of cKO mice (unpublished data). Therefore, we concluded that SOCS3 does not play an essential role in T cell development.
Reduced Th2-type response in SOCS3-cKO mice
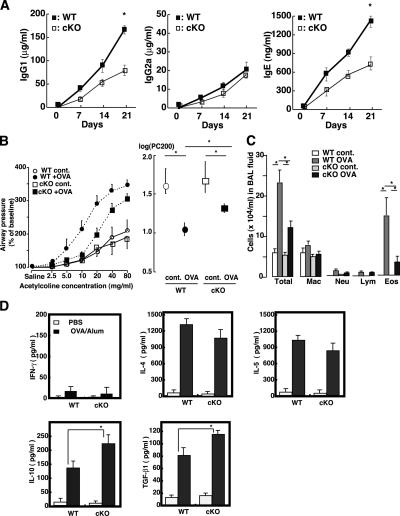
Previously, we reported that constitutive expression of SOCS3 in T cells causes preferential Th2 differentiation of CD4+ T cells, resulting in hyper IgE production and enhanced OVA-induced airway hypersensitiveness (18). Thus, we investigated the effect of SOCS3 deletion in T cells on OVA immunization. After mice were immunized with OVA and alum as an adjuvant on days 1 and 14, we examined the Ig levels and cytokine production. Total IgG1 and IgG2a levels before immunization were almost the same between WT and cKO mice (unpublished data). As shown previously (24), OVA/alum immunization significantly enhanced Th2-mediated Ig (IgG1 and IgE) production (Fig. 2 A). Interestingly, SOCS3-cKO mice produced lower levels of IgG1 and IgE than WT mice did, although Th1-mediated IgG2a production was similarly low between WT and cKO mice (Fig. 2 A). Reflecting reduced IgE levels, SOCS3-cKO mice exhibited lower sensitivity to airway responsiveness and reduced eosinophil infiltration in BAL fluids in cKO mice after OVA challenge compared with WT mice (Fig. 2, B and C). These data confirmed that SOCS3 levels in Th cells alter type 2 responses in vivo.
Figure 2.
Reduced Th2 responses of SOCS3-deficient T cells in OVA/alum immunized mice. (A) Analyses of serum OVA-specific IgG1, IgG2a, and IgE titers in cKO and WT mice. Plasma samples were taken from mice (n = 5) at indicated days after immunization with OVA/alum on days 0 and 14. Ab titers were measured by ELISA and endpoint analysis. Data indicate mean ± SD. (B) Mice (n = 9 for each group) immunized with OVA/alum were aerosol challenged with OVA. Airway responsiveness was determined by the acetylcholine-dependent change in airway pressure in saline-treated control and OVA-sensitized/challenged WT and SOCS3-cKO mice. Provocative concentration 200 (PC200), the concentration at which airway pressure is 200% of its baseline value. Data indicate mean ± SD. (C) Cell counts in bronchoalveolar lavage fluid. *, P < 0.05 by analysis of variance with Bonferroni correction. Data indicate mean ± SD. (D) Cytokine profiles of Th1 type (IFN-γ), Th2 type (IL-4 and IL-5), and TGF-β1 and IL-10. Splenic CD4+ T cells isolated from OVA-immunized mice were restimulated with or without OVA ex vivo for 48 h. Cytokine levels were determined by ELISA. Data indicate mean ± SD in one representative experiment with five mice per group out of three independent experiments.
Next, cytokine production by OVA restimulation from splenic CD4+ T cells was examined. When CD4+ T cells from OVA/alum-immunized mice were restimulated with OVA in vitro, the Th2-type signature cytokines such as IL-4 and IL-5 were highly produced, whereas in the Th1-type signature cytokine, IFN-γ levels were very low (Fig. 2 D). Levels of these Th2-type cytokines from CD4+ T cells were not significantly different between WT and SOCS3 cKO mice (Fig. 2 D). However, in several separate experiments, we always observed that T cells from cKO mice immunized with OVA/alum produced a slightly lower amount of IL-4 than those from WT mice (Fig. 2 D). We did not observe enhanced IFN-γ production in T cells from SOCS3-cKO mice, suggesting that loss of SOCS3 in Th cells did not enforce Th1 skewing. In contrast with Th2-type cytokines, TGF-β1 and IL-10 levels were higher in SOCS3-cKO mice than in WT mice (Fig. 2 D). We also confirmed high mRNA expression levels of TGF-β1 and IL-10 by RT-PCR (unpublished data). During in vitro restimulation, no significant difference in the proliferation was observed between WT and SOCS3-deficient T cells (unpublished data). These results indicate that loss of SOCS3 expression in T cells resulted in lower Th2-type immune responses, which was accompanied with reduced IL-4 levels; however, production of TGF-β1 and IL-10, but not IFN-γ, was enhanced.
Altered immune responses to L. major infection in SOCS3-cKO mice
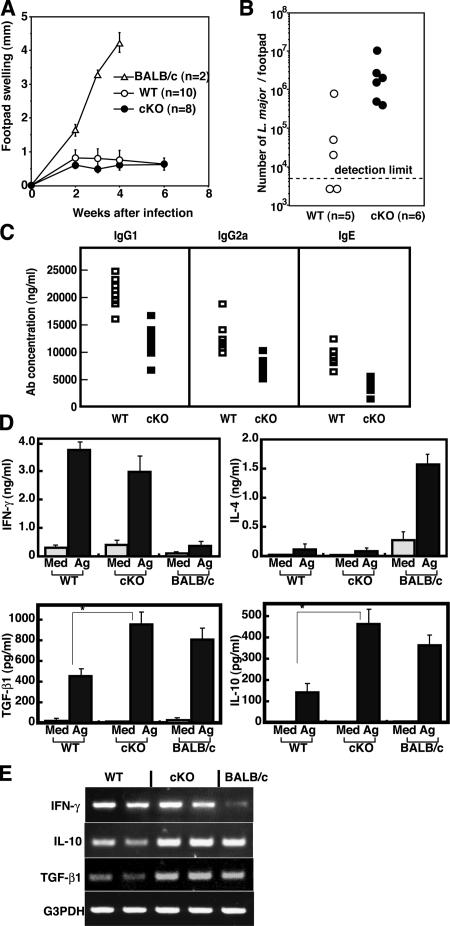
To further determine the role of SOCS3 in Th cell differentiation and function, we compared the immune responses against L. major, the intracellular protozoan parasite infection, the resolution of which is strictly dependent on Th1-type immune responses (25, 26). Because genetic background of both WT and cKO mice was 129 × C57BL/6 mixed, these mice are generally expected to be resistant to L. major compared with susceptible BALB/c mice. Mice were infected subcutaneously in the right hind footpad with 107 L. major promastigotes, and lesion development was monitored for 8 wk. Both WT and cKO mice were resistant to L. major, which was judged by footpad swelling (Fig. 3 A). However, the number of parasites remaining in the lesion at 6 wk after infection was higher in cKO mice than in WT mice (Fig. 3 B). Moreover, serum antibody levels after L. major infection were lower in cKO mice than in WT mice after infection (Fig. 3 C), suggesting that immune responses against L. major were reduced in cKO mice.
Figure 3.
Reduced Th1 responses of SOCS3-deficient T cells in L. major infection. (A) Footpad swelling after L. major infection. BALB/c (a susceptible strain), WT, and SOCS3-cKO mice were inoculated in the right hind footpad with L. major promastigotes and the size of the footpad lesion was monitored. Data shown are mean ± SD and are representative of three independent experiments. BALB/c mice were killed at 4 wk for ethical reasons. (B) The number of parasites remaining in the footpads 6 wk after infection. (C) Serum IgG1, IgG2a, and IgE levels against L. major antigen in infected mice. Samples were collected from WT (open squares) and cKO (closed squares) mice 4 wk after infection. Total IgG1, IgG2a, and IgE titers were determined by ELISA. (D) Cytokine production by CD4+ T cells of the right popliteal LN from WT and cKO mice 4 wk after L. major infection. CD4+ T cells were cultured with irradiated naive WT splenocytes with (black bar) or without (gray bar) L. major antigen for 70 h. Concentrations of IFN-γ, IL-4, IL-10, and TGF-β1 in the culture supernatant were measured by ELISA. Data indicate mean ± SD of triplicate samples from five mice per group in one representative experiment out of three independent experiments. (*, P < 0.01). (E) IFN-γ, IL-10, and TGF-β1 mRNA levels determined by RT-PCR using total RNA from CD4+ right popliteal LN 4 wk after L. major infection.
We examined cytokine production in CD4+ T cells from right popliteal LN at 4 wk after L. major infection (Fig. 3 D). After in vitro stimulation with L. major antigen, the IFN-γ level from SOCS3-deficient CD4+ T cells was comparable to that of WT CD4+ T cells, suggesting that effective Th1 differentiation occurred in cKO mice. IL-4 levels were too low to compare between WT and cKO mice. Next, we measured IL-10 and TGF-β1 levels in the same supernatant. As shown in Fig. 3 D, enhanced production of IL-10 and TGF-β1 was observed in CD4+ T cells from SOCS3-cKO mice. Similar higher expression of IL-10 and TGF-β1 in the CD4+ T cells of SOCS3-cKO mice than in WT T cells was confirmed by RT-PCR (Fig. 3 E). These data support our notion that SOCS3-deficient T cells possess higher potential to produce IL-10 and TGF-β1 than WT T cells.
Cytokine production from in vitro–differentiated SOCS3-deficient T cells
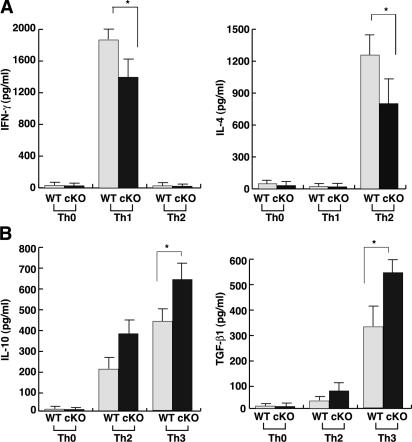
To elucidate the reason why IL-10 and TGF-β1 were elevated in CD4+ T cells from SOCS3-cKO mice, we analyzed in vitro Th cell differentiation. Purified CD4+ T cells were stimulated under Th0, Th1, and Th2 skewing conditions for 7 d and restimulated with plate-bound anti-CD3ɛ and anti-CD28 antibodies. TCR-mediated tyrosine phosphorylation of cellular proteins and ERK activation in T cells were not significantly altered in cKO mice (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20052333/DC1). Furthermore, there was no significant difference in proliferation between WT and SOCS3-deficient CD4+ T cells after TCR stimulation (unpublished data). Neither IFN-γ nor IL-4 was detected in the culture supernatant of both SOCS3-deficient and WT T cells under Th0 conditions. Under Th1 differentiating condition, the IFN-γ level was slightly reduced in SOCS3-deficient T cells compared with WT T cells (Fig. 4 A). In Th2 differentiating condition, IL-4 from SOCS3-deficient T cells was also significantly lower than that of WT T cells (Fig. 4 A). In contrast, IL-10 and TGF-β1 levels were higher in SOCS3-deficient CD4+ T cells than in WT CD4+ T cells (Fig. 4 B). In Th0 and Th1 conditions, IL-10 and TGF-β1 levels were very low in both WT and SOCS3-deficient T cells (Fig. 4 B and not depicted).
Figure 4.
Cytokine production from in vitro–differentiated CD4+ T cells. IFN-γ and IL-4 production from in vitro–differentiated Th0/Th1/Th2 cells (A), and IL-10 and TGF-β1 production from Th0/Th2/Th3 cells (B). Naive CD4+ T cells were cultured under various differentiation conditions for 7 d as described in Materials and methods. After restimulation with anti-CD3ɛ mAb and anti-CD28 mAb for 24 h for IFN-γ, and with IL-4 and IL-10 for 72 h for TGF-β1, culture supernatants were collected and analyzed by ELISA. Data indicate mean ± SD of triplicate cultures in one representative experiment out of three independent experiments. *, P < 0.01.
We next examined Th3 differentiation, which has been induced in vitro by culturing CD4+ T cells in the presence of IL-4, IL-10, and TGF-β1 (11, 27). As previously described, TGF-β1 levels were especially enhanced in the Th3 condition compared with the Th2 condition (Fig. 4 B). Under this Th3 condition, SOCS3-deficient CD4+ T cells produced higher levels of IL-10 and TGF-β1 than WT CD4+ T cells (Fig. 4 B). Collectively, SOCS3 deficiency caused enhanced production of TGF-β1 and IL-10, but reduced production of IL-4 in CD4+ T cells not only in vivo but also in vitro.
STAT3 elevates TGF-β1 promoter activity
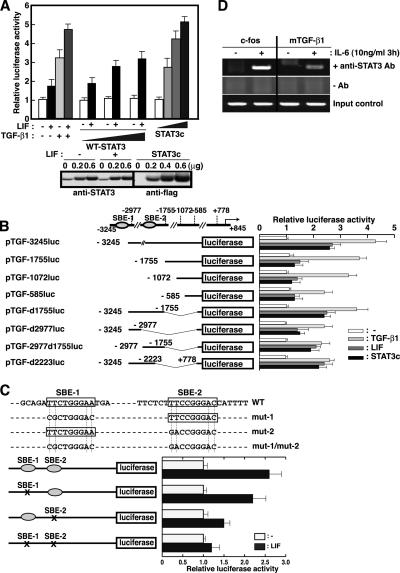
The inhibitory effect of SOCS3 is relatively specific to STAT3 among six STATs. Therefore, we next investigated whether STAT3 could directly regulate the TGF-β1 promoter activity. The 4.1-kb fragment of the 5′-flanking region of the murine TGF-β1 gene was fused to the luciferase expression vector, and promoter activity was examined in HEK293 cells by transient transfection. Luciferase gene expression was induced not only by high glucose and TGF-β1 itself as described previously (28) but also by LIF, suggesting that this 4.1-kb 5′-fragment of the TGF-β1 gene contained STAT3 responsive elements (Fig. 5 A). Co-expression of exogenous WT STAT3 enhanced LIF-mediated TGF-β1 promoter activity in a dose-dependent manner (Fig. 5 A). Furthermore, constitutive active form of STAT3 (STAT3c) (29) also enhanced TGF-β1 promoter activity similar to LIF stimulation (Fig. 5, A and B), suggesting that STAT3 positively regulates TGF-β1 promoter. We also confirmed that STAT3 elevated TGF-β1 promoter activity in the lymphoid cell line by using Jurkat cell (unpublished data).
Figure 5.
STAT3 directly enhances TGF-β1 promoter activity. (A) HEK293 cells were transfected with 0.2 μg of pTGF4.1-luc, a reporter gene containing ∼4.1 kb TGF-β1 promoter region, and 0.1 μg of β-galactosidase expression vector (β-gal) together with 0, 0.2, 0.6 μg WT-STAT3 or STAT3c expression vector. 1 d after transfection, cells were stimulated with 10 ng/ml LIF or 10 ng/ml TGF-β1 and luciferase activity was measured after 8 h. Luciferase activities were normalized by β-gal activities and expressed as fold induction to control cultures defined as 1.0. STAT3 expression levels determined by Western blotting analysis as shown (bottom). (B) Localization of the STAT3 responsive elements in the TGF-β1 promoter. HEK293 cells were transiently transfected with 0.2 μg of plasmid containing various fragments of TGF-β1 promoter region and 0.1 μg of β-gal expression vector. 1 d after transfection, cells were stimulated with 10 ng/ml LIF or 10 ng/ml TGF-β1 for 8 h and harvested. (C) Effects of point mutations introduced into the SBE-1 and SBE-2 elements. HEK293 cells were transiently transfected with WT or mutant pTGF4.1-luc plasmids and β-gal plasmid. Cells were stimulated with 10 ng/ml LIF and luciferase activities were measured. Luciferase activities were normalized by β-gal activities and expressed as fold induction to control cultures defined as 1.0. (D) ChIP assay was performed using chromatin from WT CD4+ T cells treated with IL-6 for 3 h and immunoprecipitated with antibody against STAT3. The final DNA extractions were amplified using pairs of primers that cover the STAT3 binding site (SBE-1) in the TGF-β1 or c-fos promoter region. G3PDH levels were determined by PCR using samples before immunoprecipitation as input control. Luciferase activities normalized by β-gal activity are shown as the means ± SD of three to five experiments.
As shown in Fig. 5 B, a reporter assay using a series of 5′-deletion mutants revealed that the LIF-responsive elements were present upstream of −1755. By searching for potential STAT3-binding sites with the consensus sequences, TTC/A(N)3G/TAA (30), two candidates of STAT3-binding sites were identified in the 4.1-kb TGF-β1 5′-flanking region. The two sites were at positions −3155 and −2515 upstream of the transcription initiation site in the TGF-β1 promoters designated STAT3-binding element (SBE)-1 and SBE-2, respectively. To determine the significance of these elements, mutations were introduced into the SBE-1 and/or SBE-2 sites. A mutant promoter lacking both SBE-1 and SBE-2 did not respond to LIF stimulation anymore, whereas constructs containing a single SBE site still responded to STAT3 (Fig. 5, B and C). These results indicate that the two SBE sites of the TGF-β1 promoter are important for STAT3-dependent activation.
To confirm STAT3 binding to the TGF-β1 promoter in T cells, chromatin immunoprecipitation (ChIP) assay was performed (Fig. 5 D). The chromatin–DNA complex was immunoprecipitated with anti-STAT3 antibody; then, STAT3 binding to the TGF-β1 promoter was analyzed using pairs of specific primers spanning the STAT3 binding sites. The SBE site of the c-fos promoter was used as a positive control of STAT3 recruitment (31). As shown in Fig. 5 D, STAT3 was actually bound to the TGF-β1 promoter region containing SBE-1 site in T cells in an IL-6–dependent manner. These data indicate that TGF-β1 is a direct downstream target of STAT3.
STAT3 positively regulates TGF-β1 and IL-10 induction in CD4+ T cells
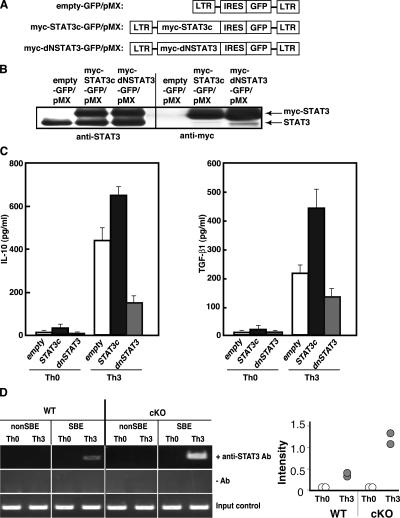
To address whether STAT3 is critical for IL-10 and TGF-β1 production in CD4+ T cells, we introduced a constitutive active form of STAT3 (STAT3c) or a dominant negative STAT3 (dNSTAT3) (32) into CD4+ T cells using a bicistronic retroviral vector pMX carrying IRES-GFP (33) (Fig. 6 A). The retrovirus vectors were infected into nonpolarized CD4+ T cells, which were stimulated with plate-bound anti-CD3ɛ mAb and anti-CD28 mAb, and on day 4, GFP-positive cells were sorted. The expression of myc-STAT3 in the sorted GFP-positive CD4+ T cells was confirmed by Western blotting (Fig. 6 B). These cells were cultured under Th0 or Th3 differentiating condition for 7 d and analyzed for cytokine production upon restimulation. We found that introduction of STAT3c into CD4+ T cells resulted in higher TGF-β1 and IL-10 production (Fig. 6 C). In contrast, dNSTAT3-GFP introduced to CD4+ T cells showed less production of TGF-β1 and IL-10 (Fig. 6 C). These data indicate that STAT3 activation is positively involved in the production of TGF-β1 and IL-10 in CD4+ T cells.
Figure 6.
Retroviral transduction of STAT3 mutants modulates TGF-β1 and IL-10 production. (A) Schematic structure of the retroviral pMX vectors containing mutant STAT3, either myc-STAT3c (constitutive active form) or myc-dNSTAT3 (dominant negative form). (B) GFP-positive cells were sorted from infected T cells and the expression levels of exogeneous myc-STAT3 were examined by Western blotting. (C) IL-10 and TGFβ1 production from infected CD4+ T cells after Th3 differentiation. GFP-positive cells were cultured in the presence of IL-4, IL-10, and TGF-β1 for 7 d and restimulated with anti-CD3ɛ mAb and anti-CD28 mAb and cytokines in the culture supernatants were measured by ELISA. Data shown are mean ± SD of triplicate samples from four independent experiments. (D) ChIP assay to compare STAT3 recruitment to TGF-β1 promoter (SBE-1 site) between Th0 and Th3 differentiated T cells from WT and cKO mice. Anti-STAT3 Ab immunoprecipitates were used as templates for PCR cells. A non-SBE region near the transcription initiation sites was amplified as a negative control. Ratios of the bands intensity of SBE-1 PCR products and those of control (G3PDH) in two independent experiments are plotted (right).
Next, STAT3 recruitment to the TGF-β1 promoter was compared between WT and SOCS3-deficient CD4+ T cells using the ChIP assay. As shown in Fig. 6 D, STAT3 was recruited to the TGF-β1 promoter region under the Th3, but not Th0, differentiating conditions. Importantly, more STAT3 was recruited to the TGF-β1 promoter in SOCS3-deficient CD4+ T cells than in WT CD4+ T cells under the Th3 differentiating condition. These data suggest that SOCS3 probably regulates the production of TGF-β1 through appropriate tuning of STAT3 activation in CD4+ T cells.
DISCUSSION
Previously, we reported that forced expression of SOCS3 in T cells resulted in Th2 skewing. SOCS3 expression levels are high in T cells from patients with asthma and atopy. Therefore, we concluded that high SOCS3 levels are related to pathological conditions, especially Th2-type diseases (18). However, the role of SOCS3 in physiological conditions has not been clarified. Here, we generated T cell–specific SOCS3-cKO mice and found that the Th2 immune responses in SOCS3-cKO mice were actually reduced. However, this is not the result of higher Th1 responses. Our SOCS3-deficient CD4+ T cells showed higher TGF-β1 and IL-10 production compared with control WT CD4+ T cells. Thus, we suspect that reduced Th2 responses in SOCS3-cKO mice may be the result of immunosuppression by these two immunoregulatory cytokines.
We proposed that SOCS3 inhibits Th1 differentiation by suppressing IL-12–mediated signaling (18). We found that IL-12–induced STAT4 phosphorylation was actually enhanced in SOCS3-deficient T cells compared with WT T cells (unpublished data). However, similar or only slightly reduced IFN-γ production occurred in CD4+ T cells from SOCS3 cKO mice compared with WT mice (Figs. 3 D and 4 A). Furthermore, delay of parasite clearance and reduced production of antibodies were observed in cKO mice during L. major infection (Fig. 3, B and C). This may be the result of immunosuppressive effect of TGF-β1 and IL-10 produced from T cells during infection. Regulatory roles of SOCS3-deficient T cells in other immune reactions should be defined in future studies.
Recently, regulatory functions of Th cells have been extensively studied. CD4+ CD25+ regulatory T (T reg) cells are recognized as naturally occurring T reg cells and exhibit immunosuppressive abilities by a mechanism that is dependent on cell-to-cell contact through the interaction of CTLA-4 with CD86 (34). Though TGF-β1 is shown to be one of the mechanisms of the immunosuppressive effects of T reg cells (10) and Foxp3 has been shown to be an essential transcription factor in the generation and function of T reg cells (35), we did not find any change in the number of CD4+ CD25+ T reg population or Foxp3 expression between SOCS3-deficient and WT T cells (unpublished data). However, regulatory function of SOCS3-deficient T reg cells remains to be investigated.
Previous studies have identified another subset of T reg cells; Tr1 cells (T reg cell 1), which are induced in vitro by repeated antigen stimulation of T cells in the presence of IL-10 (36, 37). Tr1 cells produce high levels of IL-10 rather than TGF-β1 (38). The additional subset of T reg cells is Th3, which is induced by orally administered antigens. Th3 cells can be distinguished from Th2 cells by cytokine profiles; Th3 cells produce much more TGF-β1 and IL-10 and less IL-4 than Th2 cells (11, 39). Because SOCS3-deficient CD4+ T cells produce more TGF-β1 and IL-10, they are more likely to exhibit a Th3-like phenotype.
In this study, we demonstrated that STAT3 directly binds to the promoter region of TGF-β1 and elevates TGF-β1 production in T cells. It has already been shown that IL-10 is up-regulated by STAT3 (23). We showed that constitutive active form of STAT3 enhanced TGF-β1 and IL-10 production in T cells. Furthermore, we showed that a dominant negative STAT3 suppressed TGF-β1 production (Fig. 6 C). Therefore, STAT3 could be a positive regulator of Th3-type differentiation. STAT3 being required for Th3 is unlike STAT4 and STAT6 being required for Th1 and Th2, respectively, because basal transcription of TGF-β1 and IL-10 is not completely dependent on STAT3. However, STAT3 is an important regulatory factor for Th3 differentiation because STAT3 is essential for the immunosuppressive function of IL-10 in macrophages (40) and IL-10 is usually necessary for induction of Th3 in vitro. Collectively, STAT3 seems to positively regulate induction and/or differentiation of Th3.
A question that remains unsolved is what kind of cytokines are regulated by SOCS3 during Th3-like phenotype induction. Previously, IL-4 has been shown to induce SOCS3 expression in Th2 cells (17). However, IL-4–induced STAT6 phosphorylation levels were not affected in SOCS3-deficient T cells (unpublished data). Therefore, it is unlikely that SOCS3 directly regulates IL-4 signaling. Because STAT3 is strongly activated by IL-10, we compared IL-10–induced STAT3 activation between WT and SOCS3-deficient T cells. In SOCS3-deficient T cells, however, IL-10–mediated STAT3 activation was not much affected (unpublished data). This is probably because SOCS3 does not bind to the IL-10 receptor (22). In contrast, we observed stronger and prolonged STAT3 activation in response to IL-6 and IL-27 in SOCS3-deficient T cells (Fig. 1 E and not depicted). Furthermore, STAT3 recruitment to the TGF-β1 promoter under Th3 differentiation condition was enhanced in SOCS3-deficient CD4+ T cells. Although we could not conclude that IL-6 is responsible for the Th3-like phenotype of SOCS3-deficient CD4+ T cells, these results suggest that STAT3 is hyperactivated in SOCS3-deficient T cells during T cell differentiation, and this is the result of the hypersensitivity to autocrine or paracrine cytokines that activate STAT3. Identification of these cytokines other than IL-10, which modulate TGF-β1 and IL-10 production, will be important for understanding of the regulation of Th3 differentiation.
Another possibility for answering the unsolved question is that SOCS3 affects TCR signaling. SOCS3 has been shown to be able to interact with tyrosine kinase Lck, calcineurin, and CD28 (13–16). The level of SOCS3 expression is significantly high in resting CD4+ T cells and rapidly decreased after TCR stimulation (unpublished data). Some reports have shown that the strength of TCR stimulation is an important factor for Th differentiation. Although we could not detect apparent differences in proliferation, tyrosine phosphorylation of cellular proteins, and ERK activation between SOCS3-deficient and WT T cells in response to TCR stimulation (Fig. S1 and not depicted), the absence of SOCS3 in naive CD4+ T cells may permit some stronger TCR signalings, which might lead to higher IL-10 and TGF-β1 secretion at an early stage of T cell activation, thereby leading to large differences at later stages of T cell differentiation.
Although the more detailed molecular basis of the hyperproduction of TGF-β1 and IL-10 in SOCS3-deficient T cells has remained elusive, our biochemical analyses suggest that SOCS3 regulates TGF-β1 and IL-10 production by suppressing STAT3 activity. Thus, we propose that STAT3 and SOCS3 reciprocally regulate Th2/Th3 differentiation. Therefore, suppression of SOCS3 expression in T cells may possibly be one of the ways to introduce tolerance for autoimmune diseases or to ameliorate allergic diseases.
MATERIALS AND METHODS
Generation of T cell–specific SOCS3-disrupted mice.
SOCS-3 flox/flox mice (22), and Lck-cre transgenic mice (41) have been described elsewhere. Lck-cre transgenic mice (C57BL/6 background) were bred with SOCS3 flox/flox mice (129 × C57BL/6 background) to generate mice in which SOCS3 was deleted in a T cell–specific manner. Genotypings were performed by PCR as described previously (22). Offspring carrying both lck-cre and floxed SOCS3 genes (Lck-Cre:SOCS3 flox/flox; cKO) and the floxed SOCS3 gene (SOCS3 flox/flox; WT) were used for intercrossing and further analyses. Littermate controls were used for all experiments. CD4+ T cells, splenic B cells, and DCs were isolated by MACS sorting as described previously (42). Mice were kept in specific pathogen-free facilities in the Collaborative Station Animal Facility of Kyushu University. All experiments using these mice were approved by and performed according to the guidelines of the Animal Ethics Committee of Kyushu University, Fukuoka, Japan.
OVA/alum immunization and assay for airway hyperresponsiveness.
Alhydrogel (alum; Al(OH)3gel) (LSL) was mixed with a predetermined quantity of OVA grade V (Sigma-Aldrich) and incubated at room temperature for 20 min. After centrifugation of the mixture at 14,000 g for 10 min, supernatants were used for immunization as described previously (24). Mice (8–12 wk old) were immunized with 0.1 ml of OVA (10 μg) in PBS and absorbed to alum. Boosting inoculations were performed in the same fashion 2 wk later. For airway hyperresponsiveness (AHR) and eosinophil infiltration assay, mice received aerosol challenge containing either saline or 1% OVA for 20 min/d on days 26–28 (18, 43). On day 30, 36 h after the last aerosol challenge, mice were ventilated and AHR to acetylcholine aerosol was measured. Serum levels of total and OVA-specific Ig was analyzed by ELISA with rat anti–mouse Ig (Serotec Ltd.). Ab titers were determined by endpoint analysis. For cytokine assays, splenocytes from immunized mice on day 28 were cultured ex vivo in the presence of OVA. Culture supernatants were harvested after 48 h and analyzed for IL-4, IFN-γ, TGF-β1, and IL-10 by ELISA.
L. major infection and cytokine assay.
Infection of L. major was performed as described previously (44). Mice were infected s.c. in the right footpad lesion with 107 stationary phase of L. major (MHOM/SU/73-5-ASKH). Footpad swelling was monitored weekly by a vernier caliper and compared with the thickness of the uninfected left footpad. 6 wk after infection, the footpad parasite burdens were quantified by homogenizing tissue in 3 ml of medium 199 supplemented with 10% FCS containing 2 mM glutamine, 10 mM Hepes, and 100 μl/ml gentamicin. Aliquots were diluted serially across 96-well plates and scored at 1 wk for the presence of motile promastigotes. 4 wk after L. major infection, CD4+ T cells (5 × 105/200 μl/well) from the right popliteal LN were stimulated with or without L. major antigens (equivalent to 5 × 105 promastigotes) in the presence of irradiated (30 Gy) splenocytes for 70 h. Culture supernatants were collected and analyzed for IL-4, IFN-γ, TGF-β1, and IL-10 by ELISA. Total RNA was prepared from MACS-purified CD4+ T cells of popliteal LN 4 wk after L. major infection, and the expression level of G3PDH was first evaluated as an internal control. The pair of primers for TGF-β1 was forward, 5′-TGACGTCACTGGAGTTGTACGG-3′ and reverse, 5′-GGTTCATGTCATGGATGGTGC-3′. The expression levels of IFN-γ and IL-10 were assessed using appropriate pairs of primers described previously (44).
In vitro T cell differentiation assay.
For in vitro T cell differentiation assays, CD4+ T cells (106 cells/ml) purified by MACS columns (Miltenyi Biotec) from splenocytes after depletion of red blood cells were cultured in RPMI 1640 containing 10% FCS and stimulated with 1 μg/ml of plate-bound anti-CD3ɛ mAb, 1 μg/ml anti-CD28 mAb, and 1 ng/ml mIL-2 (PeproTech) supplemented with anti–IFN-γ and anti–IL-4 antibodies for Th0 differentiation, 10 ng/ml mIL-12 (PeproTech) for Th1 differentiation, 10 ng/ml mIL-4 (PeproTech) for Th2 differentiation, or 10 ng/ml mIL-4, 10 ng/ml mIL-10 (PeproTech), and 10 ng/ml hTGF-β1 (PeproTech) for Th3 differentiation (11). Cells were collected after 7 d and washed, and an equal numbers of viable cells (106 cells/ml) were restimulated with plate-bound anti-CD3ɛ mAb and 1 μg/ml anti-CD28 mAb in the absence of any additional cytokines. Supernatants were collected 24 h later, and the production of cytokines was measured in duplicate by ELISA (Genzyme). For measurement of TGF-β1 production, secondary stimulation was done in a serum-free medium in which Nutridoma SP (Roche Diagnostic) was substituted for FCS. Supernatant was collected 72 h after secondary stimulation for TGF-β1 measurement using mTGF-β1 ELISA kit (Promega).
Construction of reporter plasmids.
PCR was done to generate the TGF-β1 promoter plasmid by using mouse genomic DNA as a template. The nucleotide sequence of the mTGF-β1 promoter has been submitted to GenBankTM/EBI Data Bank under accession no. L42456.1. A 4.1-kb XhoI–EcoRI fragment corresponding to nucleotides from −3245 to +845 relative to the determined transcriptional start site of TGF-β1 gene was subcloned into a pGV-basic2 vector (TOYOINKI), pTGF4.1-luc. Reporter plasmids, including a series of deletion mutants of the TGF-β1 promoter, were generated by excision at restriction enzyme recognition sites as follows: −2977 (NcoI), −1755 (HindIII), −1072 (SmaI), −585 (NcoI). To construct SBE-1mt, SBE-2mt, and SBE-1mt/2mt, point mutations were introduced to the two STAT binding elements (SBE-1/SBE-2) by PCR using the KOD-plus polymerase (TOYOBO) and the sequences are as follows: SBE-1mt, 5′-GCAGACGCTGGGACTGA-3′ and SBE-2mt, 5′-TTCTCTGACCGGGACCATTTT-3′ (mutated sites are underlined). The subcloned PCR products were sequenced to confirm that the products were the authentic promoter fragments.
Transfection and luciferase assay.
HEK293 (105 cells) were seeded on six-well plates, cultured for 24 h, and transfected with various amounts of an expression vector of WT-STAT3-pCDNA3 or STAT3c-pRcCMV along with 0.2 μg of TGF-β1-pGVbasic2 and 0.1 μg of β-galactosidase (β-gal) plasmid by the calcium phosphate coprecipitation method. Some of them were stimulated with LIF (10 ng/ml) or TGF-β1 (10 ng/ml) for 8 h. Cells were harvested in 40 μl lysis buffer. Luciferase assay was performed using a luciferase substrate kit (Promega) and luciferase activity was read in Packard luminometer. Luciferase activity was normalized by the internal control β-gal activity, and shown as the means ± SD of three to five experiments.
ChIP assay.
ChIP assay was performed in 107 mouse T lymphocytes. Cells were fixed with 1% formaldehyde at 37°C for 10 min after IL-6 stimulation as described previously (31). Cells were washed, suspended in SDS lysis buffer, and sonicated for 30-s pulses four times using a sonicator (Bioruptor; Cosmo Bio Co.). Samples were incubated with 5 μg anti-STAT3 antibody (C-20; Santa Cruz Biotechnology, Inc.) overnight at 4°C. After adding salmon sperm DNA and protein A–Agarose Slurry (UBI), the immunoprecipitates were sequentially washed with low-salt buffer, high-salt buffer, LiCl buffer and twice with TE buffer. The DNA–protein complex was eluted into elution by heating at 65°C for 6 h. Proteins were digested by proteinase K and RNA was removed by addition of 10 μg of RNase A. DNA was recovered by extraction with phenol and chloroform and ethanol precipitation and subjected to PCR analysis. To estimate the DNA content in the soluble chromatin samples, DNA was similarly extracted from sonicated samples and used as a template for G3PDH gene amplification. Promoter-specific primers were as follows; mTGF-β1, SBE-1 forward: 5′-TGACTAACGGCACTGAGGAGGCTGC-3′, SBE-1 reverse: 5′-TGGAAACAGGTCTATCTTCTACCTA-3′, which amplify 311-bp fragments flanking the STAT3 binding element. For negative control, 5′ franking region close to the transcription initiation site was amplified by forward: 5′-GTGCCTCCTTGTATCCGCTAAAGCTCTC-3′ and reverse: 5′-ACTACTAAAGCCGGTGACCAACCAAAG-3′. The mouse c-fos primers for the positive control of STAT3-ChIP are forward: 5′-TCTGCCTTTCCCGCCTCCCC-3′ and reverse: 5′-GGCCGTGGAAACCTGCTGAC-3′.
Retroviral constructs and transduction to primary T cells.
The STAT3c-IRES-GFP-pMX, dNSTAT3-IRES-GFP-pMX, and empty GFP-pMX plasmids (a gift from T. Kitamura, Tokyo University, Tokyo, Japan) were transfected into a packaging cell line, Plat-E (33), using FuGENE6 (Roche Diagnostic), and after incubation for 48 h, the culture supernatant were harvested. CD4+ enriched T cells were stimulated with 1 μg/ml anti-CD3ɛ mAb and 1 μg/ml anti-CD28 mAb for 24 h and infected with the viruses by adding the viral containing supernatants in the presence of 0.6 μg/ml polybrene (Sigma-Aldrich). The infected CD4+ T cells were expanded in the medium supplemented with 100 U/ml rIL-2 for 4 d. GFP-positive cells were collected by a cell sorter (EPICS ALTRA; Beckman Coulter) and restimulated with 1 μg/ml anti-CD3ɛ mAb and 1 μg/ml anti-CD28 mAb or Th3-inducing condition for 72 h. Culture supernatants were harvested after 48 h to analyze TGF-β1 and IL-10 production by ELISA.
Online supplemental material.
Fig. S1 shows tyrosine phosphorylation of cellular proteins (anti-pY blot) and ERK activation in CD4+ T cells from WT and cKO mice after TCR stimulation. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20052333/DC1.
Supplemental Material
Acknowledgments
We thank Dr. J. Takeda, Dr. M. Kubo, and Dr. S. Hori for their advice. We also thank Mr. N. Kinoshita, Ms. M. Othsu, Ms. Y. Yamada, Mr. M. Sasaki, and Ms. E. Fujimoto (Technical Support Center, Medical Institute of Bioregulation) and Ms. T. Yoshioka for technical support and Ms. Y. Nishi for manuscript preparation.
This work was supported by special grants-in-aid from the Ministry of Education, Science, Technology, Sports, and Culture of Japan, Yamanouchi Foundation for Research on Metabolic Disorders, Takeda Science Foundation and the Uehara Memorial Foundation. I. K. is supported by Postdoctoral Fellowship from the Japan Society for the Promotion of Science.
The authors have no conflicting financial interests.
Abbreviations used: ChIP, chromatin immunoprecipitation; cKO, conditional KO; LIF, leukemia inhibitory factor; SH2, Src homology 2; SBE, STAT3-binding element; SOCS, suppressor of cytokine signaling.
References
- 1.O'Shea, J.J., M. Gadina, and R.D. Schreiber. 2002. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 109:S121–S131. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, W.S., and D.J. Hilton. 2004. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 22:503–529. [DOI] [PubMed] [Google Scholar]
- 3.Murphy, K.M., and S.L. Reiner. 2002. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2:933–944. [DOI] [PubMed] [Google Scholar]
- 4.Glimcher, L.H., and K.M. Murphy. 2000. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 14:1693–1711. [PubMed] [Google Scholar]
- 5.Weiner, H.L. 2001. Induction and mechanism of action of transforming growth factor-β-secreting Th3 regulatory cells. Immunol. Rev. 182:207–214. [DOI] [PubMed] [Google Scholar]
- 6.Weiner, H.L. 2001. Oral tolerance: immune mechanisms and the generation of Th3-type TGF-β-secreting regulatory cells. Microbes Infect. 3:947–954. [DOI] [PubMed] [Google Scholar]
- 7.Shull, M.M., I. Ormsby, A.B. Kier, S. Pawlowski, R.J. Diebold, M. Yin, R. Allen, C. Sidman, G. Proetzel, and D. Calvin. 1992. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. 359:693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulkarni, A.B., and S. Karlsson. 1993. Transforming growth factor-β1 knockout mice. A mutation in one cytokine gene causes a dramatic inflammatory disease. Am. J. Pathol. 143:3–9. [PMC free article] [PubMed] [Google Scholar]
- 9.O'Garra, A., and P. Vieira. 2004. Regulatory T cells and mechanisms of immune system control. Nat. Med. 10:801–805. [DOI] [PubMed] [Google Scholar]
- 10.Annunziato, F., L. Cosmi, F. Liotta, E. Lazzeri, R. Manetti, V. Vanini, P. Romagnani, E. Maggi, and S. Romagnani. 2002. Phenotype, localization, and mechanism of suppression of CD4+CD25+ human thymocytes. J. Exp. Med. 196:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seder, R.A., T. Marth, M.C. Sieve, W. Strober, J.J. Letterio, A.B. Roberts, and B. Kelsall. 1998. Factors involved in the differentiation of TGF-β-producing cells from naïve CD4 T cells IL-4 and IFN-γ have opposing effects while TGF-β positively regulates its own production. J. Immunol. 160:5719–5728. [PubMed] [Google Scholar]
- 12.Kubo, M., T. Hanada, and A. Yoshimura. 2003. Suppressors of cytokine signaling and immunity. Nat. Immunol. 4:1169–1176. [DOI] [PubMed] [Google Scholar]
- 13.Yu, C.R., R.M. Mahdi, S. Ebong, B.P. Vistica, I. Gery, and C.E. Egwuagu. 2003. Suppressor of cytokine signaling 3 regulates proliferation and activation of T-helper cells. J. Biol. Chem. 278:29752–29759. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee, A., A.S. Banks, M.C. Nawijn, X.P. Chen, and P.B. Rothman. 2002. Cutting edge: suppressor of cytokine signaling 3 inhibits activation of NFATp. J. Immunol. 168:4277–4281. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto, A., Y. Seki, R. Watanabe, K. Hayashi, J.A. Johnston, Y. Harada, R. Abe, A. Yoshimura, and M. Kubo. 2003. A role of suppressor of cytokine signaling 3 (SOCS3/CIS3/SSI3) in CD28-mediated interleukin 2 production. J. Exp. Med. 197:425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuhara, M., H. Sakamoto, A. Matsumoto, R. Suzuki, H. Yasukawa, K. Mitsui, T. Wakioka, S. Tanimura, A. Sasaki, H. Misawa, et al. 1997. Cloning and characterization of novel CIS family genes. Biochem. Biophys. Res. Commun. 239:439–446. [DOI] [PubMed] [Google Scholar]
- 17.Egwuagu, C.E., C.R. Yu, M. Zhang, R.M. Mahdi, S.J. Kim, and I. Gery. 2002. Suppressors of cytokine signaling proteins are differentially expressed in Th1 and Th2 cells: implications for Th cell lineage commitment and maintenance. J. Immunol. 168:3181–3187. [DOI] [PubMed] [Google Scholar]
- 18.Seki, Y., H. Inoue, N. Nagata, K. Hayashi, S. Fukuyama, K. Matsumoto, O. Komine, S. Hamano, K. Himeno, K. Inagaki-Ohara, et al. 2003. SOCS-3 regulates onset and maintenance of T(H)2-mediated allergic responses. Nat. Med. 9:1047–1054. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto, K., M. Yamaguchi, N. Miyasaka, and O. Miura. 2003. SOCS-3 inhibits IL-12-induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor β2 subunit. Biochem. Biophys. Res. Commun. 310:1188–1193. [DOI] [PubMed] [Google Scholar]
- 20.Roberts, A.W., L. Robb, S. Rakar, L. Hartley, L. Cluse, N.A. Nicola, D. Metcalf, D.J. Hilton, and W.S. Alexander. 2001. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proc. Natl. Acad. Sci. USA. 98:9324–9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi, Y., N. Carpino, J.C. Cross, M. Torres, E. Parganas, and J.N. Ihle. 2003. SOCS3: an essential regulator of LIF receptor signaling in trophoblast giant cell differentiation. EMBO J. 22:372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasukawa, H., M. Ohishi, H. Mori, M. Murakami, T. Chinen, D. Aki, T. Hanada, K. Takeda, S. Akira, M. Hoshijima, et al. 2003. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat. Immunol. 4:551–556. [DOI] [PubMed] [Google Scholar]
- 23.Benkhart, E.M., M. Siedlar, A. Wedel, T. Werner, and H.W. Ziegler-Heitbrock. 2000. Role of Stat3 in lipopolysaccharide-induced IL-10 gene expression. J. Immunol. 165:1612–1617. [DOI] [PubMed] [Google Scholar]
- 24.Brewer, J.M., M. Conacher, C.A. Hunter, M. Mohrs, F. Brombacher, and J. Alexander. 1999. Aluminium hydroxide adjuvant initiates strong antigen-specific Th2 responses in the absence of IL-4- or IL-13-mediated signaling. J. Immunol. 163:6448–6454. [PubMed] [Google Scholar]
- 25.Reiner, S.L., and R.M. Locksley. 1995. The regulation of immunity to Leishmania.major. Annu. Rev. Immunol. 13:151–177. [DOI] [PubMed] [Google Scholar]
- 26.Sacks, D., and N. Noben-Trauth. 2002. The immunology of susceptibility and resistance to Leishmania major in mice. Nat. Rev. Immunol. 2:845–858. [DOI] [PubMed] [Google Scholar]
- 27.Inobe, J., A.J. Slavin, Y. Komagata, Y. Chen, L. Liu, and H.L. Weiner. 1998. IL-4 is a differentiation factor for transforming growth factor-β secreting Th3 cells and oral administration of IL-4 enhances oral tolerance in experimental allergic encephalomyelitis. Eur. J. Immunol. 28:2780–2790. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman, B.B., K. Sharma, Y. Zhu, and F.N. Ziyadeh. 1998. Transcriptional activation of transforming growth factor-β1 in mesangial cell culture by high glucose concentration. Kidney Int. 54:1107–1116. [DOI] [PubMed] [Google Scholar]
- 29.Bromberg, J.F., M.H. Wrzeszczynska, G. Devgan, Y. Zhao, R.G. Pestell, C. Albanese, and J.E. Darnell. 1999. Stat3 as an Oncogene. Cell. 98:295–303. [DOI] [PubMed] [Google Scholar]
- 30.Seidel, H.M., L.H. Milocco, P. Lamb, J.E. Darnell, R.B. Stein, and J. Rosen. 1995. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc. Natl. Acad. Sci. USA. 92:3041–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joo, A., H. Aburatani, E. Morii, H. Iba, and A. Yoshimura. 2004. STAT3 and MITF cooperatively induce cellular transformation through upregulation of c-fos expression. Oncogene. 23:726–734. [DOI] [PubMed] [Google Scholar]
- 32.Minami, M., M. Inoue, S. Wei, K. Takeda, M. Matsumoto, T. Kishimoto, and S. Akira. 1996. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc. Natl. Acad. Sci. USA. 93:3963–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066. [DOI] [PubMed] [Google Scholar]
- 34.Read, S., V. Malmstrom, and F. Powrie. 2000. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25+CD4+ regulatory cells that control intestinal inflammation. J. Exp. Med. 192:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaguchi, S. 2004. Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 22:531–562. [DOI] [PubMed] [Google Scholar]
- 36.Burkhart, C., G.Y. Liu, S.M. Anderton, B. Metzler, and D.C. Wraith. 1999. Peptide-induced T cell regulation of experimental autoimmune encephalomyelitis: a role for IL-10. Int. Immunol. 11:1625–1634. [DOI] [PubMed] [Google Scholar]
- 37.Barrat, F.J., D.J. Cua, A. Boonstra, D.F. Richards, C. Crain, H.F. Savelkoul, R.D. Waal-Malefyt, R.L. Coffman, C.M. Hawrylowicz, and A. O'Garra. 2002. In vitro generation of interleukin 10-producing regulatory CD4+ T cells is induced by immunosuppressive drugs and inhibited by T helper type 1 (Th1)- and Th2-inducing cytokines. J. Exp. Med. 195:603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundstedt, A., E.J. O'Neill, K.S. Nicolson, and D.C. Wraith. 2003. Role for IL-10 in suppression mediated by peptide-induced regulatory T cells in vivo. J. Immunol. 170:1240–1248. [DOI] [PubMed] [Google Scholar]
- 39.Chen, Y., J. Inobe, V.K. Kuchroo, J.L. Baron, C.A. Janeway Jr., and H.L. Weiner. 1996. Oral tolerance in myelin basic protein T-cell receptor transgenic mice: suppression of autoimmune encephalomyelitis and dose-dependent induction of regulatory cells. Proc. Natl. Acad. Sci. USA. 93:388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeda, K., B.E. Clausen, T. Kaisho, T. Tsujimura, N. Terada, I. Forster, and S. Akira. 1999. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 10:39–49. [DOI] [PubMed] [Google Scholar]
- 41.Takahama, Y., K. Ohishi, Y. Tokoro, T. Sugawara, Y. Yoshimura, M. Okabe, T. Kinoshita, and J. Takeda. 1998. Functional competence of T cells in the absence of glycosylphosphatidylinositol-anchored proteins caused by T cell-specific disruption of the Pig-a gene. Eur. J. Immunol. 28:2159–2166. [DOI] [PubMed] [Google Scholar]
- 42.Hanada, T., H. Yoshida, S. Kato, K. Tanaka, K. Masutani, J. Tsukada, Y. Nomura, H. Mimata, M. Kubo, and A. Yoshimura. 2003. Suppressor of cytokine signaling-1 is essential for suppressing dendritic cell activation and systemic autoimmunity. Immunity. 19:437–450. [DOI] [PubMed] [Google Scholar]
- 43.Inoue, H., R. Kato, S. Fukuyama, A. Nonami, K. Taniguchi, K. Matsumoto, T. Nakano, M. Tsuda, M. Matsumura, M. Kubo, et al. 2005. Spred-1 negatively regulates allergen-induced airway eosinophilia and hyperresponsiveness. J. Exp. Med. 201:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida, H., S. Hamano, G. Senaldi, T. Covey, R. Faggioni, S. Mu, M. Xia, A.C. Wakeham, H. Nishina, J. Potter, et al. 2001. WSX-1 is required for the initiation of Th1 responses and resistance to L. major infection. Immunity. 15:569–578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.