Abstract
It is well established that the pre–T cell receptor for antigen (TCR) is responsible for efficient expansion and differentiation of thymocytes with productive TCRβ rearrangements. However, Ptcra- as well as Tcra-targeting experiments have suggested that the early expression of Tcra in CD4−CD8− cells can partially rescue the development of αβ CD4+CD8+ cells in Ptcra-deficient mice. In this study, we show that the TCR Eδ but not Eα enhancer function is required for the cell surface expression of αβTCR on immature CD4−CD8− T cell precursors, which play a crucial role in promoting αβ T cell development in the absence of pre-TCR. Thus, αβTCR expression by CD4−CD8− thymocytes not only represents a transgenic artifact but occurs under physiological conditions.
Intrathymic development can be divided into discrete stages at which thymocytes express distinct surface markers that include CD4, 8, 25, and 44. Cells that express neither CD4 nor CD8 are called double-negative (DN) cells that consist of CD25−CD44+ (DN1), CD25+CD44+ (DN2), CD25+CD44− (DN3), and CD25−CD44− (DN4) subsets (1). Thymus development is also characterized by sequential rearrangement and expression of TCR genes (2). Rearrangement of variable γ and δTCR gene segments begins at the DN2 stage and, if productive, results in CD4−CD8− (DN) cells that express γδTCR on the cell surface. Rearrangement of variable TCRβ gene segments sets in slightly later and, also if productive, results in the surface expression of the pre-TCR consisting of a TCRβ chain that is covalently associated with the pre-TCRα chain and noncovalently associated with CD3 signal-transducing molecules (3, 4). The pre-TCR is expressed at rather low levels on DN3 and DN4 cells (5). Pre-TCR–expressing DN cells undergo several rounds of division before the rearrangement of variable TCRα gene segments sets in at the late DN4 and early CD4+CD8+ double-positive (DP) stage. This results in the surface expression of αβTCRs at the expense of the pre-TCR because TCRα chains generally compete favorably with pre-TCRα for TCRβ chains. TCRαβ-expressing DP cells then undergo positive or negative selection by intrathymic peptide–MHC complexes (6).
The temporal order of TCR V gene segment rearrangement has suggested the following scenario: DN4 cells that contain productive Tcrg and Tcrd rearrangements express γδTCR on the cell surface and, thereby, become functionally mature T cells that are ready to leave the thymus. Pre-TCR–expressing cells will eventually become DP thymocytes expressing an αβTCR, the specificity of which determines their further developmental fate. This simple scheme of T cell development was found not to be valid when T cell development was analyzed in Ptcra-deficient mice. In contrast to CD3ɛ-deficient and Tcrb plus Tcrd double-deficient mice that contain none or very few DP thymocytes, Ptcra-deficient mice were shown to harbor strongly reduced but still considerable numbers of DP thymocytes (7). Further analysis in Ptcra,Tcrd as well as Ptcra,Tcra double-deficient mice then revealed that both an early expressed γδTCR as well as an early expressed αβTCR could still rescue the development of DP thymocytes in the absence of pre-TCR (8). In the case of αβTCR in Ptcra −/− ,Tcrd −/− mice, this resulted in thymocytes of which >95% harbored TCRβ chains (i.e., DP cells that were selected by an early expressed αβTCR and therefore contained in-frame Tcrb rearrangements). In the case of an early expressed γδTCR in Ptcra −/− ,Tcra −/− mice, only 15% of DP cells contained TCRβ chains. Thus, these experiments indicated that receptors other than the pre-TCR could relieve DN3 cells from a development block resulting in the production of DP cells and that there must be a rearrangement of TCRα V gene segments at the DN3 or earlier stages of T cell development.
The temporal order of TCR V gene segment rearrangement appears especially important with regard to the Tcra and Tcrd locus. This is where the Tcra locus is embedded in the Tcra locus and where the early rearrangement of Vδ gene segments results in the formation of TCRδ chains, whereas late Vα rearrangement is accompanied by the deletion of the Tcrd locus and generation of Tcra genes (9). Two different enhancer elements have been invoked in the control of rearrangement and expression of the Tcra/Tcrd locus: the Eδ enhancer located in the Jδ-Cδ intron and the Eα enhancer 4 kb downstream of Cα (10–13). Both enhancers have been deleted by homologous recombination. In Eδ−/− animals, the development of αβ T cells appeared to proceed normally with the exception that there was a considerable reduction of thymic and peripheral γδ T cells. In this context, Eδ-deficient alleles exhibited a substantial reduction of Tcrd gene rearrangements. Eα−/− mice contained normal numbers of DP cells but reduced numbers of CD4+CD8− and CD4−CD8+ single-positive (SP) cells as a result of an almost complete block in Vα to Jα rearrangements. The Eα enhancer also controls levels of αβ as well as γδTCR expression, as evident by reduced levels of TCRα and TCRδ transcripts in Eα−/− mice. These results indicate that Eδ functions in DN thymocytes to promote Tcrd gene rearrangement and gene expression but not Jα accessibility to the V(D)J recombinase. On the other hand, Eα functions in DP thymocytes to promote TCRα gene rearrangement via Jα accessibility and controls both Tcrd and Tcra gene expression. However, because TCRα chains were still present in Eα−/− mice, it was hypothesized that in the absence of Eα, the Eδ enhancer or other elements might have a role in promoting rearrangement, possibly via the promotion of Jα accessibility and/or expression of some Vα segments (11). This would account for the TCRα chains with limited diversity (mostly Vα2) that were expressed in peripheral lymphoid tissue in Eα-deficient mice.
In this study, we have tested the hypothesis that a low level of Vα to Jα rearrangements is controlled by the Eδ enhancer and occurs in DN thymocytes earlier than the bulk of Eα-controlled Vα to Jα rearrangements, thus leading to the expression of αβTCR in DN cells, which permits some of these cells to enter the αβ lineage of DP thymocytes.
RESULTS
Early αβTCR-expressing DN cells
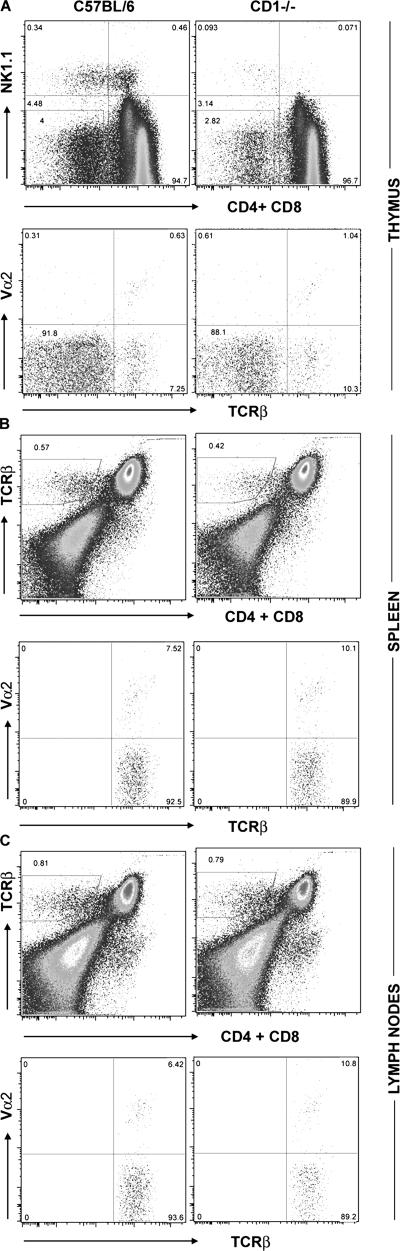
Analyzing total thymocytes and DN thymocytes from wild-type (wt) C57BL/6 mice, we find that ∼5% express TCRβ chains on the cell surface of which ∼10% are paired with Vα2-containing TCRα chains (Fig. 1 A). Because a subset of NKT cells (14) has the TCRβ+ CD4−CD8− phenotype, NK1.1 cells were excluded by using NK1.1 antibodies for depletion as well as by the analysis of NKT cell–deficient CD1−/− animals (Fig. 2). The results show that Vα+,TCRβ+ cells belong to a distinct, non–NKT cell subset of DN cells. In addition, we found TCRβ+ DN T cells in secondary lymphoid organs in both wt and CD1- deficient animals (Fig. 2).
Figure 1.
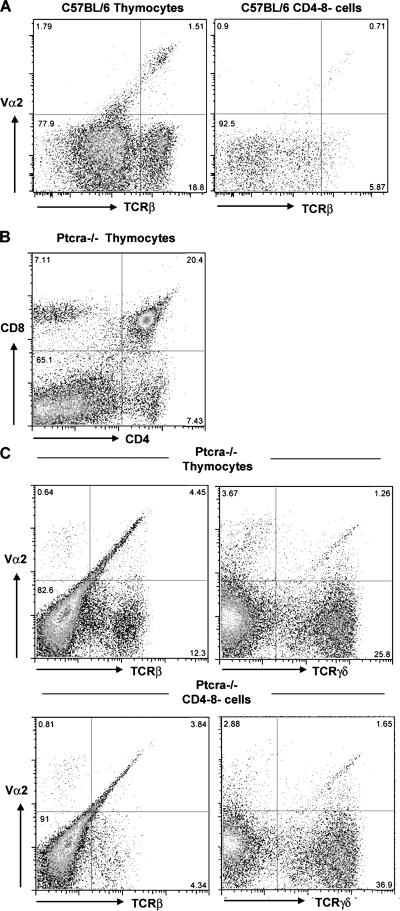
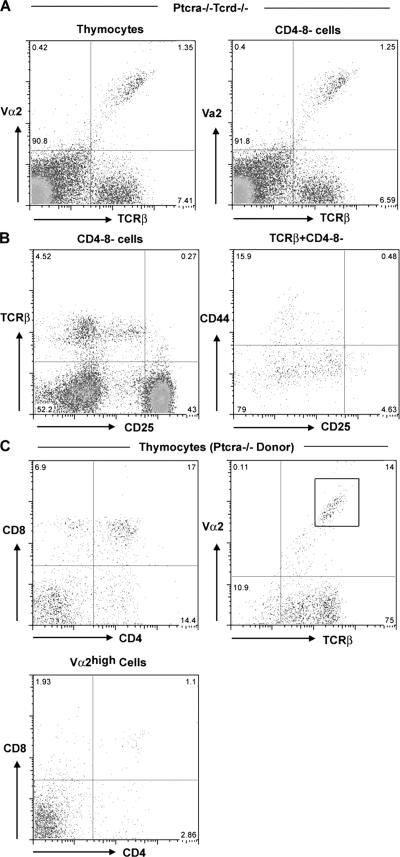
Vα2-enriched early αβ T cells in wt and Ptcra−/− mice. (A) TCRβ versus Vα2 FACS profile in thymi of wt and Ptcra −/− thymocytes. DN (CD4−CD8−) cells were electronically gated after the exclusion of CD4+, CD8+, and NK1.1 cells. (B) A CD4 versus CD8 profile of Ptcra −/− thymi. (C and D) Staining of Ptcra −/− thymocytes (total or CD4−CD8−) with TCRβ and Vα2 antibodies. Numbers in quadrants indicate the percentages of cells in that quadrant.
Figure 2.
CD4−CD8− αβ T cells in the thymus and lymphoid periphery. (A) CD4,CD8 versus NK1.1 staining in the thymus of wt and CD1−/− mice (top). TCRβ versus Vα2 analysis of NK1.1− CD4−CD8− thymocytes (bottom). (B and C) Identification of peripheral CD4−CD8− TCRβ+ cells in the spleen and lymph nodes using TCRβ, Vα2, CD4, and CD8 antibody labeling. Numbers in quadrants indicate the percentages of cells in that quadrant.
To investigate whether DN αβTCR-expressing cells are pre-TCR selected as their DP (CD4+CD8+) or SP (CD4+CD8− and CD4−CD8+) counterparts, thymocytes from Ptcra−/− mice were analyzed (Fig. 1 B). Although Ptcra−/− mice were previously shown to be deficient in NKT cells (15), we used NK1.1 antibodies in the depletion procedure that yielded DN thymocytes. In Ptcra−/− mice, ∼6–7% of DN thymocytes express TCRβ chains on the cell surface, and, in this particular experiment, 40–50% of DN cells with TCRβ proteins coexpressed Vα2,TCRα chains (Fig. 1 C). In several experiments, the proportion of Vα2+ cells among TCRβ+ DN cells in Ptcra−/− mice was variable and ranged between 10 and 50% (Fig. 4; see Fig. 7).
Figure 4.
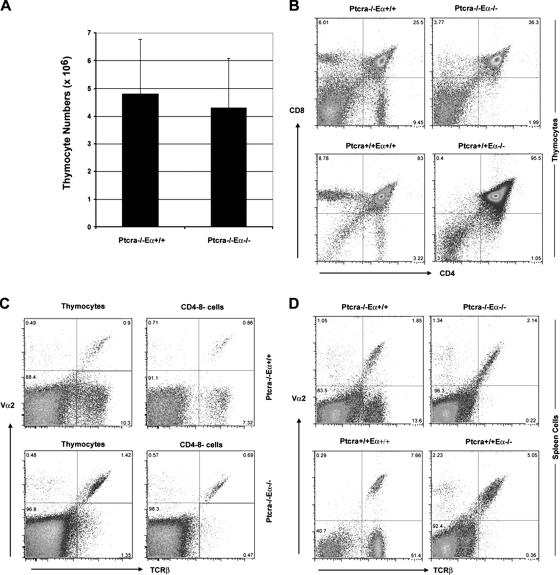
Eα controls late but not early development of αβ T cells. (A) Absolute cell numbers of Ptcra −/− and Ptcra −/− ,Eα−/− littermates. Error bars represent SD. (B) CD4 versus CD8 profiles of littermates belonging to all indicated genotypes. (C and D) Vα2 versus TCRβ antibody labeling in total DN thymocytes (C) and spleen cells (D). Numbers in quadrants indicate the percentages of cells in that quadrant.
Figure 7.
Absence of Vα2-expressing cells in the thymi of Ptcra−/−,Eγ−/− mice. TCRβ versus Vα2 staining of either total thymocytes (A) or CD4−CD8− cells (B) of littermate mice analyzed at 4 wk of age. Numbers in quadrants indicate the percentages of cells in that quadrant.
Of interest was the observation that Vα2 is not only contained in αβ but also in γδTCRs in DN thymocytes from Ptcra −/− mice (Fig. 1 C), a notion consistent with recent findings that Vα2 gene segments can join to both Jα and DδJδ sequences (16). To exclude that the existence of TCRβ+,Vα2+ DN cells is the result of Vα2 segment pairing with TCRδ diversity joining elements (16), a similar analysis was conducted in Ptcra −/− ,Tcrd −/− double-deficient mice. About 7% of CD4−CD8− cells were found to express TCRβ proteins on the cell surface, and ∼25% of the TCRβ chains were paired with Vα2-containing TCRα chains, suggesting that these TCRβ-expressing cells are bona fide αβ T cells that do not require pre-TCR selection (Fig. 3 A).
Figure 3.
Phenotype and developmental potential of early αβ T cells. (A) Vα2 versus TCRβ staining of Ptcra −/− ,Tcrd −/− thymocytes. (B) Phenotypic analysis of CD4−CD8− and CD4−CD8− TCRβ+ cells using CD44 and CD25 antibodies. (C) Embryonic day 14.5 fetal (Rag-1−/−) thymic organ culture of CD4−CD8− TCRβ+ NK1.1− Ptcra −/− cells. Cells were cultured for 7 d and were stained with Vα2, TCRβ, CD4, and CD8 antibodies. A CD4 versus CD8 staining of Vα2+ donor cells is also shown (bottom). Numbers in quadrants indicate the percentages of cells in that quadrant.
Further phenotypic analysis of the TCRαβ+ thymocytes showed that almost all “early” TCRβ-expressing CD4−CD8− cells are CD25 negative and, thus, belong to the DN4 (CD25−CD44−) subset (Fig. 3 B). The developmental potential of early αβTCR-expressing DN cells was then addressed by culturing purified LY5.2+ CD4−CD8− NK1.1− γδTCRβ+ Ptcra −/− thymocytes together with embryonic thymi from LY5.1+ in Rag1 −/− donors in fetal thymic organ cultures. After 7 d of culture, ∼40% of the donor-derived cells have up-regulated the expression of CD4,CD8 coreceptors. Moreover, >90% of the cultured thymocytes retain the surface expression of TCRβ, and ∼20% of them are Vα2+ (Fig. 3 C). Not all cells up-regulate CD4 and CD8, however, and a substantial fraction of cells expressing high levels of Vα2-containing αβTCRs remain CD4−CD8−. It is likely that these αβTCR-expressing DN cells normally exit the thymus because CD4−CD8− αβTCR+ cells can be detected in the lymph nodes and spleen of adult mice (Fig. 2), and it was shown in TCRαβ transgenic mice that DN cells with the transgenic TCR can accumulate in peripheral lymphoid tissue (6). Thus, TCRαβ-expressing DN cells can give rise to both immature DP and mature SP (CD4/8) T cells as well as DN αβ T cells that do not enter the DP αβ lineage (17).
The Eα enhancer controls late but not early development of αβ T cells in Ptcra −/− mice
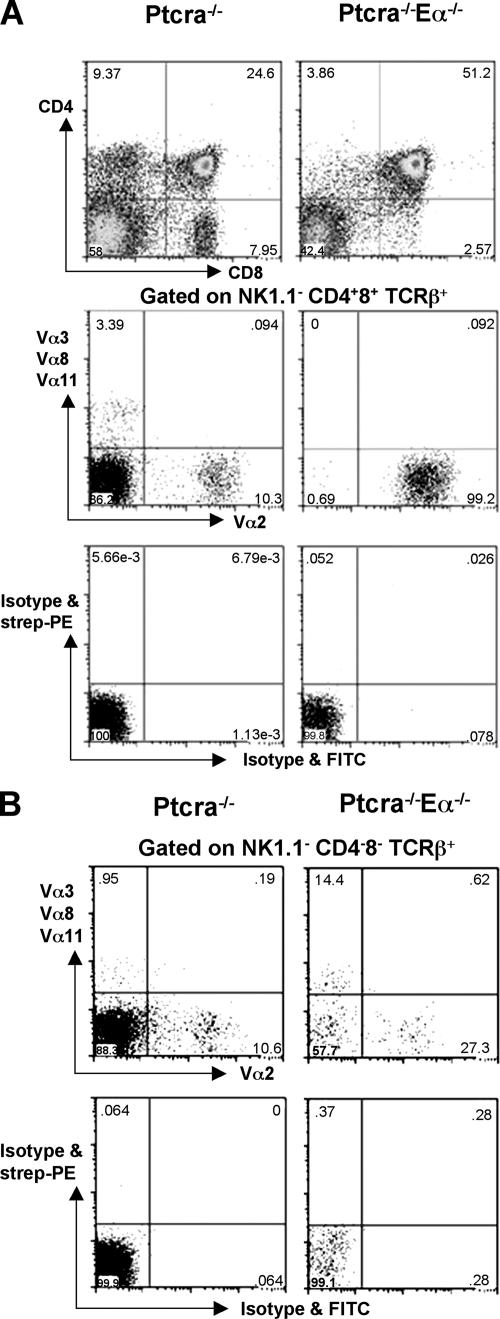
To address the role of the described TCR enhancers on early TCRα expression, we generated Ptcra −/− mice with a targeted deletion of the Eα locus (Ptcra −/− ,Eα−/−). Ptcra −/− , Eα−/− and Ptcra −/− ,Eα+/+ littermate mice contain similar numbers of thymocytes (Fig. 4 A). However, there is a clear reduction of CD4+CD8− SP thymocytes in the Ptcra −/− , Eα−/− mice because of limited TCRα diversity and/or expression levels (Fig. 4 B). This is supported by the staining of either all thymocytes or only DN thymocytes with a combination of TCRβ and Va2 antibodies: although total thymocytes from Ptcra −/− single-deficient mice contain <10% TCRβ+,Vα2+ cells, among TCRβ+ cells, this proportion is much higher (>30%) in Ptcra −/− ,Eα−/− double-deficient mice. In DN thymocytes from Ptcra −/− ,Eα+/+ mice, TCRβ+,Vα2+ cells represent 10% of all TCRβ+ cells, whereas in Ptcra −/− ,Eα−/− mice, TCRβ+,Vα2+ cells represent 50%. These data indicate that Eα is not required for the early rearrangement and expression of Tcra genes (Fig. 4 C). The data show that normally Eα predominantly contributes to the rearrangement and expression of TCRα V gene segments other than Vα2 gene segments. Consistent with this notion, the spleen of Ptcra −/− ,Eα−/− mice contains almost exclusively Vα2+,TCRβ+ cells, whereas only ∼10% of Vα2+,TCRβ+ cells among TCRβ+ cells are found in the spleen of Ptcra −/− ,Eα+/+ mice (Fig. 4 D). These observations are in line with earlier observations in Eα−/− mice (11) showing that TCRβ+ cells in the spleen of Eα−/− mice express almost exclusively Vα2 (Fig. 4 D).
DN cells but not mature T cells in Eα−/−-deficient mice express Vα2-negative Tcra genes
The data in Fig. 4 suggest that DN cells in Eα−/− mice can express Vα genes other than Vα2 and that the Vα2 dominance in the periphery of Eα−/− mice is established at a later developmental stage. This issue was addressed in more detail by analyzing the expression of TCR Vα2 genes and other TCRα V genes (Vα3, Vα8, and Vα11) in Ptcra −/− and Ptcra −/−,Eα−/− mice at various stages of development. As shown in Fig. 5 B, NKT cell–depleted CD4− and CD8− (DN) cells from both Ptcra −/− and Ptcra −/−,Eα−/− mice do express Vα genes that stain with a cocktail of Vα3, 8, and 11 antibodies. Although the proportion of cells stained with the cocktail versus Vα2-positive cells remains about the same in CD4+CD8+ cells of Ptcra −/− mice, it drastically decreases in DP cells of Eα−/− mice such that the vast majority of cells expresses Vα2 (Fig. 5 A). This trend is also evident in peripheral T cells from Eα−/− but not wt mice in which virtually all TCRβ+ cells express Vα2 (Fig. 4 D). Thus, these data indicate that the predominance of Vα2 expression in peripheral T cells of Eα−/− mice is not caused by the fact that the Eδ enhancer only allows for Vα2 rearrangement at the DN stage of T cell development. Instead, this is likely the result of the fact that Vα2 gene segments are able to sustain a sufficiently high expression of Tcra genes in the absence of the Eα enhancer such that only Vα2-positive αβ T cells can be positively selected and maintained in peripheral lymphoid tissue.
Figure 5.
Immature but not mature T cells from Eα−/− mice express diverse TCRα chains. (A) Staining of thymocytes from Ptcra −/− ,Eα+/+ and Ptcra −/− ,Eα−/− mice with CD4 and CD8 (top), and staining of CD4+CD8+ TCRβ+ cells with Vα3 + Vα8 + Vα11 antibodies versus Vα2 antibodies (middle). Bottom panel shows isotype controls for the staining in the middle panel. (B) Staining of NK1.1− CD4−CD8− TCRβ thymocytes from Ptcra −/− ,Eα+/+ and Ptcra −/− ,Eα−/− mice with Vα3 + Vα8 + Vα11 antibodies versus Vα2 antibodies (top) and with isotype controls (bottom). Numbers in quadrants indicate the percentages of cells in that quadrant.
Eγ controls Vα expression in DN cells
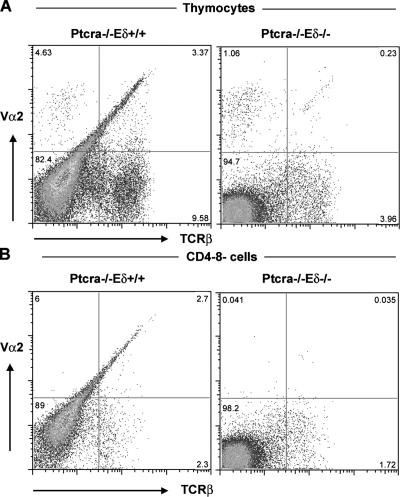
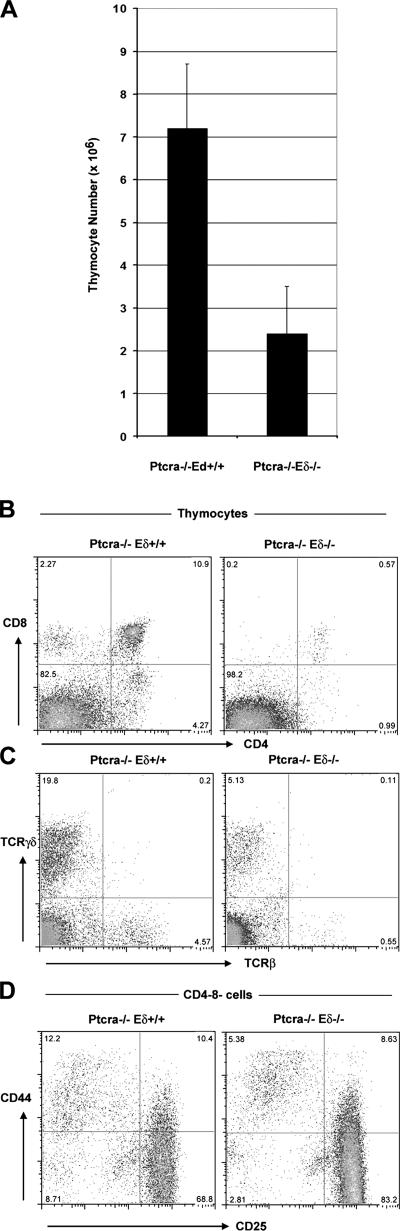
Because the analysis of Ptcra −/− ,Eα−/− mice has shown that Eα is not an enhancer element required for early TCRαβ expression and pre-TCR–independent T cell development (8, 11), we have focused our attention on the Eδ enhancer by generating and studying animals that lack both Ptcra as well as the Eδ enhancer element (Ptcra −/− ,Eδ−/− mice). When compared with Ptcra −/− ,Eδ+ thymi, it was noted that Ptcra −/− ,Eδ−/− thymi contain much-reduced numbers of thymocytes (Fig. 6 A) that are severely deficient in DP and SP cells (Fig. 6 B). Also, the number of γδTCR-expressing cells is reduced even though some γδ T cells are still present. Within the DN compartment, there is a more complete block at the DN3 stage of development in Ptcra −/− ,Eδ−/− versus Ptcra −/− ,Eδ+ mice (Fig. 6, C and D).
Figure 6.
Eγ is essential for pre-TCR–independent T cell development. (A) Absolute cell numbers of Ptcra −/− and Ptcra −/− ,Eδ−/− littermates. 4–8-wk-old mice were analyzed. Error bars represent SD. (B and C) CD4 versus CD8 (B) and TCRβ versus TCRγδ (C) profiles of littermate thymi. (D) Analysis of the DN compartment using CD25 and CD44 antibodies. Numbers in quadrants indicate the percentages of cells in that quadrant.
Ablation of Eδ in Ptrcra −/− mice has a profound effect on the percentage and absolute number of DN αβTCRs as well as of Vα2-expressing cells, as TCRβ+,Vα2+ thymocytes are virtually absent (<0.25%; Fig. 7, A and B). However, there was a small number of TCRβ-expressing cells among total thymocytes in some of the Ptcra −/− ,Eδ−/− mice (Fig. 7), suggesting some remaining level of TCRα rearrangement in the Ptcra −/− , Eδ−/− thymi (perhaps mediated by the Eα enhancer in cells) that was “rescued” by the expression of γδTCR at the DN developmental stage (8).
Thus, in the absence of Ptcra, the Eδ enhancer has a crucial role in the early assembly of TCRα chains as well as TCRδ chains in DN3 and/or earlier stages of T cell development (i.e., TCR chains that are required for the formation of γδ and αβTCR in the absence of pre-TCR). The αβTCRs (and γδTCRs) can rescue some developmental progression beyond the DN3 stage in Ptcra-deficient mice, which appropriately explains the incomplete developmental block in Ptcra −/− versus CD3−/− or Tcrb −/− × Tcrd −/− mice (i.e., mice that cannot assemble any TCR on the cell surface).
DISCUSSION
The results obtained in mice with a combined deficiency of the pre-TCRα chain plus the Eα or Eδ enhancer provide an explanation for the observation that development of DN thymocytes into αβ lineage DP cells in Ptcra −/− ,Tcrd −/− double-deficient mice can be rescued by αβTCR. The data indicate that Eδ can promote early Vα to Jα rearrangements, which results in the expression of an αβTCR on the surface of DN4 thymocytes. The early expression of an αβTCR was previously considered to represent a transgenic artifact caused by the too early expression of TCRα chains in TCR transgenic mice (18), but, as shown here, it also occurs under physiological conditions. Our experiments and earlier experiments in TCRα transgenic mice show that some but not all of the αβTCR CD4−CD8− cells can become CD4+CD8+ cells (8, 18). This developmental pathway may make only a limited contribution to the generation of DP αβ lineage cells in pre-TCR–competent mice not only because of the paucity of TCRα chains in DN cells of normal mice but also because TCRα is a bad “surrogate” for pre-TCRα. Indeed, we found that under competitive conditions, pre-TCR is far more effective in the generation of DP cells than αβTCR (19).
The fate of the DN cells that express αβTCR on the cell surface and do not become DP thymocytes needs to be further evaluated. These cells apparently can leave the thymus because they can be detected in the spleen and lymph nodes of normal (Fig. 2) as well as in exaggerated numbers in TCRα transgenic mice. Such cells were previously shown to acquire functional maturity (i.e., respond with proliferation and cytokine production to TCR ligation) and to acquire CD8α expression when antigenically stimulated. In fact, this unusual subset of T cells was analyzed in TCR transgenic mice many years ago, and it was concluded that the early expression of an αβTCR could mimic signals generated when thymocytes express γδTCR and become functionally mature (18, 20). Of interest is that such cells can express an autoreactive TCR but are not deleted because of the lack of coreceptors and can accumulate in secondary lymphoid tissue of mice, as shown in various TCR transgenic models (21). Some of these cells exhibit an activated phenotype and have an as yet undefined role in the immune system.
Recent data have, in fact, shown that the extensive accumulation of CD8αα cells in the gut of TCR transgenic mice (22) to a large extent depends on the premature expression of the transgenic TCR in DN cells because “on time” expression of the same Tcra transgene does not result in the strong accumulation of CD8αα cells in the gut (23). Another possibility is that thymic DN αβTCR+ cells represent precursors of some peripheral regulatory T cells. Perhaps these cells are akin to both mouse and human αβTCR+ CD3+ NK.1.1− CD4−CD8− DN Regulatory T cells that can suppress antigen-specific immune responses mediated by CD8+ and CD4+ T cells through a process that requires cell to cell contact and Fas–FasL interactions (24). In this regard, it is important to point out that the αβTCR+ DN studied here are different from NK1.1+ DN T cells and from αβTCR+ DN cells studied by others who concluded that all TCRβ+ DN cells were derived from DP precursors (25); either this generalization is wrong or the fate mapping approach used by the authors is not valid. However, it is clear that the TCRβ+ DN cells in Ptcrd −/− , Tcrd −/− mice are involved in the rescue of development rather than being derived from DP cells (8).
The results shown in this study also provide an adequate explanation for earlier observations in Eα-deficient mice that exhibited a TCRα repertoire limited to TCRα,Vα2 chains. In this context, on Eα-deleted alleles, other cis-acting elements such as Eδ and/or Vα2 promoters were hypothesized to promote either only Vα2 to Jα rearrangements or Vα to Jα rearrangements. These rearrangements involve a diverse array of Vα segments with assembled Vα2Jα complexes expressed in a much higher proportion in the absence of Eα. Our results indicate that in DN cells, Eδ can direct a low level of Vα to Jα rearrangements, possibly via promoting Jα accessibility that results in the expression of a variety of different Vα gene segments. The early expressed αβTCRs allow the development of some DP thymocytes with TCRs containing mostly Vα2+,TCRα chains. In the absence of Eα, only early VαJ rearrangements with a high proportion of Vα2 would continue to be expressed in DP thymocytes and mature T cells, perhaps because their promoters do not require Eα activity to drive gene expression. This pathway of differentiation observed in Eα-deficient mice might be invisible in wt mice because of continual Tcra rearrangement in DP thymocytes that will swamp out the Eδ-initiated Vα rearrangements and, thereby, lead to a much more diverse αβTCR repertoire.
MATERIALS AND METHODS
Mice.
Mice were kept in the sterile facilities of The University of Chicago, Dana-Farber Cancer Institute, and Children's Hospital. Animal protocols were approved by the Institutional Animal and Use Committees of these institutes. C57BL/6, Rag1 −/−, and Tcrd −/− mice (also on the C57BL/6 background) were purchased from Jackson ImmunoResearch Laboratories. C57BL/6 CD1−/− were provided by A. Bendelac (The University of Chicago, Chicago, IL). Eα−/− and Eδ−/− mice were generated in the laboratory of Frederick W. Alt (10, 11). Ptcra −/− mice were described previously (7).
Flow cytometric analysis and cell sorting.
Anti-CD4 (L3T4), CD8 (53–6.7), CD25 (3C7), CD44 (IM7), NK1.1 (PK136), TCRß (H57-597), γδTCR (GL3), Va2 (B20.1), Va11 (RR8-1), and Va3 (RR3-16) mAbs were purchased from BD Biosciences. The Va8 (CTVA8) antibody was purchased from CALTAG. These mAbs were directly coupled to FITC, PE, Cy-crome, APC, or biotin. Surface marker expression on thymocytes and peripheral T cells was visualized using a FACScalibur (Becton Dickinson) and analyzed with FlowJo (Tree Star) and CellQuest software (Beckton Dickinson). Cell sorting was performed using Mo-Flo (DakoCytomation) and FACS-Aria (Becton Dickinson) sorters.
Fetal thymic organ culture.
Thymi were cultured as described previously (26). In brief, (LY5.2+) TCRβ+ CD4−CD8− NK1.1− cells were FACS purified from thymi of Ptcra −/− animals. Lineage-negative (CD3, CD8, Mac-1, NK1.1, Gr-1, Ter-119, and CD19) bone marrow progenitors were also used as a reconstitution control. Isolated embryonic day 14.5 thymi from Rag1 mice (expressing LY5.1) were initially incubated in Terasaki plates (Nunc), subsequently applied on Transwell (Nunc) porous filters, and incubated in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum (Sigma-Aldrich) plus penicillin, streptomycin, and mercapto-ethanol. Cultures were maintained at 37°C for 7 d, after which LY5.2+ cells were stained and analyzed by FACS analysis.
Acknowledgments
We would like to thank A. Bendelac for the CD1−/− animals, F. Meng for expert technical assistance, R. Duggan and J. Marvin for cell sorting, and I. Apostolou for advice and technical expertise.
I. Aifantis was supported by the National Institutes of Health (NIH) grant R01 CA105129 and the V Foundation for Cancer Research. K. Sawai is supported by a University of Chicago Molecular Biology Training grant. H. von Boehmer was supported by NIH grants R01 AI45846 and R01 AI47281. F.W. Alt was supported by NIH grant R01 AI20047 and is a Howard Hughes Medical Institute Investigator.
The authors have no conflicting financial interests.
Abbreviations used: DN, double negative; DP, double positive; SP, single positive; wt, wild type.
References
- 1.Godfrey, D.I., and A. Zlotnik. 1993. Control points in early T-cell development. Immunol. Today. 14:547–553. [DOI] [PubMed] [Google Scholar]
- 2.Rodewald, H.R., and H.J. Fehling. 1998. Molecular and cellular events in early thymocyte development. Adv. Immunol. 69:1–112. [DOI] [PubMed] [Google Scholar]
- 3.Groettrup, M., K. Ungewiss, O. Azogui, R. Palacios, M.J. Owen, A.C. Hayday, and H. von Boehmer. 1993. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor beta chain and a 33 kd glycoprotein. Cell. 75:283–294. [DOI] [PubMed] [Google Scholar]
- 4.Saint-Ruf, C., K. Ungewiss, M. Groettrup, L. Bruno, H.J. Fehling, and H. von Boehmer. 1994. Analysis and expression of a cloned pre-T cell receptor gene. Science. 266:1208–1212. [DOI] [PubMed] [Google Scholar]
- 5.Aifantis, I., V.I. Pivniouk, F. Gartner, J. Feinberg, W. Swat, F.W. Alt, H. von Boehmer, and R.S. Geha. 1999. Allelic exclusion of the T cell receptor β locus requires the SH2 domain-containing leukocyte protein (SLP)-76 adaptor protein. J. Exp. Med. 190:1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Boehmer, H. 2004. Selection of the T cell repertoire: receptor- controlled checkpoints in T cell development. Adv. Immunol. 84:201–238. [DOI] [PubMed] [Google Scholar]
- 7.Fehling, H.J., A. Krotkova, C. Saint-Ruf, and H. von Boehmer. 1995. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 375:795–798. (published erratum appears in Nature. 1995. 378:419). [DOI] [PubMed] [Google Scholar]
- 8.Buer, J., I. Aifantis, J.P. DiSanto, H.J. Fehling, and H. von Boehmer. 1997. Role of different T cell receptors in the development of pre-T cells. J. Exp. Med. 185:1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien, Y.H., M. Iwashima, K.B. Kaplan, J.F. Elliott, and M.M. Davis. 1987. A new T-cell receptor gene located within the alpha locus and expressed early in T-cell differentiation. Nature. 327:677–682. [DOI] [PubMed] [Google Scholar]
- 10.Monroe, R.J., B.P. Sleckman, B.C. Monroe, B. Khor, S. Claypool, R. Ferrini, L. Davidson, and F.W. Alt. 1999. Developmental regulation of TCR delta locus accessibility and expression by the TCR delta enhancer. Immunity. 10:503–513. [DOI] [PubMed] [Google Scholar]
- 11.Sleckman, B.P., C.G. Bardon, R. Ferrini, L. Davidson, and F.W. Alt. 1997. Function of the TCR alpha enhancer in alphabeta and gammadelta T cells. Immunity. 7:505–515. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez-Munain, C., B.P. Sleckman, and M.S. Krangel. 1999. A developmental switch from TCR delta enhancer to TCR alpha enhancer function during thymocyte maturation. Immunity. 10:723–733. [DOI] [PubMed] [Google Scholar]
- 13.Balmelle, N., N. Zamarreno, M.S. Krangel, and C. Hernandez-Munain. 2004. Developmental activation of the TCR alpha enhancer requires functional collaboration among proteins bound inside and outside the core enhancer. J. Immunol. 173:5054–5063. [DOI] [PubMed] [Google Scholar]
- 14.Bendelac, A., M.N. Rivera, S.H. Park, and J.H. Roark. 1997. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu. Rev. Immunol. 15:535–562. [DOI] [PubMed] [Google Scholar]
- 15.Eberl, G., H.J. Fehling, H. von Boehmer, and H.R. MacDonald. 1999. Absolute requirement for the pre-T cell receptor alpha chain during NK1.1+ TCRalphabeta cell development. Eur. J. Immunol. 29:1966–1971. [DOI] [PubMed] [Google Scholar]
- 16.Jouvin-Marche, E., C. Aude-Garcia, S. Candeias, E. Borel, S. Hachemi-Rachedi, H. Gahery-Segard, P.A. Cazenave, and P.N. Marche. 1998. Differential chronology of TCRADV2 gene use by alpha and delta chains of the mouse TCR. Eur. J. Immunol. 28:818–827. [DOI] [PubMed] [Google Scholar]
- 17.von Boehmer, H., I. Aifantis, O. Azogui, J. Feinberg, C. Saint-Ruf, C. Zober, C. Garcia, and J. Buer. 1998. Crucial function of the pre- T-cell receptor (TCR) in TCR beta selection, TCR beta allelic exclusion and alpha beta versus gamma delta lineage commitment. Immunol. Rev. 165:111–119. [DOI] [PubMed] [Google Scholar]
- 18.von Boehmer, H., J. Kirberg, and B. Rocha. 1991. An unusual lineage of αβ T cells which contain autoreative cells. J. Exp. Med. 174:1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borowski, C., X. Li, I. Aifantis, F. Gounari, and H. von Boehmer. 2004. Pre-TCRα and TCRα are not interchangeable partners of TCRβ during T lymphocyte development. J. Exp. Med. 199:607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruno, L., H.J. Fehling, and H. von Boehmer. 1996. The αβTCR can replace the γδTCR in the development of γδ lineage cells. Immunity. 5:343–352. [DOI] [PubMed] [Google Scholar]
- 21.Terrence, K., C.P. Pavlovich, E.O. Matechak, and B.J. Fowlkes. 2000. Premature expression of T cell receptor (TCR)αβ suppresses TCRγΔ gene rearrangement but permits development of γΔ lineage T cells. J. Exp. Med. 192:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocha, B., H. von Boehmer, and D. Guy-Grand. 1992. Selection of intraepithelial lymphocytes with CD8 alpha/alpha co-receptors by self-antigen in the murine gut. Proc. Natl. Acad. Sci. USA. 89:5336–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin, T.A., M.M. Sandau, S.C. Jameson, and K.A. Hogquist. 2005. The timing of TCRα expression critically influences T cell development and selection. J. Exp. Med. 202:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford, M.S., K.J. Young, Z. Zhang, P.S. Ohashi, and L. Zhang. 2002. The immune regulatory function of lymphoproliferative double negative T cells in vitro and in vivo. J. Exp. Med. 196:261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egawa, T., G. Eberl, I. Taniuchi, K. Benlagha, F. Geissmann, L. Hennighausen, A. Bendelac, and D.R. Littman. 2005. Genetic evidence supporting selection of the Vα14i NKT cell lineage from DP thymocyte precursors. Immunity. 22:705–716. [DOI] [PubMed] [Google Scholar]
- 26.Martin, C.H., I. Aifantis, M.L. Scimone, U.H. von Andrian, B. Reizis, H. von Boehmer, and F. Gounari. 2003. Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat. Immunol. 4:866–873. [DOI] [PubMed] [Google Scholar]