Abstract
Puma is an essential mediator of p53-dependent and -independent apoptosis in vivo. In response to genotoxic stress, Puma is induced in a p53-dependent manner. However, the transcription factor driving Puma up-regulation in response to p53-independent apoptotic stimuli has yet to be identified. Here, we show that FOXO3a up-regulates Puma expression in response to cytokine or growth factor deprivation. Importantly, dysregulated Akt signaling in lymphoid cells attenuated Puma induction upon cytokine withdrawal. Our results suggest that Puma, together with another BH3 only member, Bim, function as FOXO3a downstream targets to mediate a stress response when PI3K/Akt signaling is down-regulated.
Despite its critical role in response to DNA damage treatment, p53 is dispensable for certain stress stimuli, such as cytokine withdrawal-induced apoptosis in lymphoid cells (1, 2). In contrast, the PI3K–Akt signaling pathway has been shown to mediate cell survival under these conditions (3, 4), possibly through inhibition of FOXO transcription factors. However, the critical mediators downstream of FOXOs still remain unclear.
Puma was originally identified as a p53 downstream target (5–7). Puma deficiency is known to protect cells from genotoxic stress that causes activation of p53. Additionally, cells lacking Puma are also resistant to several p53-independent death stimuli. For instance, deficiency of Puma renders myeloid progenitor cells resistant to cytokine withdrawal (1, 2, 8, 9). Interestingly, Puma mRNA levels were up-regulated under these conditions. In addition to this, a variety of growth factors, for instance, insulin-like growth factor-1 and epidermal growth factor, can suppress Puma expression in serum-starved tumor cells (6). Notably, one of the common features shared by some of these stress stimuli that cause p53-independent Puma up-regulation is that they could attenuate the PI3K–Akt signaling pathway, which in turn modulates the activity of FOXO transcription factors.
In this report, we demonstrate that upon removing survival factors in lymphoid cells or mouse embryonic fibroblast (MEF) cells, FOXO3a (one of the FOXO family members) can regulate Puma at the transcriptional level. This indicates that abnormal PI3K–Akt signaling could exert its survival effect through attenuating critical pro-apoptotic pathways involving BH3-only family members such as Puma.
RESULTS AND DISCUSSION
The PI3K–Akt pathway is involved in cytokine withdrawal-induced apoptosis in lymphoid cells
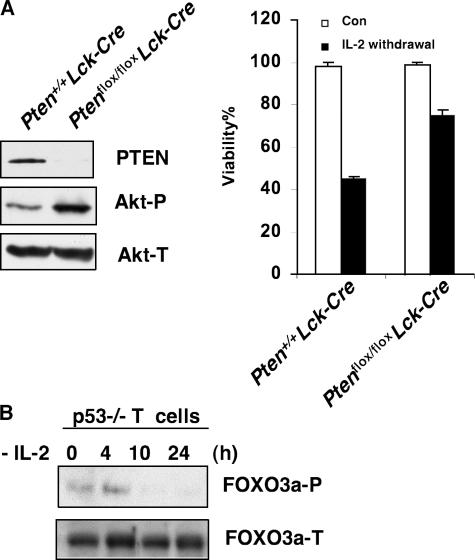
Lymphoid cells subjected to stresses such as cytokine withdrawal undergo apoptosis in a p53-independent manner (unpublished data). Our laboratory has previously reported that T cells derived from Pten flox/− Lck-Cre mice are resistant to apoptosis induced by IL-2 deprivation (10), suggesting that the mechanism mediating this cell death might involve the PI3K–Akt pathway. Pten is a critical negative regulator of the Akt pathway and frequently inactivated in human cancers (11). In this study, we confirmed that T cell–specific deletion of Pten (Pten flox/flox Lck-Cre) leads to constitutive Akt activation (Fig. 1 A, left) and that Pten-deficient T cells subjected to IL-2 withdrawal show a significant protection from IL-2 withdrawal-induced cell death (Fig. 1 A, right). These data support our hypothesis that apoptosis induced by at least some p53-independent stimuli requires the normal regulation of the PI3K–Akt pathway.
Figure 1.
PI3K/Akt/FOXO3a are involved in cytokine withdrawal-induced apoptosis in activated T cells. (A) Resistance of Pten-deficient activated T cells to apoptosis induced by IL-2 withdrawal. (left) Western blot showing levels of indicated proteins in untreated WT and Pten-deficient T cells. (right) Activated T cells were cultured in IL-2–free medium for 24 h and numbers of viable cells were determined by flow cytometric analysis. Data shown are means ± SD from three independent experiments. (B) FOXO3a phosphorylation status in IL-2–deprived activated T cells derived from p53−/− mice. Phosphorylated FOXO3a (Thr 32) (FOXO3a-P) and total FOXO3a (FOXO3a-T) were detected by Western blotting.
Activation of the survival kinase Akt leads to phosphorylation and inhibition of FOXO transcription factors. We therefore investigated whether FOXO3a is activated after removing of cytokines in lymphocytes. FOXO3a phosphorylation was markedly inhibited after removing of IL-2 (Fig. 1 B), suggesting FOXO3a is activated upon IL-2 withdrawal in primary T cells. It also indicates that compromised FOXO3a activation might account for the resistance to cytokine withdrawal-triggered apoptosis in Pten-deficient T cells.
FOXO3a directs Puma expression upon activation
As a transcriptional factor, FOXO3a has been shown to regulate cell death through several downstream targets, including p27 and Bim (3). To identify novel downstream target genes that can be induced by FOXO3a, we performed a microarray analysis. Because FOXO3a and p53 crosstalk with each other and they also share quite a few common downstream targets (12, 13), to enrich the significant hits that would be regulated by FOXO3a but independent of p53, we used inducible MEF cell lines (of the p53+/+ and p53−/− genetic backgrounds) that expressed an HA-tagged FOXO3a-TM-ER (TM-ER) construct.
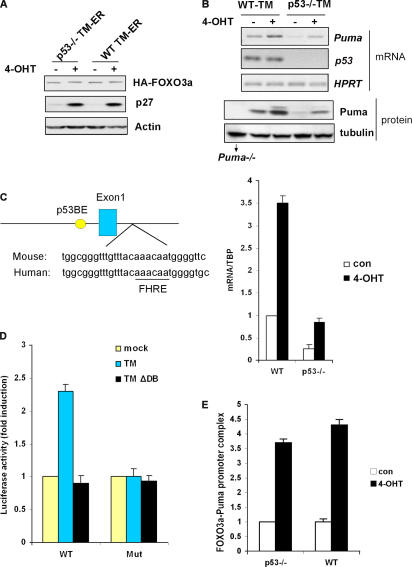
We first confirmed that the expression levels and activities of the TM-ER transgene were comparable between p53+/+ and p53−/− MEFs (Fig. 2 A). We extracted RNA from WT TM-ER and p53−/− TM-ER MEFs that had been induced for 8 h with either 4-OHT or a vehicle control. Gene expression profiles of these samples were analyzed using Affymetrix GeneChips (Mouse Genome 430 2.0). Differentially expressed candidate genes were obtained with the p-value set at 0.01 and had at least twofold induction. Consistent with previous reports, p27 was significantly up-regulated upon 4-OHT treatment (Table I). Interestingly, Puma mRNA as well as its protein levels were also induced upon TM-ER expression, even in p53−/− cells (Fig. 2 B, top). Quantification using QRT-PCR showed that the FOXO3a TM-ER induced Puma to the same degree in p53+/+ and p53−/− MEFs, taking into account the lower baseline of Puma expression in the p53−/− cells (Fig. 2 B, bottom). Our results indicate that constitutive activation of exogenous FOXO3a increases Puma expression, which might account for Puma induction under stress conditions that can activate endogenous FOXO3a.
Figure 2.
Puma is a FOXO3a transcriptional target gene. (A) p53+/+ and p53−/− FOXO3a TM-ER MEFs were exposed to 4-OHT (0.5 μM) for 6 h and lysates were subjected to Western blotting using antibodies directed against the indicated proteins. (B) Induction of Puma expression by FOXO3a-TM-ER. Levels of Puma transcripts or Puma protein in WT TM-ER and p53−/− TM-ER MEFs either left untreated (−) or treated with 4-OHT (+) for 8 h were assessed by RT-PCR or Western blotting (top) and QRT-PCR (bottom). HPRT and TBP, normalization control. Cell extracts from puma−/− MEFs were used as a negative control. QRT-PCR data are means ± SD from four independent experiments. (C) Identification of a conserved FHRE site in the human and mouse Puma promoters. p53BE, p53-binding element. (D) FOXO3a-TM activates a luciferase reporter gene driven by the Puma promoter. p53−/− MEFs were cotransfected with constructs as indicated. Luciferase assays were performed 24 h after transfection. Data shown are means ± SD from five independent experiments conducted in triplicates each time. (E) Quantification of FOXO3a association with the Puma promoter. QRT-PCR assays were conducted after chromatin IP using samples from cells that were either left untreated (con) or treated with 4-OHT. Numbers on the y-axis represent the levels of FOXO3a association with the Puma promoter region after normalizing to Ct values from input samples. Data shown are means ± SD from three independent experiments.
Table I.
Identification of FOXO3a-inducible genes by Affy-array (up-regulated: ↑; down-regulated: ↓)
| Genes | Function | Validated (QRT-PCR) |
|---|---|---|
| Bcl-2 binding component 3 (puma) |
BH-3 only family member, apoptotic factor |
+ (↑) |
| Cyclin-dependent kinase inhibitor 1B(p27) |
cell cycle regulator: G1 checkpoint |
+ (↑) |
| DnaJ (Hsp40) homologue, subfamily C, member 12 |
molecular chaperones | + (↑) |
| Insulin-like growth factor I receptor, (Igf1r) |
insulin signaling | + (↑) |
| Sestrin 1 | modulator of peroxide signaling and antioxidant defense |
+ (↑) |
| Vascular endothelial growth factor A (Vegfa) |
vasculogenesis and angiogenesis |
+ (↓) |
| Cyclin D2 | cell cycle regulator: G1/S checkpoint |
+ (↓) |
| Eph receptor A2 (Epha2) | vasculogenesis and angiogenesis |
+ (↓) |
We then determined whether FOXO3a acts directly on the murine Puma promoter. Promoter analysis identified a potential consensus FOXO-responsive element (FHRE) in intron 1 that is conserved between human and mouse (Fig. 2 C). To assess the ability of FOXO3a to regulate Puma transcription, we cotransfected p53−/− MEFs with FOXO3a-TM or FOXO3a-TMΔDB together with a reporter gene in which the Puma promoter drives the expression of a luciferase gene (14). We found that constitutively active FOXO3a efficiently induced Puma promoter-driven luciferase activity, whereas FOXO3a-TMΔDB failed to induce any reporter activity (Fig. 2 D). Replacement of the core consensus sequence AAC of the FHRE with GGG abolished FOXO3a-TM–induced luciferase activity (Fig. 2 D), indicating that this site is a functional FHRE. To test if FOXO3a could interact with the endogenous Puma promoter, a ChIP assay was performed in which the FOXO3a–TM–DNA complex was purified from p53+/+ or p53−/− FOXO3a-TM-ER cells that had been treated with 4-OHT or vehicle control. Primers flanking the putative Puma FHRE region were then used for QRT-PCR assay. We found that nuclear activated FOXO3a-TM could bind the Puma promoter regardless of p53 genotype (Fig. 2 E). Collectively, our results demonstrate that FOXO3a can act directly on the Puma promoter in a p53-independent manner.
Puma is up-regulated in lymphoid cells upon cytokine deprivation
If exogenously expressed FOXO3a could direct Puma expression through FHRE binding, one would expect that activation of endogenous FOXO3a should be able to regulate Puma. To test this, we examined Puma expression levels in activated T cells that were subjected to IL-2 withdrawal. Because Bim is a known FOXO3a downstream target under conditions of cytokine deprivation, we also analyzed the effect of IL-2 withdrawal on bim mRNA expression.
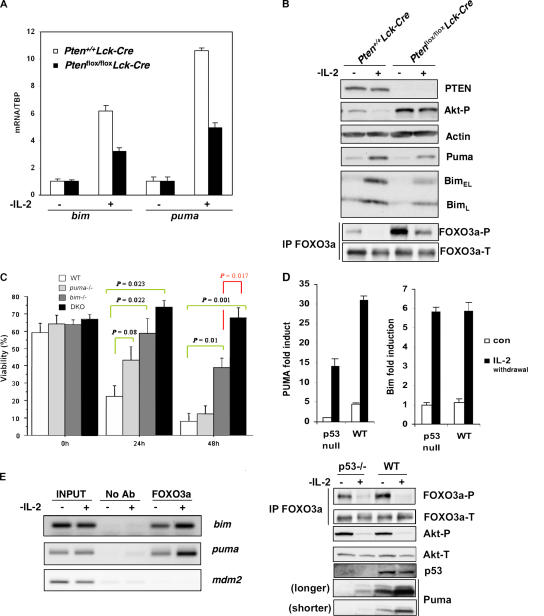
We first tested if abnormal Pten/Akt signaling had any effect on Puma and Bim expression. Using activated T cells derived from Pten +/+ Lck-Cre and Pten flox/flox Lck-Cre mice, we found the transcript levels of both bim and Puma were strikingly increased in Pten +/+ Lck-Cre cells in response to IL-2 withdrawal, whereas the loss of Pten significantly impaired Puma and bim induction in similarly treated Pten flox/flox Lck-Cre T cells (Fig. 3 A). Up-regulation of these two proteins upon IL-2 deprivation were also markedly impaired in the absence of Pten (Fig. 3 B). These results indicate that dysregulated Akt signaling may protect against cytokine deprivation-induced apoptosis partly by inhibiting the expression of proapoptotic BH3-only proteins such as Bim and Puma. Interestingly, bim deficiency confers significant protection against IL-2 withdrawal in activated T cells (15), whereas Puma deficiency confers relatively modest resistance, but nonetheless with a consistent trend, to IL-2 deprivation (Fig. 3 C, 24 h). Most importantly, lymphocytes derived from bim −/− and Puma −/− compound mutant mice are significantly resistant to cytokine withdrawal-induced apoptosis compared with lymphocytes deficient in bim alone (Fig. 3 C, 48 h, P = 0.017). This observation suggests a synergistic cooperation between these two BH3-only proteins in mediating cytokine deprivation-induced apoptosis and further indicates the total amount of activated BH3 proteins is the major determinant of cell death.
Figure 3.
FOXO3a-dependent regulation of Puma expression in lymphoid cells upon removing of IL-2. (A) Pten loss impairs Puma and bim up-regulation induced by IL-2 deprivation. RNA extracted from Pten +/+ Lck-Cre or Pten flox/flox Lck-Cre activated T cells that were either left untreated or deprived of IL-2 for 10 h was subjected to QRT-PCR. Data represent the mean and error of four independent experiments. (B) Induction of Puma and Bim protein levels was attenuated in the absence of Pten. Activated T cells were deprived of IL-2 for 24 h and cell lysates were subjected to Western blotting with antibodies as indicated. Phosphorylated FOXO3a and total FOXO3a levels were determined by immunoprecipitation and Western blotting. (C) Synergistic cooperation between Puma and Bim in mediating cytokine withdrawal-induced cell death in lymphocytes. T cells were isolated from WT, puma −/−, bim −/−, or DKO mice and expanded in the presence of IL-2 and mitogen. Activated T cells were deprived of IL-2 for the indicated times, cell viability was determined by PI staining, and FACS analysis. p-values (Student's t test) were determined by comparing indicated KO T cells to WT T cells (green line) or DKO T cells to bim −/− T cells (red line). Data shown are means ± SD from three independent experiments. (D) Induction of Puma and bim mRNA (top) and protein levels (bottom) in activated T cells. p53+/+ and p53−/− activated T cells were subjected to IL-2 withdrawal for 10 h. QRT-PCR was performed to detect Puma (top left) and bim (top right) expression levels. Fold induction was determined after normalization to TBP. Results shown are representative of three independent experiments conducted in duplicates each time. (bottom) phosphorylated FOXO3a (Thr 32) (FOXO3a-P) and total FOXO3a (FOXO3a-T) were determined by immunoprecipitation followed by Western blotting. Puma, p53, phosphorylated Akt (Ser473) (Akt-P), and total Akt (Akt-T) were detected by Western blotting. (E) FOXO3a binds to the Puma and bim promoters. Activated T cells generated from p53−/− mice were cultured in IL-2–free medium for 8 h and ChIP assays were conducted as described in Materials and methods.
We next determined Puma induction in p53+/+ and p53−/− T cells subjected to IL-2 withdrawal. Puma mRNA and protein levels were markedly increased in p53+/+ and p53−/− T cells cultured in IL-2 free medium (Fig. 3 D, left and bottom). Levels of bim mRNA were also significantly increased in T cells deprived of IL-2 (Fig. 3 D, right). Collectively, these data show that, upon cytokine withdrawal in primary lymphoid cells, Puma is up-regulated in a manner that is independent of p53 but dependent on PI3K–Akt signaling.
To confirm that the observed induction of Bim and Puma was indeed mediated by activated FOXO3a, we performed ChIP assays using FOXO3a-specific antibodies to test the physical interaction between endogenous FOXO3a and the bim or Puma promoters after IL-2 withdrawal in p53−/− lymphoid cells. This stimulation markedly increased the association of FOXO3a with the Puma or bim promoters, but not the mdm2 promoter containing p53BE (Fig. 3 E). These data suggest that FOXO3a is recruited to the Puma and bim promoters to regulate their transcription in response to stress stimuli that inactivate PI3K–Akt signaling.
FOXO3a is a direct transcriptional regulator of Puma
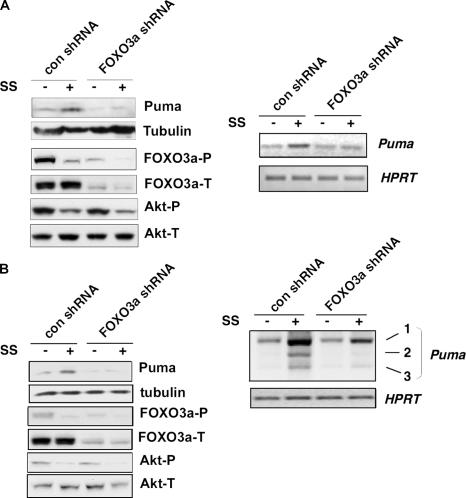
To investigate whether FOXO3a plays a direct role in Puma induction when PI3K–Akt is inactive, we ablated endogenous FOXO3a expression levels using shRNA in MEFs expressing p53QSA135V. p53QSA135V is a transcriptionally inactive form of p53 that contains two tandem mutations, L25Q and W26S, in the transactivation domain as well as a secondary point mutation, A135V, in the DNA binding domain (16, 17). Consistent with published data, we found that the mutant cells were incapable of undergoing p53-mediated apoptosis in response to DNA damage (16, 17), but still maintained a partial response to serum starvation (unpublished data). We first performed RT-PCR to examine the regulation of Puma transcripts when p53QSA135V MEFs were starved of serum. Puma transcript levels were up-regulated upon serum deprivation in p53QSA135V MEFs, but ablation of FOXO3a abrogated this induction (Fig. 4 A). Because Puma −/− MEFs are resistant to serum starvation (9), it further suggests a critical role of Puma in mediating stress responses associated with activation of FOXO transcription factors. FOXO3a-dependent induction of Puma in response to serum deprivation was also reproduced in 293T cells expressing inactive p53 (Fig. 4 B). These results implicate a direct role of FOXO3a in transcriptionally regulating Puma.
Figure 4.
Direct role of FOXO3a in transcriptional regulating Puma. (A and B) FOXO3a is required for induction of Puma upon serum withdrawal. (left) Western blots show that FOXO3a shRNA successfully blocked FOXO3a expression in p53QSA135V cells (A) or 293T cells (B). Protein levels of Puma, Akt-T/P, and tubulin before and after serum starvation (24 h) were detected by Western blotting. (right) RT-PCR shows that Puma transcripts were elevated in serum-starved p53QSA135V cells (A) or 293T cells (B) expressing control shRNA (con). Ablation of endogenous FOXO3a by FOXO3a shRNA inhibited this elevation in Puma expression in both cell types. SS, serum starvation.
Forkhead transcription factors are tightly regulated at posttranslational levels by kinases including Akt and SGK1 (12, 18, 19). Among all the FOXO downstream targets reported so far, several of them are also transcriptionally regulated by p53 (13). In this report, we identified Puma as another common downstream target of FOXO3a and p53. Induction of Puma upon oncogenic signals or DNA damage is p53-dependent and Puma functions as an essential death executer in initiating mitochondrial apoptosis. Loss of Puma can recapitulate the majority, if not all, of the apoptotic-resistant phenotype observed in p53-deficient primary cells (8, 9). Additionally, Puma can be induced in a p53-independent manner when cells are deprived of cytokines or growth factors. The transcription factor responsible for this induction has not been clearly defined in the literature. Given that cytokine and serum withdrawal down-regulate the PI3K–Akt pathway, which in turn leads to the activation of forkhead transcription factors, we hypothesized that FOXO3a may play an important role in regulating Puma and other BH3-only proapoptotic Bcl-2 family members. Indeed, our results demonstrate that FOXO3a is recruited to both Puma and bim promoter regions and that transcription of these two genes in response to cytokine deprivation is regulated by FOXO3a. Furthermore, if a critical negative regulator of the Akt pathway, Pten, is lost, then Akt activity remains constantly high. This leads to an attenuated response in Puma and bim induction upon cytokine withdrawal. This, together with other factors, is responsible for protecting activated T cells deficient in Pten from IL-2 withdrawal-induced apoptosis. In the context of IL-2 deprivation, Bim certainly contributes more compared with Puma, based on results from knockout murine models. One explanation is that, in addition to being regulated at the transcriptional level, Bim is also regulated by posttranslational modification. The kinetics of the latter are much faster and are most likely the result of kinases (such as JNK) that become activated in the absence of survival cytokines (20). Notably, active JNK can promote Bim activity by posttranslationally modifying the molecule. But in addition, active JNK can also promote the activation of FOXO3a (21, 22), which in turn can transcriptionally up-regulate downstream target genes including Puma and bim. A reasonable hypothesis is that transcriptional up-regulation of Puma and Bim by FOXO3a serves as a second wave to boost the intensity of the proapoptotic signal through Puma and Bim with subsequent activation of Bax and Bak. Unlike Bim, the regulation of Puma so far has only been attributed to transcriptional regulation. Thus, loss of Bim has a more profound effect on cells (because it is downstream of multiple signaling pathways) compared with loss of Puma in situations in which cytokines and growth factors are withdrawn or limiting.
Because BH3-only family members are involved in diverse stress responses, identifying transcription factors that are responsible for regulating their expression levels under different stress conditions is of great importance. Furthermore, because the FOXO family members share certain overlapping functions in response to some stimuli, it will be interesting to explore if other FOXOs (FOXO1 and FOXO4) could also regulate Puma expression upon aforementioned stimuli. Because FOXOs play distinct roles during embryonic development (23), their functional redundancy in stress response might be tissue-type and stress-type dependent. Interestingly, lymphocytes derived from FOXO3a-deficient mice generated by a gene trap strategy respond normally to cytokine withdrawal, but these mice develop a lymphoproliferative and inflammatory condition (24). Pten flox/− Lck-Cre mutant mice have a similar phenotype in that there is enhanced proliferation of T cells and concurrent production of cytokines, but in addition loss of Pten renders lymphocytes resistant to IL-2 withdrawal, as does transgenic overexpression of constitutively active Akt (25). Investigating the regulation of Bim or Puma expression upon germline deletion of FOXO3a may provide an answer.
MATERIALS AND METHODS
Mice, cell culture, and manipulation.
Trp53QSA135V primary MEF cells were provided by G. Wahl (The Salk Institute, La Jolla, CA). To generate FOXO3a-related stable cell lines, MEFs were cotransfected with pECE HA-FOXO3a (TM or DB) and a GFP plasmid in 15:1 ratio. Cells were selected by GFP sorting 48 h later and GFP-positive cells were pooled together for use in experiments. T cell–specific Pten-deficient mice were generated as described previously (10). Littermates carrying Lck-Cre and the floxed Pten mutation on both alleles (Pten flox/flox Lck-Cre), or Lck-Cre and the wild-type Pten gene (Pten +/+ Lck-Cre) were used for analysis as homozygous mutant and wild-type mice, respectively. puma and bim double knockout mice were generated by breeding puma +/− bim +/− double het mice together. All mice were either housed in a specific pathogen-free animal facility in the Ontario Cancer Institute (animal studies were approved by the OCI review board) or in accordance with the Austrian “Tierversuchsgesetz” and governed in Austria by the Bundesministerium für Bildung, Wissenschaft and Kultur; animal experiments were performed according to the guidelines of the Melbourne Directorate Animal Ethics Committee.
T cell purification, activation, and cell viability analysis.
Spleen and lymph nodes were isolated and single cell suspensions were made. Cells were treated with Red Cell Lysis Buffer and resuspended in PBS (with 10% FCS) containing the following biotinylated antibodies: anti–mouse B220 (RA3-6B2), anti–mouse CD11b (m1/70), anti-Ter119 (Ly-76), and anti–mouse Gr1 (RB6-8C5) (BD Biosciences). After a 30-min incubation on ice, peripheral T cells were recovered by negative depletion using BD Biosciences IMag streptavidin particles according to manufacturer's instructions. Cell culture media contained the following: Iscove media supplemented with 10% FCS, 55 μM 2-ME, 2 mM sodium pyruvate. T cells were activated by adding 10 ng/ml rmIL-2, 2 ng/ml PMA, and 100 ng/ml ionomycin for 3 d, then expanded in IL-2 alone for a further 2 d. For lymphocytes isolated from puma −/−, bim −/−, or DKO, activation of T cells were performed as described previously (15). Cell viability was determined by propidium iodide staining using FACScan (Becton Dickinson).
Constructs and antibodies.
The pECE-HA-FOXO3a-TM and TMΔDB expression plasmids were provided by M. Greenberg (Harvard Medical School, Boston, MA). TM is a triple mutant form of FOXO3a in which all three Akt phosphorylation sites are mutated to alanine (FOXO3a-TM, T32A/S253A/S315A) and TMΔDB is a form of TM that lacks the FOXO3a DNA-binding domain. In TM-ER, TM is expressed as a fusion protein with the modified estrogen receptor. When TM-ER binds the exogenously administered inducer 4-hydro-tamoxifen (4-OHT), the TM-ER protein translocates to the nucleus and remains there as a constitutively active form of FOXO3a. ShRNA sequences: 5′-GGCAAGAGCTCTTGGTGGAT-3′ (murine FOXO3a); 5′-GTGGAGCTGGACCCGGAGT-3′ (human FOXO3a). pSIRIPP retroviral vector was used for cloning and delivering shRNA by either retroviral infection (MEFs) or transfection (293T cell). Stable cells are pools of clones selected by puromycin. Antibodies were obtained from the following sources: anti-HA (12CA5, Roche); anti-phospho–Akt, anti-Bim, and anti-total–Akt (Cell Signaling); anti-PTEN (clone 6H2.1, CASCADE BioScience); and anti-Thr32-FOXO3a/anti-FOXO3a (Upstate Biotechnology), anti-Puma (ProSci), and anti-p53 (Santa Cruz Biotechnology, Inc.).
RT-PCR and real-time PCR.
Total RNA was extracted with TRIzol (Invitrogen) and purified using the RNeasy kit (QIAGEN) according to the manufacturer's protocol with the addition of “on column-DNase treatment” (QIAGEN). RNA (4 μg) was reverse-transcribed in a 20-μl reaction using the Superscript first strand RT-PCR kit (Invitrogen). After RNase H treatment at 37°C for 20 min and after inactivation by incubating samples at 95°C for 2 min, the RT reaction was diluted. cDNA was used for RT-PCR or real-time PCR. Primer sequences were as follows: murine Puma: 5′-CTGTATCCTGCAGCCTTTGC-3′, 5′-ACGGGCGACTCTAAGTGCT-3′; Taqman probe: GGACGGTCCTCAGCCCTCCCTGTCAC-3′; human Puma primer sequences were from (7); murine bim: 5′-CGACAGTCTCAGGAGGAACC-3′, 5′-CCTTCTCCATACCAGACGGA-3′.
ChIP assay.
ChIP was performed as described previously (12). Real-time PCR (for quantification of ChIP assay) conditions: 94°C, 45 s; 60°C, 45 s; and 72°C, 45 s. Ct value of each sample was normalized to the Ct value obtained from PCR reaction using the corresponding input genomic DNA as templates. Primer sequences were as follows: murine Puma FHRE: 5′-GAAGGAAGCACCTGGGACTC-3′; 5′-TCCTCCCAGGTCTCACTAGC-3′; murine bim FHRE 1: 5′-GGGCGGGTACATTCTGAGT-3′; 5′-CAGGCTGCGACAGGTAGTG-3′.
Luciferase assays.
Murine Puma luciferase reporter was a gift from T. Look (Dana-Farber Cancer Institute, Boston, MA) (for detailed information, see reference 14). Mutant Puma luciferase-reporter construct was generated using Quickchange Site-Directed Mutagenesis Kit (Stratagene). Primer sequences for mutagenesis assay were 5′-GGCGGGTTTGTTTACAGGGAATGGGGTTCGGGC-3′ and 5′-GCCCGAACCCCATTCCCTGTAAACAAACCCGCC-3′. p53−/− MEFs were seeded in 12-well plates at a density of 3 × 105/well and were cotransfected with FOXO3a-TM or TMΔDB (1 μg) and Puma promoter–driven luciferase reporter constructs (0.5 μg) together with 50 ng β-Galactosidase construct. At 24 h after transfection, cells were lysed in 100 μl lysis buffer and luciferase activity was assayed using the Luciferase Assay System kit (Promega) according to the manufacturer's protocol. β-Galactosidase activity was assayed according to the manufacturer's instructions (β-Galactosidase enzyme assay system kit; Promega). We thank M. Greenberg, T. Look, A. Strasser, and G. Wahl for reagents and mice and M. Saunders for scientific editing.
Acknowledgments
This work was supported by the Terry Fox Cancer Foundation of the National Cancer Institute of Canada. A. Villunger is supported by grants from the Austrian Science Fund -FWF (SFB021, START). H. You is a recipient of a postdoctoral fellowship from the Cancer Research Institute, New York.
The authors have no conflicting financial interests.
References
- 1.Clarke, A.R., C.A. Purdie, D.J. Harrison, R.G. Morris, C.C. Bird, M.L. Hooper, and A.H. Wyllie. 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 362:849–852. [DOI] [PubMed] [Google Scholar]
- 2.Strasser, A., A.W. Harris, T. Jacks, and S. Cory. 1994. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 79:329–339. [DOI] [PubMed] [Google Scholar]
- 3.Stahl, M., P.F. Dijkers, G.J. Kops, S.M. Lens, P.J. Coffer, B.M. Burgering, and R.H. Medema. 2002. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 168:5024–5031. [DOI] [PubMed] [Google Scholar]
- 4.Chen, W.S., P.Z. Xu, K. Gottlob, M.L. Chen, K. Sokol, T. Shiyanova, I. Roninson, W. Weng, R. Suzuki, K. Tobe, et al. 2001. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 15:2203–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu, J., L. Zhang, P.M. Hwang, K.W. Kinzler, and B. Vogelstein. 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell. 7:673–682. [DOI] [PubMed] [Google Scholar]
- 6.Han, J., C. Flemington, A.B. Houghton, Z. Gu, G.P. Zambetti, R.J. Lutz, L. Zhu, and T. Chittenden. 2001. Expression of bbc3, a proapoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc. Natl. Acad. Sci. USA. 98:11318–11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano, K., and K.H. Vousden. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell. 7:683–694. [DOI] [PubMed] [Google Scholar]
- 8.Jeffers, J.R., E. Parganas, Y. Lee, C. Yang, J. Wang, J. Brennan, K.H. MacLean, J. Han, T. Chittenden, J.N. Ihle, et al. 2003. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 4:321–328. [DOI] [PubMed] [Google Scholar]
- 9.Villunger, A., E.M. Michalak, L. Coultas, F. Mullauer, G. Bock, M.J. Ausserlechner, J.M. Adams, and A. Strasser. 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 302:1036–1038. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki, A., M.T. Yamaguchi, T. Ohteki, T. Sasaki, T. Kaisho, Y. Kimura, R. Yoshida, A. Wakeham, T. Higuchi, M. Fukumoto, et al. 2001. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 14:523–534. [DOI] [PubMed] [Google Scholar]
- 11.Cully, M., H. You, A.J. Levine, and T.W. Mak. 2006. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 6:184–192. [DOI] [PubMed] [Google Scholar]
- 12.You, H., Y. Jang, A.I. You-Ten, H. Okada, J. Liepa, A. Wakeham, K. Zaugg, and T.W. Mak. 2004. p53-dependent inhibition of FKHRL1 in response to DNA damage through protein kinase SGK1. Proc. Natl. Acad. Sci. USA. 101:14057–14062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran, H., A. Brunet, J.M. Grenier, S.R. Datta, A.J. Fornace Jr., P.S. DiStefano, L.W. Chiang, and M.E. Greenberg. 2002. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 296:530–534. [DOI] [PubMed] [Google Scholar]
- 14.Wu, W.S., S. Heinrichs, D. Xu, S.P. Garrison, G.P. Zambetti, J.M. Adams, and A.T. Look. 2005. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 123:641–653. [DOI] [PubMed] [Google Scholar]
- 15.Bouillet, P., D. Metcalf, D.C. Huang, D.M. Tarlinton, T.W. Kay, F. Kontgen, J.M. Adams, and A. Strasser. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 286:1735–1738. [DOI] [PubMed] [Google Scholar]
- 16.Jimenez, G.S., M. Nister, J.M. Stommel, M. Beeche, E.A. Barcarse, X.Q. Zhang, S. O'Gorman, and G.M. Wahl. 2000. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat. Genet. 26:37–43. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, T.M., E.M. Hammond, A. Giaccia, and L.D. Attardi. 2005. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat. Genet. 37:145–152. [DOI] [PubMed] [Google Scholar]
- 18.Brunet, A., A. Bonni, M.J. Zigmond, M.Z. Lin, P. Juo, L.S. Hu, M.J. Anderson, K.C. Arden, J. Blenis, and M.E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 96:857–868. [DOI] [PubMed] [Google Scholar]
- 19.Brunet, A., J. Park, H. Tran, L.S. Hu, B.A. Hemmings, and M.E. Greenberg. 2001. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol. Cell. Biol. 21:952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ley, R., K.E. Ewings, K. Hadfield, and S.J. Cook. 2005. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 12:1008–1014. [DOI] [PubMed] [Google Scholar]
- 21.Wang, M.C., D. Bohmann, and H. Jasper. 2005. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 121:115–125. [DOI] [PubMed] [Google Scholar]
- 22.Essers, M.A., S. Weijzen, A.M. de Vries-Smits, I. Saarloos, N.D. de Ruiter, J.L. Bos, and B.M. Burgering. 2004. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23:4802–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosaka, T., W.H. Biggs III, D. Tieu, A.D. Boyer, N.M. Varki, W.K. Cavenee, and K.C. Arden. 2004. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. USA. 101:2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, L., J.D. Hron, and S.L. Peng. 2004. Regulation of NF-κB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 21:203–213. [DOI] [PubMed] [Google Scholar]
- 25.Jones, R.G., S.D. Saibil, J.M. Pun, A.R. Elford, M. Bonnard, M. Pellegrini, S. Arya, M.E. Parsons, C.M. Krawczyk, S. Gerondakis, et al. 2005. NF-κB couples protein kinase B/Akt signaling to distinct survival pathways and the regulation of lymphocyte homeostasis in vivo. J. Immunol. 175:3790–3799. [DOI] [PubMed] [Google Scholar]