Abstract
Human natural killer (NK) cells originate from CD34(+) hematopoietic progenitor cells, but the discrete stages of NK cell differentiation in vivo have not been elucidated. We identify and functionally characterize, from human lymph nodes and tonsils, four NK cell developmental intermediates spanning the continuum of differentiation from a CD34(+) NK cell progenitor to a functionally mature NK cell. Analyses of each intermediate stage for CD34, CD117, and CD94 cell surface expression, lineage differentiation potentials, capacity for cytokine production and natural cytotoxicity, and ETS-1, GATA-3, and T-BET expression provide evidence for a new model of human NK cell differentiation in secondary lymphoid tissues.
NK cells are CD3(−)CD19(−) large granular lymphocytes that can kill infected or malignantly transformed cells and produce cytokines, such as IFN-γ, that regulate the innate and adaptive immune systems. Similar to other lymphocytes, human NK cells are thought to originate from CD34(+) hematopoietic progenitor cells (HPCs) that differentiate through discrete stages of maturation (1). Whereas T and B cell developmental intermediates have been identified within the human thymus and BM, respectively, in vivo stages of human NK cell differentiation have not been defined.
Our current understanding of NK cell development stems primarily from findings in mice and in vitro differentiation systems (1, 2). Early studies revealed that IL-15 can promote the differentiation of cytolytic NK cells from CD34(+) HPCs in vitro (3). IL-15 signals in part via the IL-2 receptor β chain (CD122), and mice lacking either IL-15 or CD122 have severe NK cell deficiencies supporting their physiological roles during NK cell development in vivo (4, 5). Other cytokines, such as flt3 ligand (FL) and c-kit ligand (KL), can induce the generation of CD122(+) NK cell precursors (NKPs) from IL-15–nonresponsive mouse BM progenitors, indicating that the acquisition of “IL-15 responsiveness” marks a critical stage in mouse NK cell development (6), and indeed the earliest committed mouse NKPs in vivo express CD122 yet lack other markers of mature NK cells (7). CD122 expression is also maintained throughout the downstream stages of mouse NK cell differentiation, which were recently elucidated by correlating integrin expression patterns with stages of proliferation, receptor expression, and acquisition of NK cell function (8).
In humans, CD122 expression on freshly isolated CD34(+) HPCs is below the limits of detection by flow cytometry, making it difficult to identify IL-15–responsive NKPs (9–11). In addition, mouse NK cells express NK1.1 and DX5, whereas human NK cells are generally defined by the expression of CD56, which is not expressed by mouse NK cells (1, 2). Therefore, it has remained a challenge to identify the orthologous NK cell developmental intermediates in humans. However, despite these differences in antigen expression between the species, results from in vitro differentiation assays initiated with human CD34(+) HPCs have revealed important general consistencies with the in vivo mouse data. Both the human in vitro data as well as the mouse in vivo data indicate that NK cell functional maturity (i.e., the ability to mediate natural cytotoxicity and produce IFN-γ) is acquired at a late stage of development, likely distal to the acquisition of surface CD56 in humans (8, 12, 13). In addition, similar to mouse NK cell differentiation, in vitro human NK cell differentiation is associated with the nonrandom, orderly acquisition of NK cell receptors (1, 2), with CD161 and NKp46 being among the first NK cell receptors to be detected, followed by CD94, and lastly CD16 and killer-cell immunoglobulin-like receptors (KIRs; references 3, 12, 14). Collectively, these results suggest that human NK cells would differentiate through discrete developmental stages in vivo; however, the precise intermediates have not yet been described.
Another major difference between mice and humans is that the latter have two NK cell subsets, with the CD56bright subset being substantially more capable of proliferation and cytokine production, whereas the fresh CD56dim subset displays a higher capacity for natural cytotoxicity ex vivo (15). Currently, the developmental relationship between mature human NK cell subsets is unknown as are their sites of differentiation in vivo (1, 15). Although ≥90% of peripheral blood (PB) NK cells are CD56dimCD16(+)KIR(+), the majority of NK cells in human LNs and tonsils are CD56brightCD16(−)KIR(−) (16, 17). We recently described a population of CD34dimCD45RA(+) HPCs that is highly enriched with NKPs and that is unique among all CD34(+) subsets by its ability to colocalize within the T cell–rich regions of human LNs near mature CD56bright NK cells (9). Based on these findings, we hypothesized that secondary lymphoid tissues (SLT) might be sites of CD56bright NK cell differentiation and would, therefore, contain all the requisite NK cell developmental intermediates spanning the continuum of differentiation from a CD34(+) progenitor to a CD56bright NK cell. In the current study, we identify and functionally characterize such intermediates from SLT, thereby providing a new model for the development of human NK cells in vivo.
RESULTS
Delineation of NK cell developmental stages within human SLT
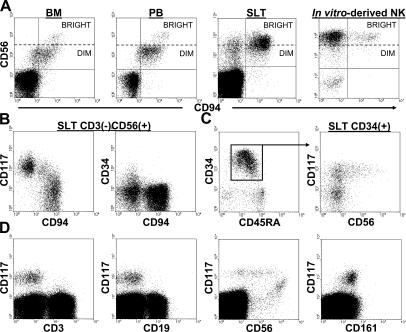
We noted from previous work that although primary CD56bright NK cells found in BM and PB coexpress CD94, the majority of CD56bright NK cells derived in vitro from CD34(+) HPCs cultured in IL-15 lack CD94 (Fig. 1 A; reference 9), suggesting that a CD94(−) intermediate stage may exist in vivo. Interestingly, fresh human SLT contain CD3(−)CD56(+)CD94(−) cells (Fig. 1 A). We speculated that the latter may be immature NK cell developmental intermediates, and additional phenotypic analyses revealed that these cells are uniformly CD117(+) and some express CD34 (Fig. 1 B). Based on these results, we made the following three predictions of in vivo human NK cell development. First, given that CD34 and CD94 are mutually exclusive antigens (Fig. 1 B, right), a developing NK cell would first lose CD34 before acquiring CD94. Second, as all CD3(−) CD56(+) cells in SLT express at least either CD117 and/or CD94 (Fig. 1 B, left), NK cell developmental intermediates with the CD34(−)CD94(−) phenotype would be identifiable by the expression of CD117. Third, the CD34(+) HPC, which represents the human IL-15–responsive NKP, would express CD117.
Figure 1.
Surface marker expression patterns of human SLT populations. (A) CD3(−)CD56brightCD94(−) cells are absent in BM and PB, yet present in human SLT and in NK cell differentiation cultures. All four dot plots were gated on CD3(−) events. (B) Distinct patterns of CD34, CD117, and CD94 expression within the CD3(−)CD56(+) fraction of human SLT. The dot plots were gated on total CD3(−)CD56(+) events. (C) A continuum of CD56 expression is observed within the CD117(+) fraction of SLT CD34(+) HPCs. The left dot plot shows CD34 versus CD45RA expression within a CD34-enriched preparation from human SLT. The right dot plot was gated on CD34(+) events as indicated by the box and arrow in the left dot plot. (D) Analysis of CD3, CD19, CD56, and CD161 expression on total CD34(−)CD117(+) cells within the mononuclear fraction of human SLT. The dot plots were gated on CD34(−) SLT preparations. No qualitative differences were observed in comparing LN and tonsil specimens. The data are representative of at least five separate analyses.
Within human SLT, CD117 expression on CD34dim CD45RA(+) HPCs identifies a CD56(+) and CD56(−) population (Fig. 1 C). The gradual up-regulation of CD56 expression seen exclusively on the CD34(+)CD117(+) population suggested that NK cells are derived from this population and that CD34(+)CD117(−) cells may be functionally distinct as it pertains to NK cell developmental potential (Fig. 1 C). Within the CD34(−) mononuclear fraction of human SLT, CD117 expression is absent on CD3(+) T cells and CD19(+) B cells, whereas CD34(−)CD117(+) cells display a similar gradual up-regulation of CD56 as that seen on CD34(+)CD117(+) HPCs and they uniformly coexpress the pan-NK cell receptor CD161 (Fig. 1 D). Thus, within both CD34(+) and CD34(−) mononuclear fractions of human SLT, CD117 expression is highly associated with the NK cell lineage.
Given these collective data, we hypothesized that human NK cells differentiate through four discrete stages within SLT: stage 1, CD34(+)CD117(−)CD94(−); stage 2, CD34(+)CD117(+) CD94(−); stage 3, CD34(−)CD117(+)CD94(−); and stage 4, CD34(−)CD117(+/−)CD94(+). Although we did not include CD56 as a definitive criterion for the intermediate populations, its progressive expression through the stages, including high-density expression within stage 4 being similar to PB CD56bright NK cells, lends additional support to this paradigm of in vivo human NK cell differentiation (Fig. 2). Through the remainder of the text, we refer to these four populations as stages 1–4.
Figure 2.
Progressive CD56 expression by in vivo stages of human NK cell differentiation. The data were obtained by first separating CD3(−)CD19(−)CD34(+) and CD3(−)CD19(−)CD34(−) fractions from SLT mononuclear cell suspensions via magnetic selection and then assessing for CD34, CD117, CD94, and CD56 expression by flow cytometry. It was confirmed that residual T cells or B cells did not contribute to the data.
Surface marker and gene expression profiles of stages 1–4
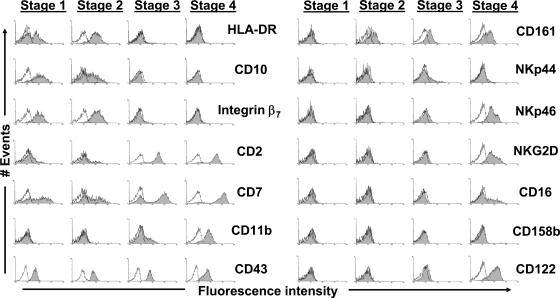
To more comprehensively characterize stages 1–4 as they exist in vivo, we first performed flow cytometric analyses on freshly isolated stages as found in SLT, with a focus on antigens expressed by either immature HPCs or mature PB NK cells (Fig. 3). In agreement with our model, antigens previously associated with immature lymphoid progenitors, including HLA-DR, CD10, and integrin β7 (9, 18), were detected at stages 1 and 2 yet not at stages 3 and 4. In contrast, similar to the gradual accumulation of CD56 expression within the stages (Fig. 2), CD2, CD7, and CD11b expression were gradually increased from stage 1 to 4. Notably, the accumulation of CD11b at stages 3 and 4 is consistent with CD11b being a surrogate marker for maturity during in vivo mouse NK cell differentiation (8). However, there were no detectable differences in terms of CD43 expression in this regard (Fig. 3).
Figure 3.
Surface marker expression profiles of stages 1–4. Data were obtained by performing flow cytomtric analyses of CD3(−)CD19(−)CD34(+) and CD3(−)CD19(−)CD34(−) cell preparations as in Fig. 2. Shaded regions represent staining for the indicated surface antigens, whereas solid lines (open regions) represent staining with appropriate isotype-matched control mAbs. The data shown are representative of at least three separate experiments for each antigen, and we did not observe any differences in comparing LN and tonsil specimens.
Analysis of NK cell receptors on fresh stages 1–4 found within human SLT from multiple donors also revealed a consistent pattern, such that a greater number of these receptors was expressed at each progressive stage of differentiation (Fig. 3). None of the NK cell receptors we analyzed were detected at stage 1. CD161 was consistently the only receptor expressed at stage 2, whereas both CD161 and NKp44 were detected at stage 3. Stage 4 cells expressed CD161, NKp44 (albeit at low levels), NKp46, and NKG2D, and we could also detect the expression of CD16 and CD158b at stage 4 on low percentages of cells, similar to the low level expression of CD16 and KIR on PB CD56bright NK cells (15). Similarly, surface expression of CD122 was first noted at stage 3 and only readily detectable at stage 4. In contrast to these data, we could not detect the expression of NKp30 at any of the four stages in SLT (unpublished data).
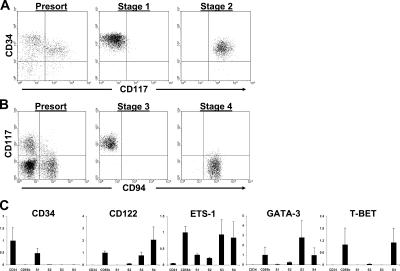
Next, we compared freshly isolated stages 1–4 for their relative messenger RNA (mRNA) expression of genes important for mouse NK cell development (19). Fig. 4 (A and B) shows representative sorts to purify these four populations directly from human SLT. As comparisons for the gene expression assay, we also analyzed each transcript in total PB CD34(+) HPCs and PB CD56bright NK cells. As a control for methodology, CD34 mRNA was highest among PB CD34(+) HPCs, lower among stages 1 and 2, and invariably undetectable in lysates from stages 3 and 4 and PB CD56bright NK cells (Fig. 4 C). Among stages 1–4, CD122 mRNA was first detectable at stage 2, more abundant in stage 3, and relatively more so in stage 4. ETS-1 mRNA is expressed by mature mouse NK cells (20), and we observed that it was detectable at stages 1 and 2, yet higher at stages 3 and 4. Mice lacking GATA-3 and T-BET display NK cell functional defects (21, 22), and it has been postulated that GATA-3 may function upstream of T-BET during mouse NK cell differentiation (19). Consistent with this hypothesis, we observed that GATA-3 mRNA expression peaked at stage 3, whereas T-BET mRNA was low or undetectable at stages 1–3, yet relatively much higher at stage 4, comparable to PB CD56bright NK cells (Fig. 4 C).
Figure 4.
Gene expression profiles of stages 1–4. (A and B) Representative sorts to purify stages 1–4 from human SLT. Stages 1 and 2 (A) and stages 3 and 4 (B) were sorted from CD3(−)CD19(−)CD34(+) and CD3(−)CD19(−)CD34(−) fresh SLT cell preparations, respectively, obtained via magnetic selection as described in Materials and methods. Additional flow cytometric analyses confirmed that sorted stage 3 and 4 cells were CD34(−) (not depicted). (C) Gene expression analysis of stages 1–4. Gene expression for each sample was normalized to 18S expression. Depending on the preferential expression of each gene of interest, the mean of normalized values from either total PB CD34(+) HPCs or total PB CD56bright NK cells was set to a value of 1, and all other normalized values were adjusted to scale. The y axis of each graph is in arbitrary units and measures the adjusted value for each population. The data are expressed as the mean of adjusted values ± SD of 3–5 separate samples tested per cell population (n = 3–5). CD34, PB CD34(+) HPCs; CD56b, PB CD56bright NK cells; S1, stage 1; S2, stage 2; S3, stage 3; S4, stage 4.
Natural killing and cytokine production of NK cell development stages
According to the model of mouse NK cell differentiation proposed by Kim et al. (8), acquisition of the capacities to mediate natural cytotoxicity and produce cytokines occurs at a late stage of maturation in vivo. Limiting cell numbers prevented us from directly assessing such functional capacities of stages 1 and 2 from human SLT. However, we were able to purify enough fresh stage 3 and 4 cells for analyses. As detected by intracellular flow cytometry, stages 1–3 lack perforin whereas perforin expression was detected at stage 4 (Fig. 5 A and not depicted). Accordingly, stage 3 cells showed no killing against either K562 or Jurkat target cells, whereas stage 4 cells displayed >30% and >90% specific lysis against these targets, respectively (E/T ratio = 20:1; Fig. 5 B). In the presence of EGTA and MgCl2, the combination of which inhibits perforin-mediated killing (13), an anti-TNF–related apoptosis-inducing ligand (TRAIL) blocking mAb had no effect on the ability of stage 4 cells to lyse Jurkat targets, whereas an anti-Fas ligand (FasL) blocking mAb did inhibit killing (Fig. 5 C). Therefore, under the conditions tested, freshly isolated stage 4 cells mediated both perforin- and FasL-dependent killing but not TRAIL-dependent killing, similar to PB NK cells (13), whereas stage 3 cells did not mediate natural cytotoxicity via any of the three mechanisms.
Figure 5.
Functional analyses of stages 3 and 4. (A) Perforin expression of freshly isolated stage 3 and 4 cells as detected by intracellular flow cytometry. Shaded regions represent staining with the anti-perforin mAb whereas solid lines (open regions) represent staining with the isotype-matched control mAb. Histograms were gated on unsorted CD3(−)CD19(−) preparations from human SLT using mAbs directed against CD34, CD117, and CD94 to identify discrete stages for analysis. Stages 1 and 2 lack detectable intracellular perforin staining (not depicted). (B) Percentages of specific lysis against K562 (black bars) and Jurkat (gray bars) targets by freshly isolated stage 3 and 4 cells. Data shown are the mean results ± SD of four separate experiments performed in triplicate at a 20:1 E/T ratio. (C) Stage 4 cells mediate FasL-dependent, but not TRAIL-dependent, killing of Jurkat target cells. Data shown are the mean results ± SD of triplicate wells from one experiment with freshly isolated stage 4 cells targeted against the Jurkat cell line as in B but in the presence of 1 mM EGTA and 2 mM MgCl2 (20:1 E/T ratio). (D) Production of IFN-γ (left) and GM-CSF (right) by stage 3 and 4 cells after 12-h stimulation with either the combination of IL-12, IL-15, and IL-18 (black bars) or the combination of PMA, ionomycin, and IL-2 (gray bars). Data shown are the combined results ± SD of four separate experiments (n = 4 for each).
Freshly purified stage 3 and 4 cells were also stimulated for 12 h in either the combination of IL-12, IL-15, and IL-18 or PMA, ionomycin, and IL-2 to assess for cytokine production ex vivo (Fig. 5 D). As predicted by the significant difference in T-BET mRNA expression (Fig. 4 C), stage 4 cells produced IFN-γ under both conditions, whereas IFN-γ was not detected in the supernatants of stimulated stage 3 cells (Fig. 5 D). Similar results were obtained using intracellular flow cytometry to detect IFN-γ, and we were also unable to detect IFN-γ production by stages 1 and 2 via this method (unpublished data). GM-CSF was detected from both stage 3 and 4 supernatants, although stage 3 cells produced less GM-CSF than stage 4 cells and did so only in the presence of PMA, ionomycin, and IL-2 (Fig. 5 D). In contrast, we could not detect the production of either IL-13 or TNF-α after 12-h stimulation of either SLT population (unpublished data). Collectively, these data suggest that functional maturity occurs within stage 4 of NK cell differentiation in vivo.
Lineage differentiation potentials of stages 1–4
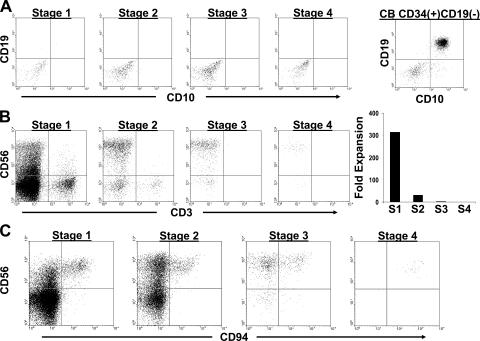
For functional assessment of the lineage potentials of stages 1–4, we used a series of in vitro differentiation assays. Stages 1–4 were first cultured in semi-solid methylcellulose medium containing KL, GM-CSF, IL-3, and erythropoietin. Whereas multiple CFUs of both erythroid and myeloid lineages were derived from a positive control of total PB CD34(+) HPCs, we did not observe any CFUs from stages 1–4 in three separate experiments (unpublished data). Similarly, we observed that in conditions promoting B cell differentiation on MS-5 stroma using control cord blood (CB) CD34(+)CD19(−) HPCs, freshly isolated stages 1–4 failed to give rise to any CD10(+)CD19(+) B lineage cells (Fig. 6 A).
Figure 6.
B and T cell differentiation potential of stages 1–4. (A) Representative analysis from one of two experiments culturing SLT-derived stages 1–4 or CB-derived CD34(+)CD19(−) cells on MS-5 stroma in FL, KL, and G-CSF for 6 wk. Dot plots were gated on live cells using a lymphocyte gate by FSC versus SSC. (B) Representative analysis from one of three experiments in which stages 1–4 were co-cultured for 4–5 wk on the OP9-DL1 cell line in FL and IL-7. The OP9-DL1 line expresses GFP. The four dot plots shown were gated on GFP(−) events within the live fraction by FSC versus SSC analysis. The graph on the right shows the fold increase in total GFP(−) cell number starting with 103 cells from each stage (S1–S4). n = 1 (representative graph from one of three experiments). (C) Generation of CD3(−)CD56brightCD94(+) stage 4 cells on OP9-DL1 stroma. Stage 1–4 cells were cultured as in B. The dot plots shown were gated on GFP(−)CD3(−) live cell events and are representative of three separate experiments.
Numerous studies indicate that NK cells are developmentally most closely related to T cells and DCs (18, 23–26). We cultured stages 1–4 in FL and IL-7 on the OP9-DL1 cell line, which supports human T cell differentiation (27). After 4–5 wk, stage 1 cells underwent profound expansions, with cell numbers increasing 1.5 × 102–5.4 × 103-fold (Fig. 6 B). In contrast, stages 2–4 displayed less than one tenth the propensity for proliferation on the OP9-DL1 line. By flow cytometry, both stage 1 and 2 cultures contained CD3(+) T cells, which expressed either TCRαβ or TCRγδ receptors, as well as CD3(−)CD4(+/−)CD8(+/−) immature T cells (Fig. 6 B and not depicted). However, comparison of the absolute numbers of CD3(+) T cells derived from stage 1 (1.37 × 105 per 103 initiating cells) versus stage 2 (5.7 × 103 per 103 initiating cells) cells revealed that there was, on average, a 24-fold difference (P = 0.024). In contrast to these results, we did not observe the generation of CD3(+) T cells or CD3(−)CD4(+/−)CD8(+/−) immature T cells in human T cell differentiation cultures initiated with purified stage 3 or 4 cells (Fig. 6 B and not depicted). We did observe CD3(−)CD56brightCD94(+) NK cells (stage 4) in these cultures when initiated with freshly isolated stage 1, 2, and 3 cells (Fig. 6 C), confirming that each of these populations is capable of giving rise to stage 4 NK cells.
To assess for DC differentiation, purified stages 1–4 were cultured in FL, KL, IL-3, IL-4, GM-CSF, and TNF-α. After 2 wk, cultures initiated with stages 1–3 maintained live cells, although there was an overall net loss in total viable cell number (mean of 0.14-, 0.35-, and 0.28-fold contractions for stage 1–3 populations, respectively). In contrast, stage 4 cells all died in these conditions. Cultured cells were harvested for analysis by flow cytometry. By forward scatter (FSC) versus side scatter (SSC) analysis, stage 1 and 2 cultures contained a distinct population that was absent from stage 3 and 4 cultures. Cells grown out from stage 1 and 2 cultures displayed a phenotype consistent with in vitro–derived DCs (HLA-DR(+)CD14(−)CD1a(+/−)) and expressed co-stimulatory molecules (Fig. 7, A and B; references 18, 25). Further, these cells displayed typical DC morphology (Fig. 7 C). In contrast, stage 3 cultures contained only smaller cells that did not display the phenotype shown in Fig. 7 B nor the typical DC morphology (unpublished data). Altogether, these results suggest that full commitment to the NK cell lineage may occur at stage 3 of development in vivo.
Figure 7.
DC differentiation potential of stages 1–4. (A and B) Representative flow cytometric analysis from one of three experiments culturing stages 1–4 in FL, KL, IL-3, IL-4, GM-CSF, and TNF-α. (A) By FSC versus SSC analysis, only stage 1 and 2 cultures contained the population (black circles) notably absent in stage 3 and 4 cultures. (B) Shaded regions in the histograms represent staining for the indicated surface antigens whereas solid lines (open regions) represent staining with isotype-matched control mAbs. The histograms were gated on the populations circled in A from stage 1 and 2 cultures. We did not observe any qualitative differences between stage 1– and stage 2–derived DCs. (C) Wright/Giemsa staining of stage 1– and stage 2–derived DCs. Note the typical DC morphology, including multiple dendrite processes and the large nucleus displaced to one side of the cell. Viable cells in stage 3 cultures did not display the DC morphology (not depicted).
Ex vivo NK cell developmental progression of stages 1–4
In addition to the phenotypic and functional assays performed on freshly isolated stages 1–4 of NK cell differentiation, we cultured these cells ex vivo to evaluate the effects of individual cytokines that have been previously shown to support NK cell differentiation in vitro (3, 11, 28) and to assess each stage for evidence of lineage progression versus reversibility. Stages 1 and 2 were first purified from human SLT and cultured in IL-15 without any other cytokines or stroma. As predicted from the CD122 mRNA expression data (Fig. 4 C), after 2 wk nearly all stage 1 cells died with an overall 0.17 ± 0.21-fold decrease in total viable cell number, whereas cultures initiated with stage 2 cells expanded 20.1 ± 9.9-fold (P = 0.0001; Fig. 8 A). These data suggest that the ability to respond to IL-15 occurs at stage 2 of differentiation in vivo. Stage 2 cells cultured in IL-15 primarily became CD3(−)CD34(−)CD56brightCD161(+) and displayed a pattern of CD117 versus CD94 expression similar to that of total SLT CD3(−)CD56(+) cells in vivo (Fig. 1 B), indicative of progression to stages 3 and 4 (Fig. 8 B and not depicted).
Figure 8.
Stage 1 and 2 NK cell developmental progression. (A) Sorted stage 1 and 2 cells were cultured in IL-15 for 2 wk and then total viable cell counts were determined. Cultures were initiated with 103 starting cells. Shown are the mean results from seven separate experiments ± SD. n = 7. (B) Representative phenotypic analysis of stage 2 cells cultured in IL-15 for 2 wk. The few viable cells remaining in stage 1 cultures after 2 wk in IL-15 displayed a similar phenotype shown for cultured stage 2 cells. (C) Limiting dilution analysis of stages 1 and 2 cultured in either IL-15 alone (◯) or IL-15 with the one time addition of FL, IL-3, and IL-7 at the initiation of culture (▪). The graphs show initiating stage 1 and 2 cell numbers versus the percentages of NK-positive wells for one of two separate experiments yielding similar results. (D) De novo generation of CD34(+)CD117(+) cells from purified stage 1 cells. Purified stage 1 and 2 cells were cultured for 4 d in FL, IL-3, IL-7, and IL-15 and then analyzed by flow cytometry for the expression CD34 and CD117. Unsorted, unstained CD34-enriched cell preparations (obtained via magnetic selection) were cultured in parallel to set voltages and compensation parameters on the flow cytometer. Results are representative of three separate experiments.
Next, we cultured stage 1 cells in IL-15 on MS-5 stroma with FL, IL-3, and IL-7 added at the initiation of culture to determine if these additional factors would promote NK cell differentiation from stage 1. In bulk culture, we observed profound expansions (37.1 ± 26.9-fold) compared with cultures in IL-15 alone (n = 4, P = 0.004), and by flow cytometry the cells became CD56brightCD94(+/−) NK cells (unpublished data). By limiting dilution analysis on MS-5 stroma, stage 1 cells cultured in IL-15 plus FL, IL-3, and IL-7 added at the beginning of culture displayed an increased NKP frequency (1/14–1/9) compared with cultures in IL-15 alone (1/44–1/37), whereas stage 2 cells displayed a high NKP frequency in IL-15 with or without the additional cytokines (<1/3.5) (Fig. 8 C). Thus, stage 1 cells require additional cytokines for differentiation into CD56bright NK cells ex vivo, consistent with the notion that stage 1 cells are less mature than stage 2 cells that can respond to IL-15.
To gain evidence for a progenitor–progeny relationship between stages 1 and 2, respectively, purified cells were cultured in FL, IL-3, IL-7, and IL-15, and then assessed by flow cytometry after 4 d. Reproducibly, we observed the appearance of CD34(+)CD117(+) cells in stage 1 cultures, whereas CD34(+)CD117(−) stage 1 cells were not detected in stage 2 cultures (Fig. 8 D). Furthermore, cultured stage 2 cells displayed lowered CD34 expression than the CD34(+)CD117(+) cells that had been derived from stage 1, suggesting that stage 2 cells were progressing to stage 3 and that the appearance of CD34(+)CD117(+) cells in stage 1 cultures was a result of the de novo generation of these cells rather than contamination by stage 2 cells at day 0. As predicted, after 10 more days of culture in IL-15, we observed the exclusive appearance of CD34(−)CD117(+)CD94(−) stage 3 and CD34(−)CD117(+/−)CD94(+) stage 4 NK cells in cultures initiated with either stage 1 or 2 cells (unpublished data). Thus, under these conditions in vitro, freshly isolated stage 1 cells gave rise to stages 2–4, whereas freshly isolated stage 2 cells gave rise to stages 3 and 4 but not stage 1.
Although the results from the OP9-DL1 co-culture experiments demonstrated that freshly isolated stage 3 cells gave rise to stage 4 cells ex vivo, those experiments were performed in the absence of exogenous IL-15 (Fig. 6 C). Therefore, we cultured freshly isolated stage 3 and stage 4 cells for 2 wk in IL-15 to determine if this cytokine was sufficient to promote stage 3 to stage 4 differentiation ex vivo (Fig. 9). Both populations proliferated indicating their responsiveness to this cytokine (Fig. 9 A). However, although neither population up-regulated CD34, we repeatedly observed that only a small fraction (≤2%) of the sorted stage 3 cells were induced to express CD94 (stage 4), whereas fresh stage 4 cells maintained their phenotype, as predicted by our previous results culturing PB CD56bright NK cells in IL-15 (Fig. 9 B; reference 9). Therefore, additional factors might be necessary to drive stage 3 to 4 differentiation in vivo. We previously observed that co-culture of total SLT CD34(+) HPCs with autologous activated SLT T cells preferentially promoted the differentiation of CD56bright NK cells that express CD94 (unpublished data), suggesting that endogenous factors produced by activated SLT T cells would promote stage 3 to 4 NK cell differentiation in vitro. Indeed, in the presence of autologous activated SLT T cells, we observed the de novo generation of CD3(−)CD34(−)CD117(+/−)CD94(+) stage 4 cells in cultures initiated with purified stage 3 cells, whereas CD3(−)CD34(−)CD117(+)CD94(−) stage 3 cells were not observed in cultures initiated with purified stage 4 cells (Fig. 9 B).
Figure 9.
Stage 3 to 4 differentiation ex vivo. (A) Proliferation of stage 3 and 4 cells cultured in IL-15. Shown are the mean results ± SD from seven separate experiments starting with 103 cells. n = 7. (B) Representative phenotypic analysis of one of three experiments with stage 3 and 4 cells cultured either in IL-15 (left dot plot for each population) or in IL-15 plus activated autologous SLT T cells (right dot plot for each population). All four dot plots were gated on total CD3(−) events, and none of the cultured cells expressed CD34 (not depicted). Cultures initiated with activated SLT T cells alone did not contain CD3(−)CD94(+) cells (reference 9).
DISCUSSION
The in vivo developmental pathways for human B and T lymphocytes have been generally understood for decades, and this has been important for studying disease processes such as HIV, childhood and adult leukemia, and lymphoma. Although in vivo stages of mouse NK cell differentiation were recently described (8), the similar populations in humans have not yet been reported (1, 2). In this study, we identified four novel populations that appeared to represent discrete stages of a human NK cell developmental continuum within SLT. Each population was isolated directly from these tissues and was shown to be capable of downstream NK cell differentiation ex vivo (i.e., stage 1 → stage 2 → stage 3 → stage 4). Further, we demonstrated that each freshly isolated population could be characterized by unique functional and phenotypic attributes and that overall progression through the stages was characterized by the gradual restriction of non-NK cell lineage differentiation potential, concomitant with the gradual acquisition of the mature CD56bright NK cell phenotype, cytokine production, and mediation of natural cytotoxicity. Collectively, these data provide evidence for a new model of in vivo human NK cell differentiation.
Many of our results are consistent with previously published mouse and human data on this subject (1, 2, 19, 23, 26). Specifically, we observed that (a) human NK cells share a common developmental origin with T cells and DCs; (b) commitment to the NK cell lineage precedes acquisition of the capacities for IFN-γ production and natural cytotoxicity; (c) the proposed progression of NK cell development in vivo is supported by the observed patterns of CD122, ETS-1, GATA-3, and T-BET mRNA expression; and (d) the expression and accumulation of NK cell receptors is nonrandom during in vivo maturation. In addition, we observed that CD94 expression in vivo correlates with the ability to produce IFN-γ, which is consistent with previous reports of in vitro human NK cell differentiation (12). However, we did observe some functional and phenotypic differences between human NK cell intermediates derived in vitro and those purified directly from human SLT. For example, CD3(−)CD56brightCD94(−) NK cells derived in vitro uniformly express NKp44 and NKp46 (9, 14), yet these expression patterns were not observed in vivo. Furthermore, whereas CD94(−) immature NK cells derived in vitro can mediate TRAIL-dependent killing and produce large amounts of IL-13 (12, 13), we did not detect these functions from stage 3 intermediates isolated ex vivo. The reasons for these differences are not yet known. However, considering that in vivo differentiation to a stage 4 cell is associated with the acquisition of the capacities for both natural killing and IFN-γ production, this process is likely to be tightly regulated to ensure self-tolerance and, therefore, may not be fully recapitulated in culture systems currently used to study human NK cell differentiation. In addition, we currently do not know the specific mechanisms of action of many cytokines, including FL, IL-3, IL-7, and IL-15, that have been previously implicated as critical in this pathway (3, 11, 28), nor do we yet know whether the cytokines themselves are present in SLT near developing NK cells at the concentrations used to promote NK cell differentiation in vitro. Therefore, these discrepancies may result from missing factors and/or the addition of nonphysiological stimuli during NK cell development in vitro.
In light of the aforementioned data, it is also noteworthy that IL-15 stimulation alone was insufficient to promote substantial stage 4 differentiation from freshly isolated stage 3 cells despite the generation of stage 4 cells in IL-15–supplemented stage 1 and 2 cultures as well as in cultures initiated with BM or PB CD34(+) HPCs (9). One interpretation of these data is that IL-15 may have induced the differentiation of stage 4 cells directly from stage 2, thus bypassing stage 3. However, this is not supported by the fact that CD34 and CD94 are mutually exclusive antigens, both in vitro and in vivo, and we would expect to find some CD34(+)CD94(+) intermediates in human SLT if this was the case. Stage 3 cells in vivo display a phenotype that is intermediate between those of stage 2 and 4 cells in terms of CD2, CD7, CD11b, CD56, CD117, CD161, and NKp44 expression. Furthermore, freshly isolated stage 3 cells failed to generate T cells or DCs, yet reproducibly gave rise to stage 4 cells when cultured with either activated SLT T cells or OP9-DL1 stroma. Therefore, we favor the hypothesis that stage 4 cells are the direct progeny of stage 3 cells in vivo and that this differentiation step may require signals other than, or in addition to, IL-15. Moreover, we speculate that signals promoting stage 4 differentiation may be common to OP9-DL1 and activated SLT T cell co-cultures as well as to cultures initiated with CD34(+) HPCs.
The work presented here provides new evidence that LNs and tonsils are sites for human NK cell development in vivo. Previous studies established that CD56bright NK cells predominate in SLT (16, 17), and we have now demonstrated that these tissues are also naturally and selectively enriched with the full complement of newly discovered intermediates spanning the continuum of NK cell differentiation from a CD34dimCD45RA(+) progenitor to a mature CD56bright NK cell. However, there are now many new questions that arise from this study. For example, it is still unclear how SLT-derived NK cells might be related to NK cells in other compartments in the body. Although SLT stage 4 cells closely resemble PB CD56bright NK cells in terms of their phenotypic and functional attributes, it is possible that SLT-resident NK cells represent a lineage distinct from that of CD56bright NK cells in the blood, as has been postulated for human decidual NK cells (29). In addition, the majority of NK cells in PB are CD56dim and express high levels of CD16 and KIRs (15). Based on their phenotypic and functional characteristics, we speculate that PB CD56dim NK cells might represent the terminally differentiated stage of human NK cell development (stage 5). However, we currently lack direct evidence to support this hypothesis and cannot yet formally exclude the possibility that PB CD56bright and/or CD56dim NK cells might be derived from distinct NKPs that differentiate in the BM.
It is also unclear to what extent stage 4 cells, or any of the other NK cell lineage populations, actually arise within SLT or merely traffic through these tissues. On the one hand, our flow cytometry data provide strong evidence for the existence of ongoing NK cell maturation within these tissues, as for example analysis of CD94 versus CD117 expression reveals a continuous pattern with cells at all points in the spectrum between stages 3 and 4 (Fig. 1 B). On the other hand, previous evidence suggests that both PB CD34(+) NKPs and PB CD56bright NK cells have the capacity to migrate into SLT, given the relatively high expression of l-selectin on these subsets in the blood versus low or undetectable expression within SLT (9, 17, 30). Therefore, some of these cells, in particular those that we have characterized as stage 4, may have recently migrated into SLT from other sites. In future studies it will be important to determine the relative capacity for NK cell developmental intermediates to recirculate in and out of multiple lymphoid organs, given the potential implications regarding self-tolerance acquisition and overall regulation of this process in vivo.
A primary goal of this study was to identify surface antigens that could be used to specifically and positively identify developing human NK cells in vivo, ideally providing an all-inclusive framework on which to base future research on the NK cell developmental pathway. In this regard, CD34, CD117, and CD94 are useful antigens because they are relatively specific to the NK cell lineage in SLT, they provide continuity for “following” developing NK cells, and they account for most if not all of the CD3(−)CD56(+) cells within these tissues. However, stages 1–4 as we defined them are likely to be functionally heterogeneous, with the potential to identify additional subsets within each of these populations. For example, our flow cytometric analyses revealed that stage 1 and 2 cells are heterogeneous with regards to the expression of CD2, CD7, CD10, and CD56. Therefore, in future experiments, it might be determined that the true NK cell progenitors within the stage 1 population are distinct from non-NK lineage cells that share the CD34(+)CD45RA(+)CD117(−) phenotype or that the stage 2 population consists of mono-committed T, DC, and NKP rather than cells with multipotency. Indeed, we cannot rule out these possibilities using the techniques we used in our study. In addition, there are expected to be differentiation steps that are downstream of the acquisition of CD94, including those involving the acquisition of CD16 and KIRs. Therefore, future studies are warranted to continue to refine the human NK cell developmental pathway in vivo and to elucidate the physiological mechanisms regulating this process in SLT.
MATERIALS AND METHODS
Cell isolation from human tissues.
All protocols were approved by the Ohio State University (OSU) Institutional Review Board. The data in this study were generated from the combined analyses of tissue specimens from >50 tonsil and 30 LN donors and we observed no appreciable differences between LNs and tonsils by phenotype or functional assays. Normal human tonsils and LNs were obtained within 24 h of elective surgery through the Tissue Procurement Shared Resource of the OSU Comprehensive Cancer Center or from brain dead tissue donors through the National Disease Research Interchange (NDRI). Tissue cells were dispersed through 70-μm cell strainers into sterile PBS followed by ficoll centrifugation. For each donor's tissues, all cells from multiple tonsil pieces or multiple LNs were pooled to obtain sufficient numbers of NK cell developmental intermediates for the experiments. Mononuclear fractions were depleted of CD3(+) and CD19(+) cells via magnetic negative selection (Miltenyi Biotec). For certain experiments, T and B cell-depleted preparations were either stimulated to detect intracellular IFN-γ production or directly stained with mAbs for phenotypic analyses. Alternatively, CD34(+) cells were enriched from CD3(−)CD19(−) preparations with the CD34 progenitor isolation kit (Miltenyi Biotech), subsequently stained with FITC-conjugated anti-CD45RA, phycoerythrin-conjugated anti-CD117, and APC-conjugated anti-CD34 mAbs (BD Biosciences), and sorted for stage 1 (CD34(+)CD45RA(+)CD117(−)) and stage 2 (CD34(+)CD45RA(+)CD117(+)) populations with a FACSVantage cell sorter (BD Biosciences). Remaining CD3(−)CD19(−)CD34(−) cells were stained with the combination of FITC-conjugated anti-CD94 mAb (clone 131412; R&D Systems), phycoerythrin-conjugated anti-CD117, APC-conjugated anti-CD3, and APC-conjugated anti-CD34 mAbs (BD Biosciences), and stage 3 (CD3 (−)CD34(−)CD117(+)CD94(−)) and stage 4 (CD3(−)CD34(−)CD117 (+/−)CD94(+)) populations were sorted to >99% purity. SLT-derived T cells and BM, PB, and CB (obtained from NDRI) mononuclear cell populations were obtained as previously described (9).
Flow cytometry.
All mAbs used in this paper were purchased from BD Biosciences, except CD16, CD56, CD122, CD158b, NKp30, NKp44, NKp46 (Beckman Coulter), and CD94 (R&D Systems). Flow cytometry and data analyses were performed as previously described (9).
Real-time PCR.
RNA extraction and reverse transcription from ≥2 × 104 sorted cells were performed as described previously (31). All primers and probes were designed using Primer Express software (Applied Biosystems) and the sequences are as follows: CD34: forward 5′-ATCGCCGCAGCTGGAG-3′, reverse 5′-CCGTTTTCCGTGTAATAAGGGT-3′, probe 5′-FAM-CCACAGGAGAAAGGCTGGGCGA-Tamra-3′; CD122: forward 5′-CCTGGCTACCTCTTGGGCA-3′, reverse 5′-GAAGCATGTGAACTGGGAAGTG-3′, probe 5′-FAM-TGCAGCGGTGAATG-MGB-3′; ETS-1: forward 5′-CGATCTGGAGCTTTTCCCC-3′, reverse 5′-TGGAGTTAATAGTGGGACATCTGC-3′, probe 5′-FAM-CCCCGGATATGGAATG-MGB-3′; GATA-3: forward 5′-AAATGAACGGACAGAACCGG-3′, reverse 5′-TGCTCTCCTGGCTGCAGAC-3′, probe 5′-FAM-CCCTCATTAAGCCCAAGCGAAGGC-Tamra-3′; T-BET: forward 5′-CAACAATGTGACCCAGATGAT-3′, reverse 5′-AATCTCGGCATTCTGGTAGG-3′, probe 5′-FAM-CCGGCTGCATATCGTTGAGGTGAAC-Tamra-3′. Real-time PCR reactions were performed in a sequence detector (ABI prism 7700; Applied Biosystems) as follows: 50°C for 2 min; 95°C for 10 min; and 45 cycles of 95°C for 15 s and 60°C for 1 min. Data were analyzed with the Sequence Detector version 1.6 software to establish the PCR cycle at which fluorescence exceeded a set threshold, CT, for each sample. Experimental CT values were used to calculate gene-specific copy numbers from a standard curve, and the gene-specific copy numbers were normalized to ribosomal 18S copy numbers from the same samples.
Cytotoxicity assays and cytokine production.
Cytotoxicity assays were performed in 100 μl RPMI-1640 medium with GlutaMAX containing 10% FBS and antibiotics (Invitrogen) in 96-well V-bottom plates (BD Biosciences) with 2 × 102 K562 or Jurkat cloned targets stably infected with mouse stem cell virus retrovirus to express the firefly luciferase enzyme. After 8-h incubations, cell pellets were lysed with passive lysis buffer (Promega) and frozen at −80°C. 25 μl of thawed lysates were transferred into Costar 96-well flat-bottom solid-white plates (Fisher Scientific) and mixed with 100 μl of Luciferase assay reagent (Promega) by a Fluoroskan Ascent FL luminometer (Thermo Electron, Inc.). Luminescence was measured as relative light units (RLU) during 10-s intervals. The percent of specific lysis for each experiment was calculated as [(mean RLU from wells containing target cells only) − (mean RLU from experimental wells)]/(mean RLU from wells containing target cells only) × 100%. Blocking experiments with the Jurkat cell line were performed with 1 mM EGTA and 2 mM MgCl2. The anti-TRAIL (clone RIK-2), anti-FasL (clone NOK-2), and isotype-matched control mAbs (BD Biosciences) were used at 10 μg/ml.
For cytokine production, 2 × 104 purified stage 3 or 4 cells were stimulated in either the combination of 10 ng/ml IL-12 (Genetics Institute), 100 ng/ml IL-15, and 100 ng/ml IL-18 (BASF) or 50 ng/ml PMA (Sigma-Aldrich), 1 μM ionomycin (Calbiochem), and 1 nM IL-2 (Hoffman LaRoche) for 12 h at 37°C. Supernatants were analyzed by ELISA for the production of IFN-γ as described previously (32) using commercially available mAbs (Endogen) or for the production of GM-CSF, TNF-α, and IL-13 using Quantikine ELISA kits (R&D Systems).
Cell culture.
Stroma-free cultures were in 96-well round-bottom plates (BD Biosciences) in 200 μl RPMI-1640 medium with GlutaMAX (Invitrogen), 10% heat-inactivated human AB serum (ICN Biomedicals), antibiotics and additional supplements previously described (9). Recombinant human IL-15 was provided by Amgen and used at 1 nM. Human KL (Amgen) and FL (Peprotech) were used at 100 ng/ml, whereas human IL-3, IL-4, IL-7, G-CSF, GM-CSF, and TNF-α (R&D Systems) were used at 10 ng/ml each. Activated T cell co-culture experiments were performed in 200 μl of medium with 104 freshly isolated SLT stage 3 or 4 cells and 2.5 × 103 autologous SLT CD3(+) T cells (purified via sorting [reference 9]) with 5 × 103 anti-CD3/CD28 beads (Dynal) and IL-15. Half the medium was replaced every 3–4 d to replenish IL-15 only. Co-cultures on MS-5 stroma (a gift of J.E. Dick, Toronto General Research Institute, Toronto, Canada) were in 96-well flat-bottom plates (BD Biosciences) in 200 μl α-MEM plus 20% FBS and antibiotics (all from Invitrogen) replacing half the medium every 3–4 d. CFU assays were performed with MethoCult GF+ from StemCell Technologies following the manufacturer's protocol. T cell differentiation cultures on the OP9-DL1 cell line (provided by J.C. Zúñiga-Pflücker, University of Toronto, Toronto, Canada) were maintained in α-MEM plus 20% FBS, antibiotics, FL, and IL-7 and were performed as described previously (27). Morphologies of in vitro–derived DCs were assessed via cytospin and Wright/Giemsa staining.
Limiting dilution analysis.
Purified stage 1 and 2 cells were cultured at 50, 25, 12.5, 6.25, 3.125, and 1.5625 cells per well, 10 replicates each, on MS-5 stroma in either IL-15 alone or IL-15 with the one time addition of FL, IL-3, and IL-7 at the initiation of culture. Half the medium was changed every 3–4 d to replenish IL-15 only. It was confirmed that all live human cells remaining in such cultures after 3 wk were CD56bright NK cells (unpublished data). Therefore, wells that contained visible growth (compared with wells not seeded with HPCs) were scored positive for NK cells, whereas wells that did not contain visible growth were harvested, stained with phycoerythrin-conjugated anti-CD56 mAb, and assessed by flow cytometry for the presence or absence of CD56bright NK cells. NKP frequencies reported in the text were calculated as the reciprocal of the concentration of cells that resulted in ≥63% positive wells using Poisson statistics and the weighted mean method, as described previously (11).
Statistical analyses.
Data were log transformed and analyzed using a paired Student's t test to obtain p-values. S-Plus version 6.0 and SAS version 8.02 software were used for the analyses.
Acknowledgments
The authors would like to thank Dr. Lewis L. Lanier for critical reading of the manuscript. We thank Dr. John E. Dick for providing the MS-5 cell line and Dr. Juan-Carlos Zúñiga-Pflücker for providing the OP9-DL1 cell line. In addition, we thank Donna Bucci, Megan Jukich, Mary McNulty, Erin Smith, and Chris Vetanovetz for their assistance in obtaining human tissue specimens.
This work was supported by the National Institutes of Health (grants P30 CA16059, CA68458, and CA95426 to M.A. Caligiuri).
The authors have no conflicting financial interests.
Abbreviations used: CB, cord blood; FasL, Fas ligand; FL, flt3 ligand; FSC, forward scatter; HPC, hematopoietic progenitor cell; KIR, killer-cell immunoglobulin-like receptor; KL, c-kit ligand; mRNA, messenger RNA; NKP, NK cell precursor; PB, peripheral blood; RLU, relative light units; SSC, side scatter; SLT, secondary lymphoid tissues; TRAIL, TNF-related apoptosis-inducing ligand.
References
- 1.Colucci, F., M.A. Caligiuri, and J.P. Di Santo. 2003. What does it take to make a natural killer? Nat. Rev. Immunol. 3:413–425. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama, W.M., S. Kim, and A.R. French. 2004. The dynamic life of natural killer cells. Annu. Rev. Immunol. 22:405–429. [DOI] [PubMed] [Google Scholar]
- 3.Mrozek, E., P. Anderson, and M.A. Caligiuri. 1996. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 87:2632–2640. [PubMed] [Google Scholar]
- 4.Kennedy, M.K., M. Glaccum, S.N. Brown, E.A. Butz, J.L. Viney, M. Embers, N. Matsuki, K. Charrier, L. Sedger, C.R. Willis, et al. 2000. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15–deficient mice. J. Exp. Med. 191:771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki, H., G.S. Duncan, H. Takimoto, and T.W. Mak. 1997. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor β chain. J. Exp. Med. 185:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams, N.S., T.A. Moore, J.D. Schatzle, I.J. Puzanov, P.V. Sivakumar, A. Zlotnik, M. Bennett, and V. Kumar. 1997. Generation of lytic natural killer 1.1+, Ly-49− cells from multipotential murine bone marrow progenitors in a stroma-free culture: definition of cytokine requirements and developmental intermediates. J. Exp. Med. 186:1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosmaraki, E.E., I. Douagi, C. Roth, F. Colucci, A. Cumano, and J.P. Di Santo. 2001. Identification of committed NK cell progenitors in adult murine bone marrow. Eur. J. Immunol. 31:1900–1909. [DOI] [PubMed] [Google Scholar]
- 8.Kim, S., K. Iizuka, H.S. Kang, A. Dokun, A.R. French, S. Greco, and W.M. Yokoyama. 2002. In vivo developmental stages in murine natural killer cell maturation. Nat. Immunol. 3:523–528. [DOI] [PubMed] [Google Scholar]
- 9.Freud, A.G., B. Becknell, S. Roychowdhury, H.C. Mao, A.K. Ferketich, G.J. Nuovo, T.L. Hughes, T.B. Marburger, J. Sung, R.A. Baiocchi, et al. 2005. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 22:295–304. [DOI] [PubMed] [Google Scholar]
- 10.Shibuya, A., H. Kojima, K. Shibuya, K. Nagayoshi, T. Nagasawa, and H. Nakauchi. 1993. Enrichment of interleukin-2-responsive natural killer progenitors in human bone marrow. Blood. 81:1819–1826. [PubMed] [Google Scholar]
- 11.Yu, H., T.A. Fehniger, P. Fuchshuber, K.S. Thiel, E. Vivier, W.E. Carson, and M.A. Caligiuri. 1998. Flt3 ligand promotes the generation of a distinct CD34(+) human natural killer cell progenitor that responds to interleukin-15. Blood. 92:3647–3657. [PubMed] [Google Scholar]
- 12.Loza, M.J., and B. Perussia. 2001. Final steps of natural killer cell maturation: a model for type 1-type 2 differentiation? Nat. Immunol. 2:917–924. [DOI] [PubMed] [Google Scholar]
- 13.Zamai, L., M. Ahmad, I.M. Bennett, L. Azzoni, E.S. Alnemri, and B. Perussia. 1998. Natural killer (NK) cell–mediated cytotoxicity: differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J. Exp. Med. 188:2375–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivori, S., C. Cantoni, S. Parolini, E. Marcenaro, R. Conte, L. Moretta, and A. Moretta. 2003. IL-21 induces both rapid maturation of human CD34+ cell precursors towards NK cells and acquisition of surface killer Ig-like receptors. Eur. J. Immunol. 33:3439–3447. [DOI] [PubMed] [Google Scholar]
- 15.Cooper, M.A., T.A. Fehniger, and M.A. Caligiuri. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22:633–640. [DOI] [PubMed] [Google Scholar]
- 16.Fehniger, T.A., M.A. Cooper, G.J. Nuovo, M. Cella, F. Facchetti, M. Colonna, and M.A. Caligiuri. 2003. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood. 101:3052–3057. [DOI] [PubMed] [Google Scholar]
- 17.Ferlazzo, G., D. Thomas, S.L. Lin, K. Goodman, B. Morandi, W.A. Muller, A. Moretta, and C. Munz. 2004. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J. Immunol. 172:1455–1462. [DOI] [PubMed] [Google Scholar]
- 18.Galy, A., M. Travis, D. Cen, and B. Chen. 1995. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 3:459–473. [DOI] [PubMed] [Google Scholar]
- 19.Vosshenrich, C.A., S.I. Samson-Villeger, and J.P. Di Santo. 2005. Distinguishing features of developing natural killer cells. Curr. Opin. Immunol. 17:151–158. [DOI] [PubMed] [Google Scholar]
- 20.Barton, K., N. Muthusamy, C. Fischer, C.N. Ting, T.L. Walunas, L.L. Lanier, and J.M. Leiden. 1998. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 9:555–563. [DOI] [PubMed] [Google Scholar]
- 21.Samson, S.I., O. Richard, M. Tavian, T. Ranson, C.A. Vosshenrich, F. Colucci, J. Buer, F. Grosveld, I. Godin, and J.P. Di Santo. 2003. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 19:701–711. [DOI] [PubMed] [Google Scholar]
- 22.Townsend, M.J., A.S. Weinmann, J.L. Matsuda, R. Salomon, P.J. Farnham, C.A. Biron, L. Gapin, and L.H. Glimcher. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 20:477–494. [DOI] [PubMed] [Google Scholar]
- 23.Lanier, L.L., H. Spits, and J.H. Phillips. 1992. The developmental relationship between NK cells and T cells. Immunol. Today. 13:392–395. [DOI] [PubMed] [Google Scholar]
- 24.Mebius, R.E., P. Rennert, and I.L. Weissman. 1997. Developing lymph nodes collect CD4+CD3- LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 7:493–504. [DOI] [PubMed] [Google Scholar]
- 25.Shen, H.Q., M. Lu, T. Ikawa, K. Masuda, K. Ohmura, N. Minato, Y. Katsura, and H. Kawamoto. 2003. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J. Immunol. 171:3401–3406. [DOI] [PubMed] [Google Scholar]
- 26.Spits, H., B. Blom, A.C. Jaleco, K. Weijer, M.C. Verschuren, J.J. van Dongen, M.H. Heemskerk, and P.C. Res. 1998. Early stages in the development of human T, natural killer and thymic dendritic cells. Immunol. Rev. 165:75–86. [DOI] [PubMed] [Google Scholar]
- 27.De Smedt, M., I. Hoebeke, and J. Plum. 2004. Human bone marrow CD34+ progenitor cells mature to T cells on OP9-DL1 stromal cell line without thymus microenvironment. Blood Cells Mol. Dis. 33:227–232. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J.S., V. McCullar, M. Punzel, I.R. Lemischka, and K.A. Moore. 1999. Single adult human CD34(+)/Lin-/CD38(-) progenitors give rise to natural killer cells, B-lineage cells, dendritic cells, and myeloid cells. Blood. 93:96–106. [PubMed] [Google Scholar]
- 29.Koopman, L.A., H.D. Kopcow, B. Rybalov, J.E. Boyson, J.S. Orange, F. Schatz, R. Masch, C.J. Lockwood, A.D. Schachter, P.J. Park, and J.L. Strominger. 2003. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 198:1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frey, M., N.B. Packianathan, T.A. Fehniger, M.E. Ross, W.C. Wang, C.C. Stewart, M.A. Caligiuri, and S.S. Evans. 1998. Differential expression and function of L-selectin on CD56bright and CD56dim natural killer cell subsets. J. Immunol. 161:400–408. [PubMed] [Google Scholar]
- 31.Trotta, R., R. Parihar, J. Yu, B. Becknell, J. Allard II, J. Wen, W. Ding, H. Mao, S. Tridandapani, W.E. Carson, and M.A. Caligiuri. 2005. Differential expression of SHIP1 in CD56bright and CD56dim NK cells provides a molecular basis for distinct functional responses to monokine costimulation. Blood. 105:3011–3018. [DOI] [PubMed] [Google Scholar]
- 32.Cooper, M.A., T.A. Fehniger, S.C. Turner, K.S. Chen, B.A. Ghaheri, T. Ghayur, W.E. Carson, and M.A. Caligiuri. 2001. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 97:3146–3151. [DOI] [PubMed] [Google Scholar]