Abstract
Regulatory T (T reg) cells are critical regulators of immune tolerance. Most T reg cells are defined based on expression of CD4, CD25, and the transcription factor, FoxP3. However, these markers have proven problematic for uniquely defining this specialized T cell subset in humans. We found that the IL-7 receptor (CD127) is down-regulated on a subset of CD4+ T cells in peripheral blood. We demonstrate that the majority of these cells are FoxP3+, including those that express low levels or no CD25. A combination of CD4, CD25, and CD127 resulted in a highly purified population of T reg cells accounting for significantly more cells that previously identified based on other cell surface markers. These cells were highly suppressive in functional suppressor assays. In fact, cells separated based solely on CD4 and CD127 expression were anergic and, although representing at least three times the number of cells (including both CD25+CD4+ and CD25−CD4+ T cell subsets), were as suppressive as the “classic” CD4+CD25hi T reg cell subset. Finally, we show that CD127 can be used to quantitate T reg cell subsets in individuals with type 1 diabetes supporting the use of CD127 as a biomarker for human T reg cells.
Over the past decade, there have been tremendous advances in our understanding of the basic processes that control immune tolerance. The identification of CD4+CD25+ regulatory T (T reg) cells as an important component of self-tolerance has opened a major area of investigation in immunology and numerous studies have demonstrated the potent influence of T reg cells in suppressing pathologic immune responses in autoimmune diseases, transplantation, and graft-versus-host disease (for reviews see references 1–6). T reg cells have a unique and robust therapeutic profile. The cells require specific TCR-mediated activation to develop regulatory activity, but their effector function appears to be nonspecific, regulating local inflammatory responses through a combination of cell–cell contact and suppressive cytokine production (7–9). Moreover, there are several therapeutic interventions that appear to promote T reg cell development and function (10, 11). This so-called “adaptive” T reg cell population shares many of the attributes of thymic-dependent, natural T reg cells, but can differ in critical cell surface biomarkers and functional attributes (12). For instance, Tr1 and Th3 cells have been described that produce IL-10 and TGFβ, respectively (13, 14). These results have led to novel approaches in immunotherapy just as the ability to isolate and expand this cell subset in mice has led to novel therapeutic interventions in immunological diseases (6, 15). However, a major obstacle to the study and application of T reg cells in the human setting has been the lack of specific cell surface biomarkers to define and separate T reg cells from other regulatory or effector T cell subsets.
Although many studies indicate that CD25 is a crucial cell surface marker for the regulatory subset (16, 17), unlike the mouse, several studies have suggested that only the CD4+ T cell subset expressing the highest levels of CD25 (termed CD25hi) have in vitro suppressive activity (16). Moreover, the addition of other markers such as HLA-DR suggest that even a lower percentage (often <1%) of CD4+ T cells comprise the suppressive T cell subset. Finally, some markers such as CTLA-4 and GITR, which have been reported to be expressed on T reg cells (18–21), are also expressed on potent effector T cells and as such make immunophenotyping and determination of their functional role problematic (22, 23). This has led to several disparate reports of T reg cell quantification in disease settings. For instance, some studies suggest that the quantity of CD4+CD25hi T reg cells is deficient in type 1 diabetes (T1D) (24), whereas others suggest that the number and function of these cells is normal in T1D (25). Moreover, the ability to isolate only limited numbers of these cells from peripheral blood has made expansion of this regulatory cell population problematic.
One significant advance in the study of mouse and human T reg cells has been the discovery of the transcription factor, FoxP3, as a major marker and functional regulator of T reg cell development and function (26–29). In a series of elegant mouse and human genetic studies, investigators demonstrated that mutations in the FoxP3 gene were linked to the autoimmune manifestations observed in the Scurfy mouse and humans with immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) disease (28). Subsequent studies in the mouse showed that FoxP3-deficient animals lack T reg cells, whereas overexpression of the FoxP3 protein leads to profound immune suppression (30). Although recent studies have questioned whether all T reg cells are FoxP3+ or whether all FoxP3+ T cells are regulatory, FoxP3 protein remains the best and most specific marker of T reg cells to date (30). In this regard, flow cytometric and immunohistochemical analyses that FoxP3 is expressed in significantly more T cells than previously identified using the other available cell surface markers, including CD25. FoxP3 protein is found in CD25low and negative CD4+ T cells and under certain conditions some CD8+ T cells (30, 31). Thus, it is likely that many of the natural and adaptive T reg cells are missed in current biomarker studies, calling into question the conclusions related to deficiencies or defects in certain autoimmune settings. Importantly, as FoxP3 is an intracellular protein, it cannot be used to separate human T reg cells for functional studies or in vivo expansion for cellular therapy, thereby limiting its use in the human setting.
In an effort to define new biomarkers of human T reg cells, we have combined gene expression microarray, flow cytometry, and functional assays to identify new cell surface proteins that distinguish human T reg cells. We observed that IL-7R (CD127) is down-regulated on all human T cells after activation. In contrast with the reported reexpression of CD127 on the majority of effector and memory T cells (32–35), FoxP3+ T cells remain CD127lo/−. In fact, the CD127lo/−, FoxP3+ T cells accounted for a significant percentage of CD4+ T cells in the peripheral blood. We demonstrate that FoxP3 interacts with the CD127 promoter and, given its purported repressor function, likely contributes to the reduced expression of CD127 in T reg cells. Finally, we show that the isolated CD4+ CD127lo/− T cell subset is anergic and suppresses alloantigen responses in vitro. Together, these data suggest a dichotomy between memory T cells, namely IL-2RloIL-7Rhi and regulatory FoxP3+ T cells, which in most instances up-regulate IL-2R while remaining IL-7Rlo/− (30). Thus, the CD127 biomarker can be used to selectively enrich human T reg cells for in vitro functional studies and potentially in vivo therapy.
RESULTS
Lack of correlation between FoxP3 and CD25 in human CD4+ T cells
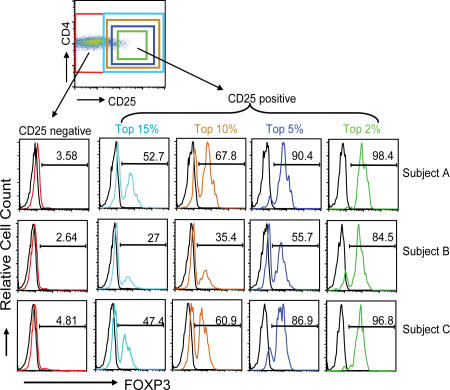
Previous studies in mouse using FoxP3-GFP knockin mice have demonstrated that FoxP3 does not always correlate with CD25 expression (36). Because current efforts in humans have focused on the use of CD25 to isolate and quantify T reg cells, we analyzed the expression of FoxP3 in the various CD4+ T cell subsets. Peripheral blood cells from normal subjects were purified on Ficoll gradients and cell surfaces stained with anti-CD4 and anti-CD25 mAbs. This staining was followed by cell membrane permeabilization and intracellular staining with a monoclonal anti-FoxP3 mAb. As seen in Fig. 1, although the majority of CD4+CD25hi cells (top 2% of gate) were FoxP3+ (ranging from 84.5–96.8% in three individuals), there were considerable numbers of FoxP3+ cells that were CD25 dull or even negative. In fact, if FoxP3 expression is assessed on all the CD4+CD25+ T cells, independent of level of CD25 expression, between 27 and 52.7% of the cells were FoxP3+. Thus, up to 7.5% of the CD4+ T cells express this putative T reg cell marker. No significant staining was observed using an isotype control IgG-Alexa 488, whereas a second anti-FoxP3 mAb from Biolegend gave similar results (unpublished data). An analysis of FoxP3 expression in the CD4+CD25− T cell subset showed that <5% of the CD25−CD4+ T cells expressed FoxP3, although that percentage was probably an overestimate as the result of some background staining using the isotype control Ig. However, given the large numbers of cells in this gate, there are likely to be at least some CD4+CD25− T cells that are FoxP3+. Thus, rather than <2% of the CD4+ T cells falling into a putative T reg cell subset, as many as 8–10% of the CD4+ T cells may be regulatory in nature.
Figure 1.
FoxP3 is expressed on a significant percentage of CD4+ T cells independent of CD25 expression. Human PBMCs were cell surface stained using a combination of anti-CD4 and anti-CD25 mAbs. Once fixed, the cells were stained additionally with anti-FoxP3 mAb. Data are representative of >20 independent individuals and >10 experiments. The numbers in the histograms indicate the percentage of FoxP3+ cells.
Analysis of novel T reg cell–specific cell surface molecules
To identify additional cell surface markers associated with function and phenotype of T reg cells, microarray analysis was performed comparing mRNA expressed by CD4+CD25hi T cells with CD4+CD25neg T cells isolated from healthy donor PBMCs. mRNA was prepared from three blood donors and cRNA was prepared and tested on Affymetrix U133A GeneChips. The sorting parameters were based on published studies in which the top 1–2% of CD4+CD25+ T cells was selected as the prototypic T reg cell subset (16, 37). Among the genes that differed between the two subsets, IL-7R (CD127) expression was noted to be expressed at 2.4-fold lower levels in CD4+CD25hi T cells as compared with CD4+CD25neg T cells. To confirm the findings, mRNA isolated from three independent CD4+CD25hi and CD4+CD25neg T cell preparations was examined by quantitative real-time PCR (qPCR). Expression of CD127 mRNA was inversely correlated with CD25 expression. In fact, the level of expression was 3.14 lower in the CD4+CD25hi T cells as compared with CD4+CD25− T cells (range: 2.26–4.21-fold).
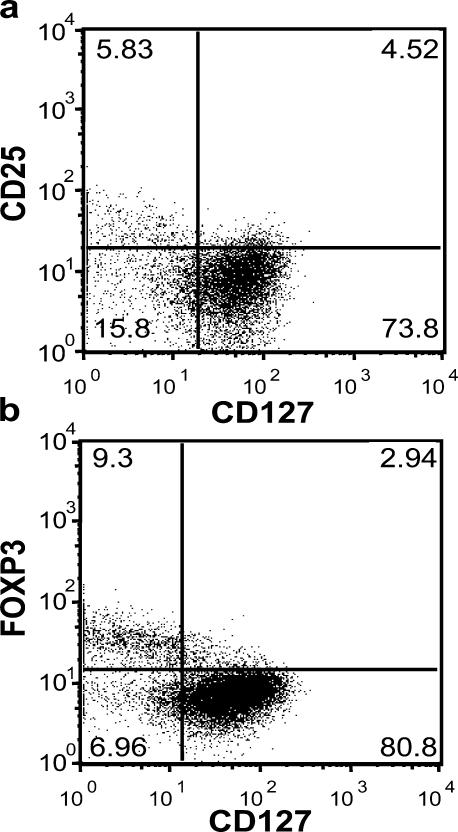
As predicted by the gene expression studies, the majority of the CD4+CD25+ cells, especially the CD4+CD25hi T cells had low expression of CD127 (Fig. 2 a). However, not all of the CD4+CD127lo/− T cells were CD25+. In fact, a significant percentage of CD4+CD127− T cells (15.8% in this individual) was CD25 negative. That said, the majority of the CD4+CD25− T cells were CD127 bright (73.8%), accounting for the differential expression observed in the gene array analyses. More importantly, flow cytometric analysis of FoxP3 expression in CD127 positive and negative T cell subsets showed that the majority of FoxP3+ T cells were in the CD127lo/– T cell subset (Fig. 2 b). Interestingly, the relative expression of CD127 was inversely correlated with FoxP3 with the highest FoxP3-expressing CD4+ T cells expressing the lowest levels of CD127. These results were uniformly observed in >20 individuals examined.
Figure 2.
Expression of FoxP3 on different CD4+CD127+/− human T cell subsets. (a) PBMCs were harvested from human peripheral blood and stained with CD4, CD25, CD127 as well as intracellularly with FoxP3-specific mAbs, then analyzed on a Becton-Dickinson FACSCalibur. (b) Human PBMCs were stained for cell surface expression of CD4 and CD127. The stained cells were fixed and stained intracellularly for FoxP3. For analysis, the PBMCs were gated on lymphocytes (based on forward and side light scatter) and analyzed for CD127 and FoxP3 expression. The numbers in the dot plot indicate the percentage of gated cells expressing the relevant marker. Data are representative of >20 independent individuals and >10 experiments.
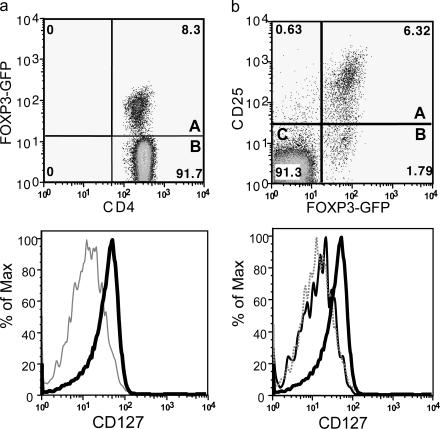
Similar results were observed in mice. CD4+ T cells isolated for FoxP3-GFP knockin mice were stained for CD127 (36, 38). The vast majority of the mouse CD4+FoxP3+ T cells were CD127lo/− (Fig. 3 a). Additional analyses of these mice showed that CD4+CD25+FoxP3+ T reg cells were CD127lo/−, however, as in humans, CD127 was a better marker than CD25 because all the CD4+ T cells were CD127lo/− independent of CD25 expression (Fig. 3 b).
Figure 3.
Expression of FoxP3 on different CD4+CD127+/− mouse T cell subsets. Mouse spleen and lymph node cells were stained for cell surface expression of CD4 and CD127. For analysis, the spleen cells from FoxP3-GFP mice were gated on lymphocytes (based on forward and side light scatter) and analyzed for CD127 and FoxP3 (GFP) expression. The numbers in the dot plot indicate the percentage of gated cells expressing the relevant marker. The letters A and B in the quadrants represent the gray and thick solid lines (bottom). (b) Spleen and lymph node cells isolated from FoxP3 transgenic mice were stained for cell surface expression of CD4, CD25, and CD127. For analysis, the spleen cells were gated on CD4+ lymphocytes (based on forward and side light scatter) and analyzed for CD127 and FoxP3 expression. The numbers in the dot plot indicate the percentage of gated cells expressing the relevant marker. The letters A, B, and C in the quadrants represent the dotted, thin solid, and thick solid lines (bottom).
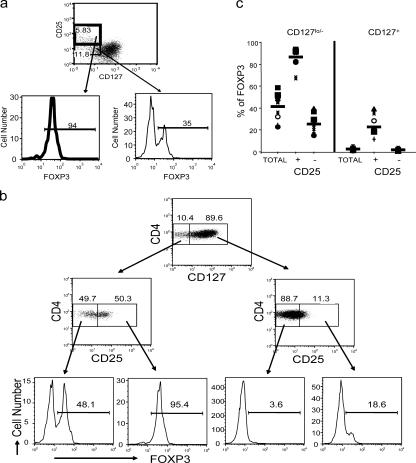
Further studies were conducted to determine the relationship of CD4, CD127, FoxP3, and CD25 using multiparameter flow cytometry (Fig. 4). The overwhelming majority of CD4+CD25+CD127lo/− T cells express FoxP3 (94% in this individual) (Fig. 4 a). However, a significant percentage of CD4+CD25−CD127lo/− cells are also FoxP3+ (35% in this individual), although the mean fluorescence is often less than the CD4+CD25hi cells. In sharp contrast, there were few FoxP3+ T cells in the CD4+CD127+ subset except a small percentage among those that expressed CD25 (Fig. 4 b). Interestingly, back-gating of the CD4+CD127+FoxP3+ subset showed that the expression of CD25 in these T cells was intermediate and distinct from the CD25hi subset described as “classic” T reg cells (unpublished data), suggesting that they may be a transitional cell. Finally, it is important to note that in this individual (which is representative of most individuals tested), there was a significant percentage of FoxP3+ T cells in the CD4+CD25−CD127lo/− T cell subset. Back-gating showed that these cells fell within the CD25int populations (unpublished data). Similar results were observed in PBMCs obtained from 10 healthy donors stained for cell surface expression of CD4, CD127, CD25 followed by intracellular staining with FoxP3-specific mAb (Fig. 4 c and Table I). Examination of multiple individuals confirmed that the majority of the CD4+FoxP3+ T cells were within the CD25+CD127lo/− subset; however, in some individuals, a significant percentage of the CD25−CD127lo/− and/or CD25+CD127+ T cells were FoxP3+. As seen in Table I, ∼40% of the CD4+CD127lo/− cells were FoxP3+ as compared with only 2.5% of the CD4+CD127+. Significantly, on average >85% of the CD4+CD25+CD127lo/− cells were FoxP3+. Thus, CD127 is a better cell surface marker than CD25 for the identification of CD4+FoxP3+ T cells; however, the best combination of cell surface markers is CD4+CD25+CD127lo/−, which accounts for ∼80% of the FoxP3+ depending on the individual. Thus, a broad gating strategy of CD4+CD25+CD127lo/− results in a highly purified FoxP3+ T cell population as compared with the other subsets (Fig. 4 b).
Figure 4.
Expression of FoxP3 on different CD4+ T cell subsets. (a) Human PBMCs were stained for cell surface expression of CD4, CD25, and CD127. The stained cells were fixed and stained intracellularly for FoxP3. For analysis, the PBMCs were gated on CD4+ lymphocytes (based on forward and side light scatter and CD4 staining) and analyzed for CD127 and FoxP3 expression. The boxes represent arbitrary designations of CD25+ versus CD25− cells. The numbers in the histograms indicate the percentage of gated cells expressing the relevant marker. (b) Human PBMCs were stained for cell surface expression of CD4, CD25, and CD127. The stained cells were fixed and stained intracellularly for FoxP3. For analysis, the PBMCs were gated on lymphocytes (based on forward and side light scatter) and analyzed for CD4, CD25, CD127, and FoxP3 expression. The boxes represent arbitrary designations of CD127+ versus CD127lo/− cells. The numbers in the dot plot indicate the percentage of gated cells expressing the relevant marker. (c) Similar staining and analysis was performed on whole blood obtained from 10 healthy individuals. Each symbol represents an individual person and the narrow bar represents the mean percentage of FoxP3+ T cells on either CD4+ T cells gated based on CD25 and/or CD127 expression.
Table I.
Percent of FoxP3+ T cells in CD127 subsets
| Mean % of FoxP3 | Range | |
|---|---|---|
| CD4+ CD127lo/− | 41.5 | 22.8–58.5 |
| CD4+ CD127+ | 2.5 | 1.2–5.0 |
| CD4+ CD127lo/−CD25+ | 86.6 | 67.4–93.6 |
| CD4+ CD127lo/−CD25− | 25.5 | 14.8–39.5 |
| CD4+ CD127+ CD25+ | 22.9 | 11.5–39.2 |
| CD4+ CD127+ CD25− | 1.8 | 1.0–3.3 |
The average CD4+ CD127 lo/− gate was 14% (range 10–19%).
ChIP-chip analysis of FoxP3 interaction with CD127
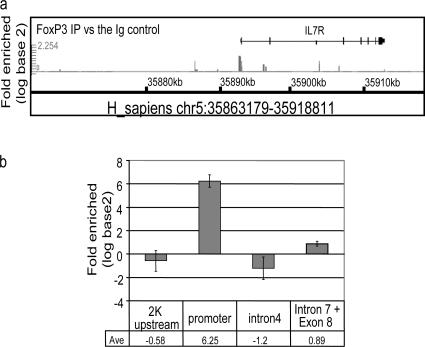
The data clearly showed a “general” relationship between FoxP3 expression and CD127 down-regulation. However, we were struck with the apparently inverse correlation between FoxP3 and CD127 protein (Fig. 2 b). These results suggested that there may be a direct structural relationship between the transcription factor, FoxP3, and CD127 transcription. This was especially attractive given previous studies suggesting that FoxP3 represses gene expression (39). Chromatin immunoprecipitation (ChIP) of transcription factor-bound genomic DNA followed by microarray hybridization (chip) of IP-enriched DNA is a new technology that allows genome-wide analysis of transcription factor binding. ChIP-chip data are different than classical microarray gene expression data obtained by measuring mRNA levels in that they examine direct control of gene transcription, not just potentially indirect downstream regulation. We performed ChIP-chip experiments on anti-CD3/anti-CD28–expanded CD4+CD25hi human T reg cells (37). Anti-FoxP3 or control rabbit Ig was used to precipitate cross-linked protein–DNA complexes from nuclear lysates. The cross-linking of the immunoprecipitated material was removed and protease treated, and the DNA was purified and amplified. The resultant material was hybridized to the whole genome using GeneChip Human Tiling 1.0R Array Set (Affymetrix 900774) to identify the locations of binding sites for FoxP3. Statistical analysis was performed to determine sites that were selectively associated with FoxP3 protein and not rabbit Ig. The IL-7R promoter region scored among the various sites bound by the FoxP3 protein immunoprecipitates (unpublished data). The CD127 promoter binding was confirmed by qPCR. Oligonucleotide primers spanning the CD127 promoter region were used on anti-FoxP3 immunoprecipitated DNA from CD4+CD25hi human T reg cells (Fig. 5). There was a strong enrichment of CD127 DNA amplified from the anti-FoxP3 immunoprecipitates as compared with the rabbit Ig immunoprecipitates specifically in the IL-7R promoter region, but not other DNA sequences surrounding this area on the genome. These data support the direct regulation of CD127 by FoxP3.
Figure 5.
ChIP-chip and ChIP-qPCR analysis of FoxP3 bound DNA from CD4+CD25hi human T reg cells. Anti-FoxP3 or control rabbit Ig was used to precipitate cross-linked protein–DNA complexes from expanded CD4+CD25hi human T reg cells lysate. The cross-linking of the immunoprecipitated material was removed and protease-treated and the DNA was purified and amplified. The resultant material was hybridized to the whole genome using GeneChip Human tiling 1.0R array set to identify the locations of binding sites for FoxP3. Two sets of graphs: FoxP3 IP versus the Ig control and FoxP3 IP versus Input DNA were generated on the hs.NCBIv35 version of the genome essentially following the method described in Cawley et el. (reference 50). (a) Signal enrichment graphs of IL-7R locus (chr5:35863179-35918811). Several regions in IL-7R locus are predicted to be positive (chr5:35892564-35892809 promoter) and negative (chr5:35890618-35890846 2K upstream; chr5:35907667-35907852 Intron 4; chr5:35911721-35911888 intron 7 and exon 8). (b) SYBR green qPCR of IL-7R chromosomal regions. FoxP3 IP versus the IgG fold enrichment ratio was determined from duplicate ChIP assay evaluated in duplicate by real time PCR.
Suppression of allogeneic mixed lymphocyte response (MLR) using different CD4+ T cell subsets
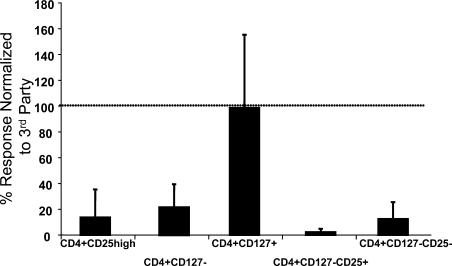
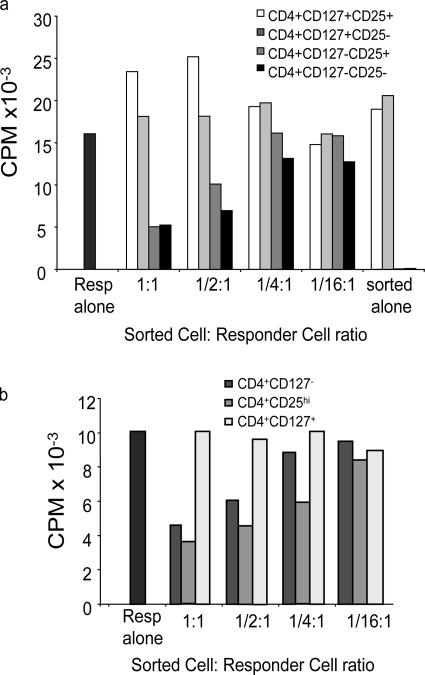
Although the low expression of CD127 correlated with FoxP3 expression, several studies have questioned whether FoxP3 is always a marker of T reg cells in humans. Thus, we examined the ability of CD4+CD25+CD127lo/− T cells and other subsets to suppress an allogeneic MLR. PBMCs were sorted into four subsets based on CD127, CD25, and CD4 expression. First, we examined the ability of the individual subsets to respond to the allogeneic APCs. As can be seen in Fig. 6, as previously reported, the CD4+CD25hi T cell subset was anergic when stimulated with alloantigen consistent with the fact that these cells were FoxP3+ and comprise the “classical” T reg cell subset. Similarly, neither of the CD127lo/− subsets, CD25+ or CD25− CD4+ T cells or the bulk of CD4+CD127lo/− T cells responded in the allogeneic MLR, suggesting that like the CD4+CD25hi T cells, these cells were anergic (37). In contrast, the CD4+CD127+ T cell subset responded normally to alloantigen consistent with publications suggesting that these cells represented naive and memory T cell compartments (32, 34, 40). Similar results were observed when the cells were stimulated with anti-CD3 and anti-CD28 (unpublished data), suggesting that like “classical” T reg cell subsets, the FoxP3-expressing cells were anergic. Next, the various subpopulations were added to an allogeneic MLR and compared for their ability to suppress T cell proliferation. The CD4+CD127lo/−CD25+ T cell subset suppressed the MLR as well or better that CD4+CD25hi T cells (Fig. 7). This is significant because this subset represents at least threefold more CD4+ T cells including both CD25 intermediate and negative subsets. Thus, CD127 is more than just another marker of CD4+CD25hi T reg cells, but allows for the identification and isolation of a significantly more inclusive suppressive T cell subset. In fact, suppressive activity was independent of CD25 as both the CD4+CD127lo/−CD25+ and CD4+CD127lo/−CD25− T cell subsets suppressed the MLR, although in multiple studies the CD4+CD127lo/−CD25+ T cells suppressed responses more effectively than the CD4+CD127lo/−CD25− T cell subset, especially at lower T reg cell:T responder ratios. These results are consistent with the lower percentage and level of expression of FoxP3+ cells in this T cell subset. Neither of the CD127+ cells suppressed the MLR reproducibly (n = 9). These results suggested that CD127 is a sufficient marker for defining the CD4+ T reg cell subset. To demonstrate this directly, PBMCs were sorted based only on the expression of CD4 and CD127 and examined in an allogeneic MLR. The CD4+CD127lo/− T cell subset was anergic (Fig. 6) and suppressed the MLR almost as efficiently as the CD4+CD127lo/−CD25+ or CD4+CD25hi T cells (Fig. 7 b).
Figure 6.
Proliferative response of isolated T cell subsets. Buffy coat samples were sorted based on CD4, CD127, and CD25 expression. 30,000 sorted cells were put into culture with allogeneic anti-CD3–depleted, irradiated, third-party PBMCs as stimulators. T cells were incubated for 7 d at 37°C in 5% CO2. 16 h before the end of the incubation, 1 μCi 3H-thymidine was added to each well. Plates were harvested and data were analyzed. Data are representative of nine separate experiments.
Figure 7.
Suppression of allogeneic MLR by individual T cell subsets. Buffy coat samples were sorted based on CD4, CD127 and CD25 expression. 30,000 sorted cells were combined with 100,000 autologous PBMCs as responders, and 100,000 allogeneic anti-CD3–depleted, irradiated third-party PBMCs as stimulators. T cells were incubated for 7 d at 37°C in 5% CO2. 16 h before the end of the incubation, 1 μCi 3H-thymidine was added to each well. Plates were harvested and data were analyzed. Data representative of nine separate experiments sorting seven different subpopulations of CD4+ cells indicated (a) CD127+CD25+, CD127+CD25−, CD127lo/−CD25+, CD127lo/−CD25− and (b) CD127lo/−, CD25hi, CD127+. 100,000 responders are present in each well with decreasing numbers of sorted cells added at 1:1 ratio (30,000:100,000), 1:1/2 (15,000 sorted cells), 1:1/4 (7,500 sorted cells), 1:1/16 (1,875 sorted cells) in comparison to sorted cells alone. Results are represented as counts per minute (CPM).
Frequency of CD4+CD25+CD127lo/− T cells from patients with T1D
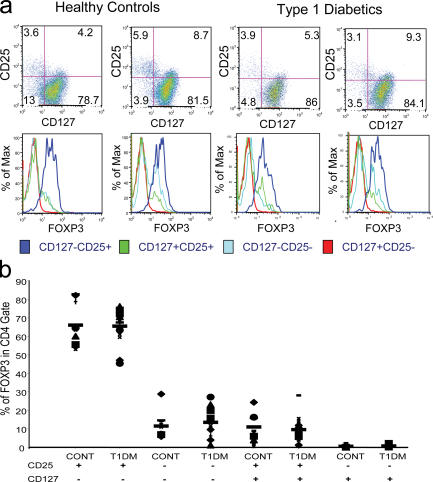
Previous studies have suggested that T reg cell numbers might be deficient in patients with T1D (24). To investigate quantitative differences in the T reg cell populations in patients with T1D versus control subjects, peripheral blood T cells were stained with CD4, CD25, CD127, and FoxP3. Representative examples of the staining of healthy controls and patients with T1D are shown in Fig. 8 a. The frequency of FoxP3+ T cells in each subset was not significantly different between T1D and control subjects. This included the putative CD4+CD25+CD127lo/− and CD4+CD25−CD127lo/− T reg cells. Additional patients and controls were analyzed and a scatter plot of all the data is shown in Fig. 8 b. Although the percentages of FoxP3+ T cells among the CD4+ T cells in the two groups were higher than those reported for the CD4+CD25hi T reg cell subset, the patterns matched those previously observed. There was no significant difference in the percentage of FoxP3+ T cells in any of the T cell subsets between control and T1D samples. As an example, the CD4+CD25+CD127lo/− subset in controls had a mean of 66.1% FoxP3+ cells (SD 11.3%; range 53.7–82.8%) versus the healthy control individuals (mean 65.5%; SD 8.66%; range 45.4–76.2%). These results are in contrast with some reports that found differences in the percentage of CD4+CD25hi T cells in T1D subjects when analyzed as a percentage of CD4+ T cells.
Figure 8.
Frequency of various T cell subsets in patients with T1D versus healthy control subjects. (a) FACS data are depicted from two healthy control and two individuals with T1D. Individual histograms are depicted to show the FoxP3 expression in CD4+ T cells subdivided into the various CD25 and CD127 subsets. (b) Individual FoxP3 expression data is depicted from total 10 healthy control individuals and 16 patients with T1D. Each symbol represents an individual subject and the narrow bar represents the mean percentage of FoxP3+ T cells in various T cell subsets (CD4+ T cells gated based on CD25 and/or CD127 expression as indicated at the bottom of the graph).
DISCUSSION
The emergence of T reg cells as an essential pathway in maintaining immune tolerance has opened the opportunity for a better understanding of immune homeostasis and the potential for therapeutic intervention. However, unlike the mouse, the human phenotyping of T reg cells has been complex. Typically, investigators have noted that the most suppressive T reg cells coincide with the CD4+ T cells with the brightest CD25 staining. Unfortunately, the ability to accurately gate for CD25 is rather arbitrary as no other cell surface marker can be used to definitively identify the subset. Baecher-Allen has suggested that other markers such as HLA-DR allows for subdividing the CD4+CD25hi subset to enrich T reg cell activity even further (41). However, this additional marker suggests that the number of T reg cells is even less than previously suggested although a recent paper published by this same group showed that DR− T reg cells are suppressive as well but with a different kinetics and different mechanism (42). The identification of FoxP3 as a specific transcription factor that marks these cells suggests that there may be a larger population of T reg cells in human peripheral blood than previously appreciated, although this has been controversial as the result of unanticipated expression in several activated CD25− T cell populations (30, 38). However, these studies have been compromised by the absence of cell surface markers that can be used to isolate these and other T cell subsets to examine T reg cell activity because FoxP3 cannot be used as a means to purify the cells for function. In this paper, we demonstrate that CD127 expression is an excellent biomarker of human T reg cells, especially when combined with CD25. The combination of these markers identifies T reg cells that account for up to 7–8% of CD4+ T cells, a significantly greater percentage than identified by previous approaches. Moreover, these cells suppress the proliferative response of alloreactive T cells in an MLR and are themselves anergic to the same stimuli, characteristics shared by CD4+CD25hi human T reg cells.
These results raise several critical issues. First, because the majority of CD4+FoxP3+ T cells may fall outside the typical gate for human T reg cells, studies used for functional and immunophenotypic analyses are potentially missing a large number of putative T reg cells. This has important implications in determining quantitative differences in patients with a variety of diseases. Second, the fact that CD4+CD127−CD25− T cells suppress an allogeneic MLR calls into question those studies suggesting that FoxP3 is not a “good” marker for T reg cell activity. It may be that FoxP3 is an excellent marker and that the small populations that arise during normal T cell activation are indeed adaptive T reg cells expanding as a consequence of suboptimal or supraoptimal TCR signaling. It should be emphasized, however, that not all FoxP3+ T cells are necessarily T reg cells and their activity may depend on the level of FoxP3 expression and isoforms of the protein expressed. However, these CD4+CD25+CD127lo/−, once isolated, may be treated in vitro with TGFβ or other factors to enhance T reg cell function in these cells. Third, efforts to select T reg cells for in vitro expansion may be hindered by the underestimation of T reg cells in any separation strategy based on CD25 expression. The ability to identify and select a significantly greater number of T reg cells circulating in the peripheral blood of humans, especially those with autoimmune diseases, is likely to make it easier to expand sufficient cell numbers for immunotherapy. Finally, the identification of CD127 as a marker that distinguishes effector/memory from T reg cells may suggest that anti-CD127 therapy might be appropriate for the treatment of autoimmune diseases such as T1D, systemic lupus erythematosis, or multiple sclerosis.
The identification of CD127 as a useful marker was heralded by genetic observations. First, microarray analysis of mRNA from individual T cell subsets showed that CD127 was expressed at significantly lower levels in CD4+CD25hi versus CD4+CD25− T cells. Unlike the majority of activated T cells, which rapidly reexpress CD127 and memory T cells that express high levels of CD127, the T reg cell population remains CD127lo/−. There may be two reasons for this. First, T reg cells may be constantly undergoing antigenic stimulation that is CD28 dependent, resulting in continued signaling that shuts down CD127 mRNA transcription. In this regard, it is interesting to note that activation of naive T cells by anti-CD3 plus anti-CD28 but not anti-CD3 alone led to a rapid down-regulation of CD127 (unpublished data), suggesting that CD28 signals are uniquely involved in regulating CD127 down-regulation. Alternatively, and not mutually exclusively, is the possibility that FoxP3 expression in this T cell subset controls CD127 expression. There are several reasons that this may be the case. As illustrated in the flow cytometric staining profiles, the more FoxP3 expression, the less CD127 (Fig. 2 b). In addition, overexpression of FoxP3 in transgenic mice results in a uniformly CD127lo/− population of cells with suppressive activity. Finally, data generated using CHiP analysis (first by ChIP-Chip followed by ChIP-qPCR) suggested that the CD127 promoter is a target for FoxP3 binding. It will be critical to determine whether the low expression of CD127 is indeed a consequence of constant antigen exposure in vivo or FoxP3 up-regulation resulting in CD127 gene repression, although these are not mutually exclusive.
The CD4+CD127lo/−CD25− T cells suppress quite effectively, although the percentage of FoxP3+ T cells in this subset can be quite variable. These results suggested that the CD127 marker may be useful in identifying different subtypes of T reg cells including Tr1 and TH3 cells. In this regard, there are currently several settings, including the treatment of humans with T1D with anti-CD3 that induces T cells with a regulatory phenotype (11, 31, 43, 44). These studies, which mimic similar results in T reg cell–deficient mice treated with nonmitogenic anti-CD3 (10), indicate that it may be possible to identify an “adaptive” T reg cell response using CD127 as a biomarker in addition to lower CD25 expression previously observed on these cells.
One of the more intriguing aspects of the results is the seeming dichotomy in cytokine receptor expression in memory T cells versus T reg cells. Although a high percentage of T reg cells now appear to be IL-7R low and IL-2R positive, memory T cells have the opposite phenotype, expressing high levels of IL-7R and low levels of IL-2R. The theoretical basis for this differential expression is unclear but it may reflect the evolution of distinct pathways for cell survival and expansion of these T cell subsets. For instance, it is possible that T reg cells may play a critical role in normal homeostasis. Thus, the cells attempt to regulate the earliest immune perturbation that may occur in the absence of a pathogenic response. Because IL-2 is an “early cytokine” produced rapidly by activated T cells in the draining lymph nodes, IL-2 may be a critical signal for awakening the T reg cell response that can effectively suppress T cell expansion in these lymphoid tissues (45). In contrast, IL-7 is commonly produced locally in sites of inflammation leading to increased survival and expansion of effector cells. If this localized IL-7 expression promoted T reg cell expansion, it might be counterproductive. Moreover, avoiding competition for the use of the common γ chain by these receptors would enhance the functionality of the cytokine function. Finally, it should be noted that the situation might be quite distinct in thymus when all the pre–T and immature T cells are CD127+ and CD25+. At this stage in development, other factors might come into play to determine the differentiation pathways that determine whether a T cell becomes a T reg cell or a naive conventional T cell.
Several studies have examined the number and function of T reg cell cells in humans with autoimmune diseases. In some settings, such as multiple sclerosis, T1D, and autoimmune polyglandular syndrome II, the data, based on the number and function of CD4+CD25hi T reg cells, suggest that there are either fewer T reg cells or less functional T reg cells in diseased individuals (24, 46–48). However, in T1D and other autoimmune diseases, there have been contradictory results (25, 49). In the present study, we reevaluated T reg cells in patients with T1D as compared with normal individuals. Using the new markers, FoxP3 and CD127, we analyzed the frequency of CD4+CD25+FoxP3+CD127lo/− T cells. In this study, it is clear that human T reg cells as defined by CD4, CD25, CD127, and FoxP3 expression are present within the same range of percentages as control individuals with no autoimmunity (Fig. 8, a and b). Moreover, the functionality of the T reg cells isolated from the patients with T1D cannot be distinguished from healthy control subjects (unpublished data). We cannot explain the basis for differences between our studies and those of others in the T1D field. It has been suggested that the discrepancy might be the result of subtle differences in flow cytometry–based techniques for cell separation or different mAbs used. However, our use of distinct markers that identify the overwhelming bulk of T reg cells in human peripheral blood is likely to be a more definitive assessment of the T reg cell numbers and functional potential in this patient population. Lastly, it is interesting to note that several of the T1D patients had high T reg cell numbers as compared with the bulk of the control and T1D subjects. This is consistent with some studies in other autoimmune diseases in which the frequency of CD4+CD25hi T cells was reported to be increased as compared with controls. Moreover, these results fit with mouse studies, demonstrating increased T reg cell number at the time of T1D disease onset as well as other immune disease settings (unpublished data). We hypothesize that rather than a T reg cell deficiency being the cause of disease precipitation, there is actually increased T reg cell activity in an attempt to stem the increasingly aggressive effector cells that may indeed become T reg cell resistant.
In summary, we have determined that CD127 is an excellent marker of T reg cells in human peripheral blood. The cell surface marker is expressed at low levels on an overwhelming majority of T reg cells and distinguishes up to 10% of CD4+ T cells as potential T reg cells. Moreover, the cell surface marker can be used, in the absence of CD25 to separate a suppressive T cell subset and will thus be a useful tool for the selection and expansion of T cell for diagnostics and therapeutic applications.
MATERIALS AND METHODS
Antibodies.
The human antibodies (PE-conjugated anti-CD127, APC-conjugated anti-CD25, PerCP-conjugated anti-CD4) were used for staining and in sorting and were provided by Becton Dickinson (BD Biosciences). Alexa488-conjugated anti-FoxP3 was purchased from BioLegend and intracellular staining was performed according to the manufacturer's instructions and modified as follows: 5 × 105 cells were stained with cell surface markers for 30 min at 4°C and fixed for 30 min using 1x Fix/Perm buffer. After three washes, cells were permeabilized in Perm buffer with DNase I (Sigma-Aldrich) for 30 min followed by three washes. Then cells were blocked with human IgG and stained with anti–human FoxP3 Alexa488-conjugated (BioLegend; clone 206D). The following anti–mouse antibodies were purchased from the indicated sources: anti-CD4, anti-CD25, and mouse IgG1 (isotype control) (BD Biosciences) and CD127 (eBioscience).
Subjects.
A total of 16 patients with longstanding T1D were studied. Patients (age range 16–56 yr, mean age 34, with a disease duration >5 yr) were recruited from the Barbara Davis Center for Childhood Diabetes. Diagnosis of T1D was made primarily by the presence of biochemical autoantibodies or presentation of hyperglycemia with ketosis in childhood. None of the diabetic subjects had severe nephropathy or neuropathy. As controls, 10 subjects (age range 20–50 yr, mean age 29) with no family history of diabetes mellitus were also tested. Blood samples were obtained with informed consent under Institutional Review Board approved protocols at either the University of Colorado Health Sciences Center or UCSF as needed.
Sorting of CD4+ T cell subsets for flow cytometry and functional studies.
Human T cells were isolated from leukopacs (Blood Centers of the Pacific). In some cases, negative selection using RosetteSep human CD3 depletion Cocktail (StemCell Technologies, Inc.) was performed. 100–120 × 106 PBMCs were washed once, counted, and resuspended in sorting buffer (PBS + 0.1% BSA + 1 mM EDTA) at 100 × 106 per ml in a 15-ml conical tube. After addition of 1 μl/1 million cell volume PerCP-conjugated anti-CD4, 1 μl/1 million cells PE-conjugated anti-CD127, and 0.7 μl/million cells APC-conjugated anti-CD25 antibodies, the cell suspension was mixed gently and incubated at 4°C for 30 min. Cold FACS sorting buffer was added up to a volume of 15 ml and T cells were pelleted and resuspended at 20 × 106 per ml. Labeled T cells were sorted using a FACSAria (Becton Dickinson). The sorted gates for the various T cell subsets were set to include only those events exhibiting the CD4-specific fluorescence that were also within the lowest density region of a scatter plot. This amounted to between 11.4% and 33.9% (mean 20.87%) (n = 22) of the total number of events for the CD4+ T cells. Based on the CD4 gate, cells were further gated based on CD127 and/or CD25 expression (CD4+CD127+/−CD25+/− and CD4+CD127+/− alone independent of CD25 as well as CD25hi conventional T reg cells). Cells were collected into 100% human AB serum (Cambrex) and washed once with media (RPMI 1640/5% human serum) until they were ready to be plated in suppression assay. Sorted T reg cells were 95–98% CD4+CD25hi with a typical yield of 5–12 × 105 T cells per sort, whereas CD4+CD127−CD25+ cells had a typical yield of 0.9–1.2 × 106 and 98% purity.
Isolation of CD4+CD25hi and CD4+CD25− cells for GeneChip arrays.
Human CD4+ T cells were isolated by negative selection from leukopacs (Stanford University Blood Center) using RosetteSep Human CD4+ T cell Cocktail (StemCell Technologies, Inc.). 0.75–1.25 × 108 CD4+ T cells (>90% purity by FACS) were washed once, counted, and resuspended in FACS staining buffer (PBS + 0.1% BSA) at 10 × 106 per ml in a 50-ml conical tube. After addition of 1/90 volume Cy-5–conjugated anti-CD4 (BD Biosciences) and 1/100 FITC-conjugated anti-CD25 (DakoCytomation) antibodies, the cell suspension was mixed gently and incubated on ice for 45 min. Cold FACS staining buffer was added to a volume of 50 ml and T cells were pelleted and resuspended at 20 × 106 per ml. Labeled T cells were incubated on ice for 45 min and submitted to flow sorting on a DakoCytomation MoFlo high-speed cell sorter. The sort gates were set to include only those events exhibiting the highest levels of CD25-specific fluorescence (CD4+CD25hi cells) or lowest levels (CD4+CD25neg cells) that were also within the lowest density region of a scatter plot. This amounted to between 0.8% and 1.4% of the total number of events for the CD4+ T cells for each subset.
RNA isolation.
Total RNA was isolated from T reg cells using the total RNA isolation protocol from the RNA RT-PCR Miniprep kit (Stratagene) with the following modifications: 100,000 cells were lysed in 150 μl of lysis buffer. To digest DNA, 2 U of DNase was added/μg nucleic acid, and phenol/CHCl3 (Sigma-Aldrich) was used to purify total RNA followed by ethanol precipitation. The quantity of total RNA was measured using Nanodrop ND 100 (Nanodrop Technologies). 100 ng of each RNA sample was used for target labeling by a two-round amplification protocol. This protocol was modified from the Affymetrix eukaryotic small sample prep by using 6 pMol of T7 primer and 3 μg/μl of the random primer.
GeneChip arrays and data analysis.
A total 16 of human HG-U133A GeneChip arrays were used in this study (Affymetrix). 10 μg of fragmented cRNA per GeneChip hybridization were processed on the Affymetrix Fluidic station 450 and GeneChip scanner GCS2500 (Hewlett-Packard Company). Gene expression profile was analyzed with MAS5.0 (Microarray Suite version 5.0; Affymetrix) and used for data acquisition and normalization. Present genes were defined by selecting genes that were present in three out of four arrays. Signal intensities of all present genes for the activated and control groups were combined and analyzed by Student's t test. Significant genes were selected with P < 0.05. The "signal log ratio" (SLR) and "increase" or "decrease" call was generated by comparison analysis MAS5.0 and used for calculating fold changes between groups. We selected 9 out of 16 pair-wise comparisons for particular genes that showed increase or decrease at fold change >2.0. In the second step, signal intensities of present genes were analyzed with Significance Analysis of Microarrays (SAM) and applied for analyzing and determining the gene list based on the number of significant genes that were identified by Student's t test and fold change. The final significant genes were combined from the aforementioned two steps. Two-dimensional hierarchical clusters are generated using GeneSpring 6.0 software (Silicon Genetics).
Real-time PCR analysis.
RNA was isolated using RNeasy mini kits (QIAGEN) according to the manufacturer's instructions. For cDNA synthesis, 500 ng total RNA was transcribed with cDNA transcription reagents using SuperScriptIII reverse transcriptase and oligo(dT)12-18 (Invitrogen), according to the manufacturer's instructions. Gene expression was measured in real-time with the GeneAmp 7900 Sequence Detection System (Applied Biosystems) using primers and QuantiTect SYBR green PCR Kit purchased from QIAGEN. The expression level of a gene in a given sample was represented as 2−ΔΔCt where ΔΔCT = [ΔCT(experimental)] − [ΔCT(medium)] and ΔCT = [CT(experimental)] − [CT(housekeeping)]. Data are presented and normalized to the glyceraldehyde phosphate dehydrogenase (GADP).
Suppression assays.
Suppression assays were performed in round-bottom 96-well microtiter plates. 100,000 responder PBMCs from the same cell source as sorted populations, 30,000 sorted cells (one of seven different sorted subtypes based on CD25 and/or CD127 expression), 100,000 allogeneic irradiated CD3-depleted PBMCs were added as indicated. Responder ratio indicated refers to T reg cell to responder where 1 sorted:1 responder is 30,000 sorted cells: 100,000 PBMC responder cells. APCs consisted of allogeneic PBMCs depleted of T cells using StemSep human CD3+ T cell depletion as per the manufacturer's recommendations (StemCell Technologies, Inc.) and irradiated with 40 Gy. Cells were plated in the following order in 50 μl per well: sorted cells, responders, and APCs. No additional stimulus was added to the wells; however, additional media was added to each well so the final volume was 200 μl per well. Wells surrounding culture wells were filled with PBS to prevent evaporation. T cells were incubated for 7 d at 37°C in 5% CO2. 16 h before the end of the incubation, 1 μCi 3H-thymidine was added to each well. Plates were harvested using a Tomtec cell harvester and 3H-thymidine incorporation was determined using a 1450 microbeta Wallac Trilux liquid scintillation counter.
Antibody staining and FACS analysis.
5 × 104 T cells per sample were washed once with FACS staining buffer (PBS + 0.1% BSA) and resuspended in 100 μl buffer. 1 μl of fluorescence-conjugated specific antibodies (1 μg/million T cells) was added and T cells were lightly vortexed and incubated on ice for 20 min. 500 μl of staining buffer was added to each sample and T cells were pelleted, resuspended in 200 μl buffer, and analyzed on a flow cytometer (Becton Dickinson, FACScalibur). Intracellular staining was conducted using the recommended procedure obtained from BD Biosciences or eBioscience where indicated.
Chromatin immunoprecipitation–DNA microarray (ChIP-chip).
Human CD4+CD25hi T reg cells were expanded in vitro as described previously (37). Chromatin fixation and immunoprecipitation were performed using chromatin immunoprecipitation assay kit (Upstate Biotechnology) as recommended by the manufacturer. Expanded human T reg cell were fixed in 1.1% formaldehyde. Protein–DNA cross-linked cell pellets were resuspended in SDS-Lysis Buffer (1 ml per 108 cells) and incubated for 10 min on ice. Lysates were sonicated to shear DNA to lengths between 200 and 1,000 basepairs and centrifuged for 10 min at 13,000 revolutions/min at 4°C to remove debris. The sonicated cell supernatant was diluted 10-fold in ChIP Dilution Buffer with protease inhibitors (Upstate Biotechnology) to reduce nonspecific background and the diluted cell supernatant was precleared with 40 μl of a Protein A Agarose-50% slurry per 1 ml lysate for 30 min at 4°C with agitation. Cross-linked protein–DNA complexes were immunoprecipitated using control rabbit Ig or affinity-purified rabbit polyclonal anti–human FoxP3 (a gift from R. Khattri and F. Ramsdell, Celltech, Lt., Seattle, WA). The cross-linking of the material was reversed and proteinase K treated to remove protein from the DNA. The remaining DNA was purified with QIAquick PCR Purification Kit (QIAGEN) and amplified by LMPCR (ligation-mediated PCR) as described previously (50). Array hybridization and analysis were performed at Affymetrix as described previously (51). SYBR green qPCR was performed to verify the binding sites predicted by the arrays. Primers for the PCR reactions included: IL-7R promoter: 5′-primer, 5′-CAGGGAATATCCAGGAGGAA-3′; 3′-primer, 5′-TGTGTGAGCCAGTGTGTATGAA-3′; IL-7R 2K upstream: 5′-primer, 5′-TTTGGGATTTCTCCTTGAACA-3′; 3′-primer 5′-TCTCTGGGCATTTCAAAACC-3′; IL-7R intron 4:5′ primer, 5′-GAGGTGGCAGAAGAGTGGAG-3′; 3′-primer, 5′-TGCATCACACTGCAAACAAA-3′; and IL7-R intron 7 and exon 8:5′-primer, 5′-ACATGCTGGCAATTCTGTGA-3′; 3′-primer, 5′-TCTGGCAGTCCAGGAAACTT-3′.
Acknowledgments
The authors would like to thank Q. Tang, H. Bour-Jordan, and A. Abbas for technical assistance and review of the manuscript. We also thank A. Rudensky for the FoxP3-GFP mice.
This study was supported by National Institutes of Health grant nos. P30 DK63720, U19 DK61934, AI0 48779; grants from the Juvenile Diabetes Research Foundation (JDRF 4-2005-1168, JDRF 1-2004-98); and funding from the Becton-Dickinson Corporation.
The authors have no conflicting interests.
Abbreviations used: ChIP, chromatin immunoprecipitation; MLR, mixed lymphocyte response; T1D, type 1 diabetes; T reg, regulatory T.
W. Liu and A.L. Putnam contributed equally to this work.
References
- 1.Sakaguchi, S., N. Sakaguchi, J. Shimizu, S. Yamazaki, T. Sakihama, M. Itoh, Y. Kuniyasu, T. Nomura, M. Toda, and T. Takahashi. 2001. Immunologic tolerance maintained by CD25+ CD4+ regulatory T cells: their common role in controlling autoimmunity, tumor immunity, and transplantation tolerance. Immunol. Rev. 182:18–32. [DOI] [PubMed] [Google Scholar]
- 2.Chatenoud, L., B. Salomon, and J.A. Bluestone. 2001. Suppressor T cells–they're back and critical for regulation of autoimmunity! Immunol. Rev. 182:149–163. [DOI] [PubMed] [Google Scholar]
- 3.Wood, K.J., and S. Sakaguchi. 2003. Regulatory T cells in transplantation tolerance. Nat. Rev. Immunol. 3:199–210. [DOI] [PubMed] [Google Scholar]
- 4.Singh, B., S. Read, C. Asseman, V. Malmstrom, C. Mottet, L.A. Stephens, R. Stepankova, H. Tlaskalova, and F. Powrie. 2001. Control of intestinal inflammation by regulatory T cells. Immunol. Rev. 182:190–200. [DOI] [PubMed] [Google Scholar]
- 5.Curotto de Lafaille, M.A., and J.J. Lafaille. 2002. CD4(+) regulatory T cells in autoimmunity and allergy. Curr. Opin. Immunol. 14:771–778. [DOI] [PubMed] [Google Scholar]
- 6.Tang, Q., K.J. Henriksen, M. Bi, E.B. Finger, G. Szot, J. Ye, E.L. Masteller, H. McDevitt, M. Bonyhadi, and J.A. Bluestone. 2004. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J. Exp. Med. 199:1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Boehmer, H. 2005. Mechanisms of suppression by suppressor T cells. Nat. Immunol. 6:338–344. [DOI] [PubMed] [Google Scholar]
- 8.Shevach, E.M. 2002. CD4+ CD25+ suppressor T cells: more questions than answers. Nat. Rev. Immunol. 2:389–400. [DOI] [PubMed] [Google Scholar]
- 9.Thornton, A.M., C.A. Piccirillo, and E.M. Shevach. 2004. Activation requirements for the induction of CD4+CD25+ T cell suppressor function. Eur. J. Immunol. 34:366–376. [DOI] [PubMed] [Google Scholar]
- 10.Belghith, M., J.A. Bluestone, S. Barriot, J. Megret, J.F. Bach, and L. Chatenoud. 2003. TGF-β-dependent mechanisms mediate restoration of self-tolerance induced by antibodies to CD3 in overt autoimmune diabetes. Nat. Med. 9:1202–1208. [DOI] [PubMed] [Google Scholar]
- 11.Apostolou, I., and H. von Boehmer. 2004. In vivo instruction of suppressor commitment in naive T cells. J. Exp. Med. 199:1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bluestone, J.A., and A.K. Abbas. 2003. Natural versus adaptive regulatory T cells. Nat. Rev. Immunol. 3:253–257. [DOI] [PubMed] [Google Scholar]
- 13.Levings, M.K., and M.G. Roncarolo. 2005. Phenotypic and functional differences between human CD4+CD25+ and type 1 regulatory T cells. Curr. Top. Microbiol. Immunol. 293:303–326. [DOI] [PubMed] [Google Scholar]
- 14.Weiner, H.L. 2001. Oral tolerance: immune mechanisms and the generation of Th3-type TGF-β-secreting regulatory cells. Microbes Infect. 3:947–954. [DOI] [PubMed] [Google Scholar]
- 15.Tarbell, K.V., S. Yamazaki, K. Olson, P. Toy, and R.M. Steinman. 2004. CD25+ CD4+ T cells, expanded with dendritic cells presenting a single autoantigenic peptide, suppress autoimmune diabetes. J. Exp. Med. 199:1467–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baecher-Allan, C., J.A. Brown, G.J. Freeman, and D.A. Hafler. 2001. CD4+CD25high regulatory cells in human peripheral blood. J. Immunol. 167:1245–1253. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi, T., Y. Kuniyasu, M. Toda, N. Sakaguchi, M. Itoh, M. Iwata, J. Shimizu, and S. Sakaguchi. 1998. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 10:1969–1980. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu, J., S. Yamazaki, T. Takahashi, Y. Ishida, and S. Sakaguchi. 2002. Stimulation of CD25(+)CD4(+) regulatory T cells through GITR breaks immunological self-tolerance. Nat. Immunol. 3:135–142. [DOI] [PubMed] [Google Scholar]
- 19.Salomon, B., and J.A. Bluestone. 2001. Complexities of CD28/B7: CTLA-4 costimulatory pathways in autoimmunity and transplantation. Annu. Rev. Immunol. 19:225–252. [DOI] [PubMed] [Google Scholar]
- 20.McHugh, R.S., M.J. Whitters, C.A. Piccirillo, D.A. Young, E.M. Shevach, M. Collins, and M.C. Byrne. 2002. CD4(+)CD25(+) immunoregulatory T cells: gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity. 16:311–323. [DOI] [PubMed] [Google Scholar]
- 21.Kataoka, H., S. Takahashi, K. Takase, S. Yamasaki, T. Yokosuka, T. Koike, and T. Saito. 2005. CD25(+)CD4(+) regulatory T cells exert in vitro suppressive activity independent of CTLA-4. Int. Immunol. 17:421–427. [DOI] [PubMed] [Google Scholar]
- 22.Tang, Q., E.K. Boden, K.J. Henriksen, H. Bour-Jordan, M. Bi, and J.A. Bluestone. 2004. Distinct roles of CTLA-4 and TGF-β in CD4+CD25+ regulatory T cell function. Eur. J. Immunol. 34:2996–3005. [DOI] [PubMed] [Google Scholar]
- 23.Ronchetti, S., O. Zollo, S. Bruscoli, M. Agostini, R. Bianchini, G. Nocentini, E. Ayroldi, and C. Riccardi. 2004. GITR, a member of the TNF receptor superfamily, is costimulatory to mouse T lymphocyte subpopulations. Eur. J. Immunol. 34:613–622. [DOI] [PubMed] [Google Scholar]
- 24.Kukreja, A., G. Cost, J. Marker, C. Zhang, Z. Sun, K. Lin-Su, S. Ten, M. Sanz, M. Exley, B. Wilson, et al. 2002. Multiple immuno-regulatory defects in type-1 diabetes. J. Clin. Invest. 109:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putnam, A.L., F. Vendrame, F. Dotta, and P.A. Gottlieb. 2005. CD4+CD25high regulatory T cells in human autoimmune diabetes. J. Autoimmun. 24:55–62. [DOI] [PubMed] [Google Scholar]
- 26.Fontenot, J.D., M.A. Gavin, and A.Y. Rudensky. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336. [DOI] [PubMed] [Google Scholar]
- 27.Khattri, R., T. Cox, S.A. Yasayko, and F. Ramsdell. 2003. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4:337–342. [DOI] [PubMed] [Google Scholar]
- 28.Gambineri, E., T.R. Torgerson, and H.D. Ochs. 2003. Immune dysregulation, polyendocrinopathy, enteropathy, and X-linked inheritance (IPEX), a syndrome of systemic autoimmunity caused by mutations of FOXP3, a critical regulator of T-cell homeostasis. Curr. Opin. Rheumatol. 15:430–435. [DOI] [PubMed] [Google Scholar]
- 29.Hori, S., T. Nomura, and S. Sakaguchi. 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science. 299:1057–1061. [DOI] [PubMed] [Google Scholar]
- 30.Ziegler, S.F. 2006. FOXP3: Of Mice and Men. Annu. Rev. Immunol. 24:209–226. [DOI] [PubMed] [Google Scholar]
- 31.Bisikirska, B., J. Colgan, J. Luban, J.A. Bluestone, and K.C. Herold. 2005. TCR stimulation with modified anti-CD3 mAb expands CD8+ T cell population and induces CD8+CD25+ Tregs. J. Clin. Invest. 115:2904–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuller, M.J., D.A. Hildeman, S. Sabbaj, D.E. Gaddis, A.E. Tebo, L. Shang, P.A. Goepfert, and A.J. Zajac. 2005. Cutting edge: emergence of CD127high functionally competent memory T cells is compromised by high viral loads and inadequate T cell help. J. Immunol. 174:5926–5930. [DOI] [PubMed] [Google Scholar]
- 33.Boettler, T., E. Panther, B. Bengsch, N. Nazarova, H.C. Spangenberg, H.E. Blum, and R. Thimme. 2006. Expression of the interleukin-7 receptor α chain (CD127) on virus-specific CD8+ T cells identifies functionally and phenotypically defined memory T cells during acute resolving hepatitis B virus infection. J. Virol. 80:3532–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huster, K.M., V. Busch, M. Schiemann, K. Linkemann, K.M. Kerksiek, H. Wagner, and D.H. Busch. 2004. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. USA. 101:5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, J., G. Huston, and S.L. Swain. 2003. IL-7 promotes the transition of CD4 effectors to persistent memory cells. J. Exp. Med. 198:1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fontenot, J.D., J.P. Rasmussen, L.M. Williams, J.L. Dooley, A.G. Farr, and A.Y. Rudensky. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341. [DOI] [PubMed] [Google Scholar]
- 37.Earle, K.E., Q. Tang, X. Zhou, W. Liu, S. Zhu, M.L. Bonyhadi, and J.A. Bluestone. 2005. In vitro expanded human CD4+CD25+ regulatory T cells suppress effector T cell proliferation. Clin. Immunol. 115:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontenot, J.D., J.L. Dooley, A.G. Farr, and A.Y. Rudensky. 2005. Developmental regulation of Foxp3 expression during ontogeny. J. Exp. Med. 202:901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schubert, L.A., E. Jeffery, Y. Zhang, F. Ramsdell, and S.F. Ziegler. 2001. Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J. Biol. Chem. 276:37672–37679. [DOI] [PubMed] [Google Scholar]
- 40.Okada, E., M. Yamazaki, M. Tanabe, T. Takeuchi, M. Nanno, S. Oshima, R. Okamoto, K. Tsuchiya, T. Nakamura, T. Kanai, et al. 2005. IL-7 exacerbates chronic colitis with expansion of memory IL-7Rhigh CD4+ mucosal T cells in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 288:G745–G754. [DOI] [PubMed] [Google Scholar]
- 41.Baecher-Allan, C., E. Wolf, and D.A. Hafler. 2005. Functional analysis of highly defined, FACS-isolated populations of human regulatory CD4+ CD25+ T cells. Clin. Immunol. 115:10–18. [DOI] [PubMed] [Google Scholar]
- 42.Baecher-Allan, C., E. Wolf, and D.A. Hafler. 2006. MHC class II expression identifies functionally distinct human regulatory T cells. J. Immunol. 176:4622–4631. [DOI] [PubMed] [Google Scholar]
- 43.Herold, K.C., W. Hagopian, J.A. Auger, E. Poumian-Ruiz, L. Taylor, D. Donaldson, S.E. Gitelman, D.M. Harlan, D. Xu, R.A. Zivin, and J.A. Bluestone. 2002. Anti-CD3 monoclonal antibody in new-onset type 1 diabetes mellitus. N. Engl. J. Med. 346:1692–1698. [DOI] [PubMed] [Google Scholar]
- 44.Chen, W., K.C. Herold, and J.A. Bluestone. 2005. Achieving antigen-specific tolerance in diabetes: regulating specifically. Int. Rev. Immunol. 24:287–305. [DOI] [PubMed] [Google Scholar]
- 45.Tang, Q., J.Y. Adams, A.J. Tooley, M. Bi, B.T. Fife, P. Serra, P. Santamaria, R.M. Locksley, M.F. Krummel, and J.A. Bluestone. 2006. Visualizing regulatory T cell control of autoimmune responses in nonobese diabetic mice. Nat. Immunol. 7:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox, A.L., S.A. Thompson, J.L. Jones, V.H. Robertson, G. Hale, H. Waldmann, D.A. Compston, and A.J. Coles. 2005. Lymphocyte homeostasis following therapeutic lymphocyte depletion in multiple sclerosis. Eur. J. Immunol. 35:3332–3342. [DOI] [PubMed] [Google Scholar]
- 47.Viglietta, V., C. Baecher-Allan, H.L. Weiner, and D.A. Hafler. 2004. Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. J. Exp. Med. 199:971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kriegel, M.A., T. Lohmann, C. Gabler, N. Blank, J.R. Kalden, and H.M. Lorenz. 2004. Defective suppressor function of human CD4+CD25+ regulatory T cells in autoimmune polyglandular syndrome type II. J. Exp. Med. 199:1285–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Kleer, I.M., L.R. Wedderburn, L.S. Taams, A. Patel, H. Varsani, M. Klein, W. de Jager, G. Pugayung, F. Giannoni, G. Rijkers, et al. 2004. CD4+CD25bright regulatory T cells actively regulate inflammation in the joints of patients with the remitting form of juvenile idiopathic arthritis. J. Immunol. 172:6435–6443. [DOI] [PubMed] [Google Scholar]
- 50.Oberley, M.J., J. Tsao, P. Yau, and P.J. Farnham. 2004. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 376:315–334. [DOI] [PubMed] [Google Scholar]
- 51.Cawley, S., S. Bekiranov, H.H. Ng, P. Kapranov, E.A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A.J. Williams, et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 116:499–509. [DOI] [PubMed] [Google Scholar]