Abstract
The chemokine stromal cell–derived factor (SDF-1; also known as chemokine ligand 12 [CXCL12]) regulates many essential biological processes, including cardiac and neuronal development, stem cell motility, neovascularization, angiogenesis, apoptosis, and tumorigenesis. It is generally believed that SDF-1 mediates these many disparate processes via a single cell surface receptor known as chemokine receptor 4 (CXCR4). This paper characterizes an alternate receptor, CXCR7, which binds with high affinity to SDF-1 and to a second chemokine, interferon-inducible T cell α chemoattractant (I-TAC; also known as CXCL11). Membrane-associated CXCR7 is expressed on many tumor cell lines, on activated endothelial cells, and on fetal liver cells, but on few other cell types. Unlike many other chemokine receptors, ligand activation of CXCR7 does not cause Ca2+ mobilization or cell migration. However, expression of CXCR7 provides cells with a growth and survival advantage and increased adhesion properties. Consistent with a role for CXCR7 in cell survival and adhesion, a specific, high affinity small molecule antagonist to CXCR7 impedes in vivo tumor growth in animal models, validating this new receptor as a target for development of novel cancer therapeutics.
Stromal cell–derived factor (SDF-1), a member of the superfamily of chemoattractant cytokines known as chemokines, is a key regulator of B cell lymphopoiesis (1, 2), hematopoietic stem cell mobilization (3), and leukocyte migration. In mice genetically deleted of SDF-1 (1), early stage embryos exhibit profound defects in the formation of large vessels, as well as other morphological anomalies such as septal malformation during cardiac development and abnormal brain patterning, including a disorganized cerebellum (1). Ultimately, embryonic lethality is observed typically between days 15 and 18 of gestation. Several other reports demonstrate that SDF-1 can induce angiogenesis in a variety of ex vivo and in vivo models (4, 5).
SDF-1 has been thought to mediate all of these functions exclusively via a single cell surface receptor known as chemokine receptor 4 (CXCR4) (6). There are multiple shared biological functions of SDF-1 and CXCR4, with animals bearing CXCR4 −/− and SDF-1 −/− genetic mutations manifesting overlapping phenotypic features and exhibiting embryonic lethality at approximately the same point in fetal development (1, 2, 7). CXCR4 is a coreceptor for HIV (8, 9), and SDF-1 blocks HIV viral entry (6). Moreover, both SDF-1 and CXCR4 have been implicated in tumor cell metastasis (10) and proliferation (11–13), and CXCR4 antagonism blocks in vivo growth of several tumors (13).
We have previously reported (14) that SDF-1 binds a second receptor, which we assigned the temporary designation of CCX-CKR2 until further characterization was obtained. The present study reports on the use of cellular, molecular, biological, and pharmacological approaches to extensively characterize CCX-CKR2. The results reveal a previously unknown mechanism by which SDF-1 functions that is separate from the classical SDF-1–CXCR4 pathway. With an expanded understanding of CCX-CKR2 properties, we now rename this receptor CXCR7 and provide evidence for its role in oncologic processes.
RESULTS
Evidence for a novel SDF-1 binding protein
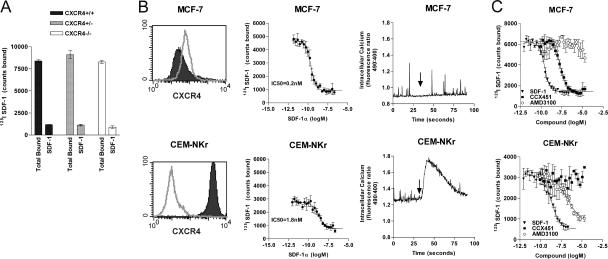
SDF-1– and CXCR4-deficient mice share the phenotype of embryonic lethality occurring around embryonic days 15–18 (E15–18) of gestation (1, 2, 7), clearly establishing a critical role for both proteins in embryonic development. To better understand the role of SDF-1/CXCR4 in early development, we further explored the consequences of CXCR4 gene deletion in mice before the E15–18 lethality event. However, experiments designed to characterize the wild-type, heterozygous, and homozygous mice in our CXCR4 −/− colony (7) unexpectedly revealed comparable binding of radiolabeled SDF-1 by E13 fetal liver cells obtained from all three mouse groups (Fig. 1 A).
Figure 1.
Evidence for a novel SDF-1 binding protein. (A) 125I SDF-1 binds to E13 fetal liver from wild-type (CXCR4 +/+), heterozygous (CXCR4 +/−), and homozygous (CXCR4 −/−) mouse embryos. In each case, binding specificity is demonstrated by the ability of 100 nM of unlabeled SDF-1α to effectively compete radiolabeled ligand binding to fetal liver calls. Data represent the means of three determinations ± SD. (B) FACS analysis of CXCR4 expression with the anti-CXCR4 mAb 12G5 (left), assessment of radiolabeled 125I SDF-1 binding (middle), and 10 nM SDF-1–induced calcium mobilization (right) using MCF-7 cells (top) or CEM-NKr cells (bottom). FACS histogram indicates isotype control (open) and specific staining (shaded). Arrows indicate ligand (SDF-1α) addition to cells. This set of analyses has been reproduced in at least 10 independent studies, and error bars represent SEM. (C) SDF-1 binding site on MCF-7 cells is pharmacologically distinct from CXCR4. 125I SDF-1 binding to CEM-NKr and MCF-7 cells is performed in the presence of SDF-1α, CXCR7 antagonist CCX451, or CXCR4 antagonist AMD3100. Error bars represent SEM.
Consistent with these observations using cells from CXCR4-deficient mice, separate experiments revealed discrepancies between CXCR4 expression and SDF-1 binding on different cell lines. Several tumor cell lines, including the breast tumor cell line MCF-7, bound 125I SDF-1 with exceptionally high affinity (∼200 pM; Fig. 1 B, top middle) but did not stain with the commercially available anti-CXCR4 mAb 12G5 (Fig.1 B, top left), nor did cells mobilize calcium (Fig. 1 B, top right) or migrate (not depicted) in response to SDF-1. In contrast, a transformed T cell line, CEM-NKr, exhibited more typical characteristics of CXCR4 expression, namely high affinity binding to 125I SDF-1 (IC50 ∼1.8 nM; Fig. 1 B, bottom middle), good reactivity with anti-CXCR4 mAb (Fig.1 B, bottom left), and both a robust increase in cytosolic calcium levels (Fig. 1 B, bottom right) and specific migration (not depicted) in response to SDF-1. These data suggested that functional CXCR4 protein was not expressed on the MCF-7 cell surface and revealed a previously unrecognized pattern of SDF-1 activity. In addition, the data indicated that, in some cases, SDF-1 interacts differently with different cell types, with CEM-NKr and MCF-7 exemplifying the contrasting behaviors.
We tested ∼100,000 “small molecule” organic compounds for their ability to inhibit binding of 125I SDF-1 to either CEM-NKr or MCF-7 cells. One compound that inhibited SDF-1 binding to MCF-7 cells, but not to CEM-NKr cells, was optimized chemically, yielding an SDF-1 binding antagonist designated CCX451 (14). CCX451 inhibited binding of 125I SDF-1 to MCF-7 cells with high affinity (∼5 nM; Fig.1 C, top, closed squares) but did not inhibit binding of 125I SDF-1 to CEM-NKr cells (Fig. 1 C, bottom, closed squares). In contrast, another small molecule, AMD3100, known to specifically inhibit SDF-1 binding to CXCR4 (15) was shown to have the inverse pattern of inhibition; it inhibited binding of 125I SDF-1 to CEM-NKr cells but not to MCF-7 cells (Fig. 1 C, open circles). Consistent with these binding data, AMD3100 inhibited all functions of SDF-1 on CEM-NKr cells (i.e., calcium mobilization and cell migration), whereas CCX451 had no effect (unpublished data). CCX451 did not affect the binding of any other known CXCR to its specific chemokine ligand (CXCL; unpublished data).
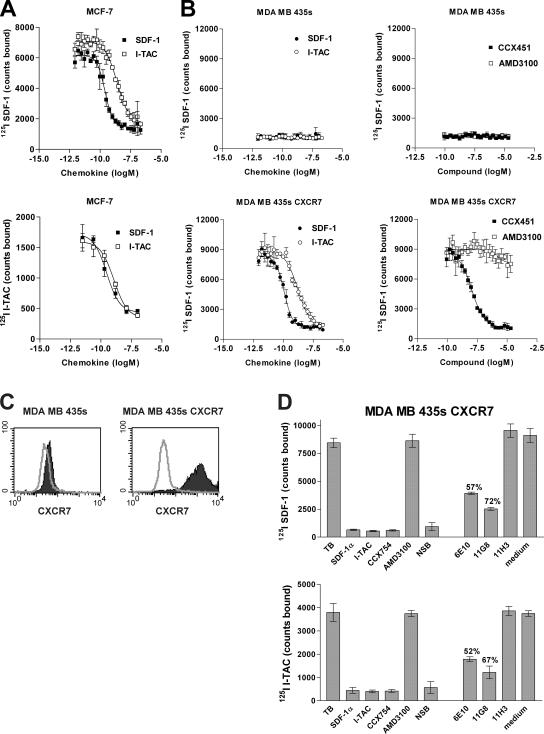
To explore the specificity of SDF-1 binding to the MCF-7 and CEM-NKr cell lines, we used a comprehensive competition binding approach described in detail elsewhere (16). Using 125I SDF-1 as the “signature” ligand, the ability of >90 discrete chemokines and chemokine variants to displace the binding of SDF-1 to CEM-NKr and MCF-7 cells was investigated. On CEM-NKr cells, only SDF-1 and the human herpes virus 8–encoded chemokine vMIP-II (known to bind CXCR4) (17) were effective competitors (unpublished data). In contrast, a previously unknown binding pattern was observed on MCF-7 cells, where in addition to SDF-1 and vMIP-II, the CXC chemokine interferon-inducible T cell α chemoattractant (I-TAC; also known as CXCL11) strongly displaced 125I SDF-1 binding (Fig. 2 A, top). Similarly, when 125I I-TAC was used as the signature ligand, it bound to and was displaced from MCF-7 cells by unlabeled SDF-1 (Fig. 2 A, bottom).
Figure 2.
The novel SDF-1/I-TAC binding protein is RDC1. (A)Binding of 125I SDF-1 or 125I I-TAC to MCF-7 cells was effectively competed using either unlabeled SDF-1 or unlabeled I-TAC. Error bars represent SEM. (B) Binding of 125I SDF-1 to either human breast tumor MDA MB 435s wild-type cells (top) or MDA MB 435s transfected with CXCR7 (bottom) in the presence of increasing concentrations of SDF-1α, I-TAC, CXCR7 antagonist CCX451, and CXCR4 antagonist AMD3100. Total counts bound for each condition are shown and represent the mean of four determinations ± SD. RDC1 Homo sapiens is available from GenBank/EMBL/DDBJ under accession no. P25106. (C) Mouse mAb to human CXCR7 (11G8) binds MDA MB 435s cells transfected with CXCR7 (right) but not MDA MB 435s wild-type cells (left). FACS histogram indicates isotype control (open) and specific staining (shaded). (D) Mouse anti–human CXCR7 mAbs 11G8 and 6E10 but not irrelevant isotype control, 11H3, largely inhibit binding of radiolabeled SDF-1 (top) or I-TAC (bottom) to CXCR7 transfectants. Error bars represent SEM. Data shown in (C) and (D) have been obtained in at least five experiments, and the percent inhibition relative to untreated controls (total bound) is shown.
Previously I-TAC, as well as two other chemokines, Mig (CXCL9) and IP10 (CXCL10), had been thought to bind only CXCR3 (18, 19). However, the MCF-7 cells used in these studies expressed neither CXCR3 protein nor mRNA (unpublished data). Furthermore, IP10 and Mig had no inhibitory effect on the binding of 125I SDF-1 or 125I I-TAC to the MCF-7 cells (unpublished data). Like 125I SDF-1, 125I I-TAC binding to MCF-7 was inhibited by CCX451 but not AMD3100 (unpublished data), and I-TAC failed to induce either calcium mobilization in or migration of MCF-7 cells. CCX451 did not inhibit the binding of 125I I-TAC to a cell transfectant expressing CXCR3 and lacking SDF-1 binding sites (unpublished data). Competitive binding analysis of SDF-1 on the CEM-NKr cells showed low nM affinities in all cases (Figs. 1 C and 2 A). However, SDF-1 consistently bound to the MCF-7 cells with a ∼20-fold higher affinity than that of I-TAC binding to MCF-7 cells or SDF-1 binding to CEM-NKr cells; i.e., ∼100–200 pM versus 2–5 nM, respectively (Fig. 1 B, middle panels).
The novel SDF-1/ITAC-1 binding protein is the 7-transmembrane receptor RDC1
To identify the molecular nature of the new SDF-1/I-TAC binding site, we used the signature binding characteristics defined in the previous section; i.e., 125I SDF-1 binding that could be inhibited by I-TAC and CCX451 but not by AMD3100. Among other strategies, our search included evaluation of “orphan” G protein–coupled receptors (GPCRs), which by predicted amino acid sequence looked structurally similar to CXCRs (for review see reference 20). One orphan, RDC1 (21, 22), was introduced into the human breast tumor–derived cell line MDA MB 435s, which lacks both CXCR4 and RDC1 mRNA (not depicted) and also lacks the ability to bind 125I SDF-1 (Fig. 2 B). In contrast to the wild-type line, the RDC1 transfectant possessed the capacity for specific high affinity SDF-1 binding; this binding could be blocked by I-TAC and CCX451 but not by AMD3100 (Fig. 2 B), exhibiting affinities commensurate with those seen on MCF-7 cells (Fig. 2 A). Identical ligand binding properties were observed using a second polyclonal RDC1 transfected cell line, as well as 293 cells that had been transfected with RDC1 but not parental 293 cells (not depicted).
The RDC1 gene product was serologically distinct from both CXCR4 and CXCR3, as specific antibodies to these receptors failed to bind RDC1-transfected cells (unpublished data). In contrast, mAbs produced by genetic immunization of mice with a plasmid-encoding human RDC1 specifically bound to the RDC1 transfectant but not to the wild-type MDA-MB 435s cells (Fig. 2 C). Separate experiments showed the anti-RDC1 antibodies bound to two separate polyclonal lines of MBA-MB 435s cells transfected with RDC1, as well as 293 cells transfected with RDC1, but did not bind to MBA-MB 435s cells or 293 cells that had been mock transfected or transfected with another CXCR (unpublished data). Anti-RDC1 hybridoma supernatants 11G8 or 6E10 but not an unrelated isotype control antibody, 11H3, specifically inhibited binding of both 125I SDF-1 and 125I I-TAC to the RDC1 transfectants in a manner similar to that of the small molecule antagonist CCX754, a CCX451 analogue (Fig. 2 D). In subsequent experiments, higher concentrations of purified preparations of 11G8 and 6E10 mAbs completely neutralized ligand binding, further validating the reagents' specificity (unpublished data). Collectively, these data indicate that the RDC1-encoded protein contains an SDF-1 binding site with properties that are indistinguishable from the novel SDF-1/I-TAC binding site expressed on MCF-7 cells (Figs. 1 and 2). We provisionally designate the RDC1 gene product as CXCR7 for the seventh receptor identified to date for chemokines belonging in the CXC class.
Preferential expression of CXCR7 by transformed cells and during embryonic development
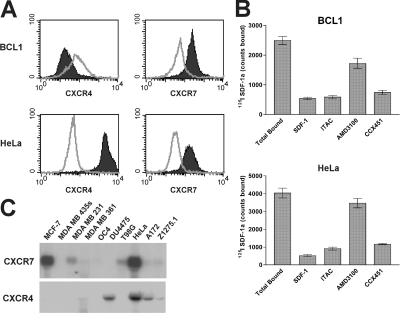
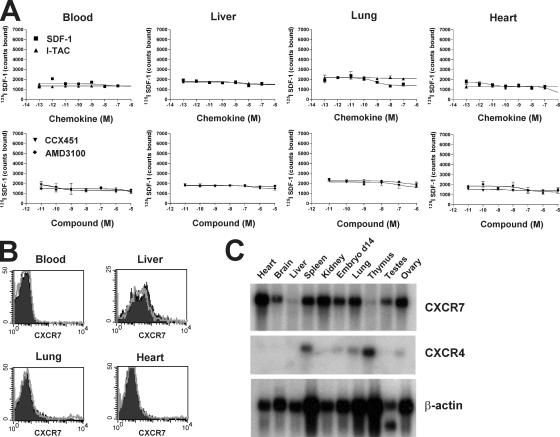
Our initial characterization of CXCR7 expression revealed that many of the human and mouse transformed cell types we tested (e.g., MCF-7 breast tumor, HeLa cervical carcinoma, and BCL1 lymphoma are reported in this paper, whereas T98G glioma, A549 lung carcinoma, and 4T1 breast tumor are unpublished data) were abundantly positive for expression of CXCR7 protein (Fig. 3). This conclusion was evident when CXCR7 expression was evaluated by any of several techniques; e.g., flow cytometric staining with the anti-CXCR7 mAb (Fig. 3 A), 125I SDF-1 binding that could be inhibited by ITAC and CCX451 but not by AMD3100 (Fig. 3 B), and by Northern blot analysis of CXCR7 mRNA levels (Fig. 3 C). Importantly for these experiments, the anti–human CXCR7 antibody also bound mouse CXCR7 on mouse tumor lines and cell lines transfected with mouse CXCR7 (Fig. 3 A and not depicted). CXCR7 was observed on cell lines both coexpressing or lacking CXCR4 (Fig. 3, A and C). In contrast, little or no expression of CXCR7 was observed on nontransformed adult mouse tissues surveyed as defined by the 125I SDF-1 binding assay (representative examples out of many are shown in Fig. 4 A) or staining by the anti-CXCR7 mAb (Fig. 4 B). Interestingly, many of the same nontransformed tissues that lacked surface CXCR7 expression expressed CXCR7 mRNA (Fig. 4 C), suggesting that CXCR7 can be regulated in a posttranslational manner. Collectively, the data in Figs. 3 and 4 suggest that membrane CXCR7 protein is frequently expressed on transformed cells but not on normal cells.
Figure 3.
Expression of CXCR7 by transformed human and mouse cell lines. (A) Membrane CXCR7 and CXCR4 expression on mouse B cell lymphoma, BCL1, and human cervical carcinoma HeLa cells is demonstrated by anti-CXCR7 antibody 11G8 staining and anti-CXCR4 antibody 12G5. Staining is represented as shaded versus open for isotype control. (B) 125I SDF-1 binding profile reveals membrane CXCR7 expression on BCL1 and HeLa cell surfaces. CXCR7 binding is defined by inhibition of the 125I SDF-1 binding with 100 nM of nonradiolabeled ligand or 10 μM CXCR7 antagonist CCX451, but not by 10 μM CXCR4 antagonist AMD3100. Error bars represent SEM. (C) Northern blot analysis of mRNA expression of CXCR7 and CXCR4 in a panel of human transformed cell lines.
Figure 4.
Nontransformed human and mouse tissues express little membrane CXCR7 but frequently express CXCR7 by Northern blot analysis. (A) Binding of 125I SDF-1 to various tissue cells obtained from the mouse. Error bars represent SEM (B) Anti-CXCR7 antibody 11G8 fails to bind mouse blood, liver, lung, or heart cells. FACS histogram indicates isotype control (open) and specific staining (shaded). (C) Northern blot analysis shows CXCR7-specific mRNA in normal mouse tissues, but this does not correlate with cell surface protein expression.
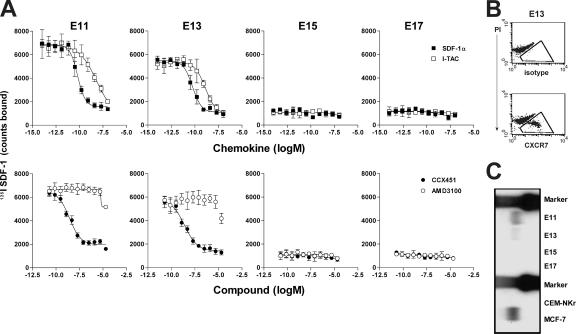
Our initial observation of SDF-1 binding to fetal liver cells from normal and CXCR4-deficient mice (Fig. 1 A) suggested that CXCR7 is expressed during normal embryonic development. Indeed, the SDF-1 binding site expressed by E13 embryos of CXCR4−/− mice was inhibited by I-TAC and CCX451 but not by AMD3100 (Fig. 5 A), confirming its identity as CXCR7. CXCR7 was found to be expressed abundantly during embryonic development, but its expression was transitory, being detected on mouse fetal liver cells at E11 and E13 but not on E15 and E17 (Fig. 5, A–C). This pattern of CXCR7 expression in fetal development was demonstrated by ligand binding (Fig. 5 A), antibody staining (Fig. 5 B), and Northern blot analysis (Fig. 5 C). Preliminary flow cytometric and immunohistochemical analysis suggest that this subset of CXCR7 positive cells comprises primitive erythrocytes in transit through the liver (unpublished data). Further studies are ongoing to characterize this population of cells. Interestingly, this loss of CXCR7 expression coincides with the lethal consequence of SDF-1 or CXCR4 genetic deletion occurring between E15 and E17 (1, 2, 7), suggesting that CXCR4 becomes critical in embryonic development at a stage when CXCR7 expression diminishes.
Figure 5.
CXCR7 is expressed in early mouse embryonic development. (A) Binding of 125I SDF-1 to embryonic fetal liver (E11–17) in the presence of increasing concentrations of SDF-1α, I-TAC, the CXCR7 antagonist CCX451, or the CXCR4 antagonist AMD3100. Error bars represent SEM. (B) A subset of E13 mouse fetal liver cells stain positive using anti-CXCR7 antibody 11G8, as indicated by the boxed areas. (C) Northern blot analysis shows expression of CXCR7 mRNA in E11 and E13 but not E15 and E17 fetal liver. Total RNA from CXCR7-positive MCF7 cells and CXCR7-negative CEM-NKr cells are included as controls. Data shown were obtained in at least five experiments.
Role for CXCR7 in cell survival in vitro
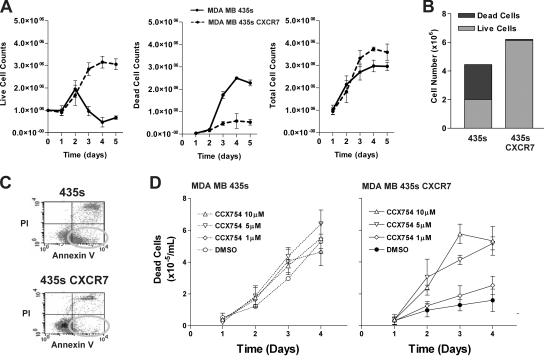
A role of CXCR7 in cell growth/survival was first indicated by an observation that CXCR7-transfected MDA MB 435s cells expanded more rapidly in culture than wild-type MDA MB 435s cells, especially in suboptimal tissue culture medium (e.g., medium supplemented with only 1% fetal calf serum). To explore this phenomenon, both cell types were cultured in 1% serum-containing medium for 5 d in replicate culture dishes, with separate dishes harvested on each day for cell count evaluation. As shown in Fig. 6 A, most wild-type cells died during culture in the decreased serum environment (continuous line); in contrast, the CXCR7 transfectants expanded exponentially for the first few days, reaching a plateau of live cells by 4–5 d after plating (dashed line). Similar results were obtained using several different CXCR7 transfectant lines, as well as wild-type cells that were mock transfected. The difference in live cell numbers in cultures of CXCR7 transfectants versus wild-type lines appeared to reflect increased cell survival rather than increased proliferation of the CXCR7 transfected cells, because the total cell recovery of transfected and wild-type cells after 5 d of culture was similar (Fig. 6 A, right; and Fig. 6 B), whereas the proportion of dead cells was dramatically different (Fig. 6 A, middle; and Fig. 6 B). Indeed, use of Annexin V staining to identify apoptotic cells showed a substantial proportion (∼40%) of apoptotic cells in wild-type 435s cells after a 5-d culture, whereas the CXCR7 transfectant cell culture contained relatively few apoptotic cells (Fig. 6 C). The specificity of this effect was demonstrated by the fact that CXCR7 antagonist CCX754 could reverse this protection in a dose-related fashion (Fig. 6 D). The same compound had no effect on wild-type cells (Fig. 6 D) or other CXCR7-negative cell lines (unpublished data). Collectively these data indicate that expression of CXCR7 confers a survival advantage to cells that becomes experimentally evident using tissue culture conditions that are suboptimal for cell growth.
Figure 6.
Introduction of CXCR7 into human breast tumor cell line MDA MB435s confers a growth advantage to these cells. In all experiments, wild-type or CXCR7-transfected MB435s cells were cultured in vitro in media containing a suboptimal concentration of serum (1% instead of the standard 10%). (A) Cell counts of live, dead, and total wild-type or CXCR7-transfected cells cultured over time. (B) Summary of the day 4 time point from growth experiments in histogram form to emphasize the fact that the wild-type and CXCR7-transfected cells have similar total cell numbers but are distinguished by how many of these total cells are dead. (C) Apoptotic cells in these cultures were identified (see oval) as cells excluding propridium iodide and that stained with the Annexin V marker for apoptotic cells. (D) CCX754 inhibits CXCR7-mediated growth advantage in a dose-dependant manner. Wild-type or CXCR7-transfected cells were incubated with various concentrations of CXCR7 compound throughout assay.
Role for CXCR7 in cell adhesion in vitro
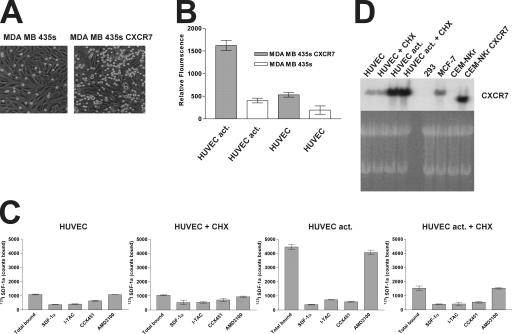
We additionally tested the ability of CXCR7 to promote adhesion to activated endothelial cell monolayers. Fluorescently labeled wild-type MDA MB 435s cells or CXCR7 transfectant cells were applied to human umbilical vein endothelial cell (HUVEC) monolayers after stimulation of the endothelium with the activating cytokines TNF-α and IL-1β, and adherence was measured by counting fluorescent cells (Fig. 7 A) or quantitating fluorescence (Fig. 7 B). The CXCR7 transfectant adhered to the activated HUVECs markedly more than did the wild-type cells (Fig. 7, A and B). Interestingly, the wild-type line showed some adherence to activated HUVECs, and the CXCR7 transfectant showed some adherence to unstimulated HUVECs (Fig. 7 B). In contrast, no adherence was observed when wild-type cells were added to unstimulated HUVECs (Fig. 7 B). Similar experiments using a separate polyclonal CXCR7-transfected cell line instead of the clonal CXCR7-transfected MDA MB 435s cells produced similar results (unpublished data). Further insight into the possible mechanism of this effect was provided by 125I SDF-1 binding assays, which revealed that the HUVECs up- regulate CXCR7 expression after in vitro stimulation with TNF-α and IL-1β (Fig. 7, C and D). Preincubation of the HUVEC monolayer with cyclohexamide before TNF-α and IL-1β stimulation substantially reduced expression of CXCR7 (Fig. 7, C and D), suggesting that the activated HUVECs synthesize new CXCR7 receptors instead of using preexisting (but nonbinding) CXCR7 receptors. Increased CXCR7 mRNA expression was also observed by Northern blot analysis after HUVEC activation (Fig. 7 D). Similar results were obtained using endothelial cells derived from lung, heart, and various other tissues (unpublished data). Thus, in addition to its marked expression in many transformed cells and early fetal liver cells (Figs. 3–5 ), CXCR7 can also be expressed by activated endothelium. Moreover, although expression of CXCR7 on one cell type can promote some cell–cell interactions, CXCR7 expression on both activated HUVECs and tumor cells concomitantly produced maximal adherence in vitro.
Figure 7.
CXCR7 mediates cell adhesion in vitro. Adherence is measured (A) by capturing a brightfield image to visualize both the HUVEC monolayer and the labeled adherent cells (bright circles) and (B) by fluorescence to quantitate adhesion. Error bars represent SEM. (C) Activated endothelium expresses CXCR7 by radiolabeled binding assay. HUVECs were stimulated with TNF-α and IL-1β (HUVEC act.) or sham-treated in the presence or absence of cycloheximide (CHX) and incubated with radiolabeled SDF-1. Unlabeled SDF-1α and I-TAC (100 nM each), as well as CCX451 and AMD3100 (10 μM each), were examined for the ability to compete with 125I SDF-1 binding. (D) Northern blot analysis of CXCR7-specific mRNA expressed by unstimulated (HUVEC) or TNF-α/IL-1β (HUVEC act.) cells cultured in the presence or absence of Cycloheximide (CHX). MCF7 (native expression) and CEM-NKr (transfectant) CXCR7 transcripts are shown as controls. Data for A–C were reproduced in at least five experiments. Data for D were obtained twice.
Effect of CXCR7 antagonism in mouse tumor models
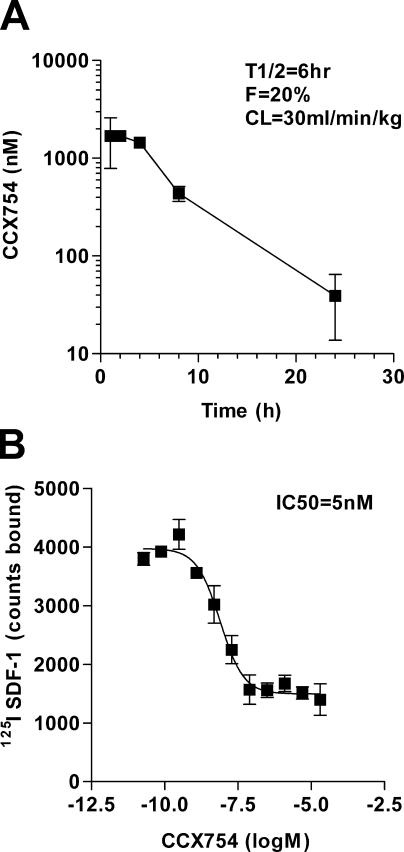
The role of CXCR7 on cell survival and adhesion, coupled with the pronounced expression of the receptor on transformed cells and activated endothelial cells, prompted us to examine its role in mammalian models of tumor formation. To this end, we tested the CXCR7 antagonist CCX754 (14), an analogue of CCX451 that has superior in vivo pharmacokinetic properties, in mouse models engrafted with various tumors. CCX754's pharmacokinetic properties permitted once daily dosing (Fig. 8 A), bound to CXCR7 with low nM affinity (Fig. 8 B), and demonstrated high selectivity for CXCR7 over other CXCRs (Table I). Efficacy of CCX754 was evaluated in immunodeficient or syngeneic mice engrafted with either human B lymphoma IM9 (Fig. 9 A), human lung carcinoma A549 (Fig. 9 C), or mouse lung carcinoma LL/2 (Fig. 9 D). Each of these tumors was shown to express CXCR7 by both the signature 125I SDF-1 binding profile and flow cytometric staining with the anti-CXCR7 mAb (unpublished data).
Figure 8.
Pharmacological properties of CXCR7 antagonist (CCX754) used in in vivo experiments. The experiments are shown in Fig. 9. (A) Pharmacokinetic properties of CXCR7 antagonist in mice show a serum half-life (T1/2) of 6 h, bioavailability of 20% (F), and an acceptable liver clearance rate of 30 ml/min/kg (CL). These pharmacokinetic properties are compatible with once a day dosing in mouse animal models. (B) CCX754 inhibits binding of 125I SDF-1 to MCF-7 human breast tumor cells with an IC50 of 5 nM. Error bars represent SEM in both panels.
Table I.
Cross-reactivity of CCX754 for other CXCRs
| Receptor | Affinitya | Receptor | Affinity |
|---|---|---|---|
| CCR1 | >100 μM | CXCR1 | >100 μM |
| CCR2 | >100 μM | CXCR2 | 30 μM |
| CCR3 | >100 μM | CXCR3 | 60 μM |
| CCR4 | 60 μM | CXCR4 | >100 μM |
| CCR5 | >100 μM | CXCR6 | >100 μM |
| CCR6 | >100 μM | CX3CR1 | 90 μM |
| CCR7 | >100 μM | ||
| CCR8 | 90 μM | CXCR7 | 5 nM |
| CCR9 | >100 μM | ||
| CCR12 | >100 μM |
CCX754 is highly selective for CXCR7 inhibition. Data report the concentration of CCX754 required to inhibit specific CXCL binding to the receptors indicated.
Numbers represent IC50 in radiolabeled binding assays.
Figure 9.
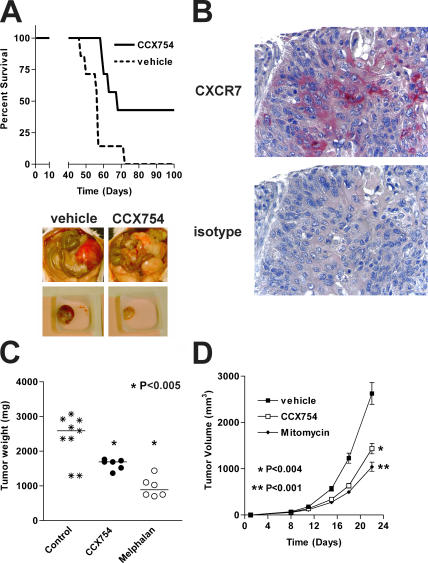
In vivo efficacy of CXCR7 antagonist. Efficacy of CCX754 was evaluated in syngeneic or xenograft mouse models engrafted with (A) human B lymphoma IM9 cells, (C) human A549 lung carcinoma cells, or (D) mouse LL/2 Lewis lung carcinoma cells. Control groups receiving vehicle alone were included in all experiments. Horizontal bars in C indicate means, and error bars in D represent SEM. Images in A show peritoneal cavity of mice bearing IM9 tumor cells and receiving either CXCR7 antagonist (right) or vehicle alone (left). (B) Immunohistochemical analysis of CXCR7 expression on a human biopsy sample of a malignant lung carcinoma. n = 8 in all groups. Statistical differences between treatment groups were calculated using survival curve statistics and the Student's t test using GraphPad Prism software.
In the first tumor model, IM9 human B lymphoma cells were introduced into the peritoneal cavity of immunodeficient mice; animals were randomized and treated with either CCX754 or an otherwise identical vehicle formulation that lacked drug. Levels of CCX754 in sera collected throughout the experiment were quantitated by mass spectrometry and found to exceed the IC50 of this compound determined in binding studies (unpublished data). By day 70, all of the mice in the vehicle-treated cohort had succumbed to the tumor (Fig. 9 A). In contrast, the cohort receiving CCX754 fared significantly better (P < 0.012), with almost half the cohort surviving at 100 d (Fig. 9 A). In a second experiment of this type we found that, on necropsy, all the mice treated with vehicle had large, encapsulated, vascularized tumors, whereas more than half of the animals treated with CCX754 developed no tumors at all, and those tumors that did form were not encapsulated, poorly organized, and frequently had minimal if any vascularization (Fig. 9 A).
We were particularly interested in evaluating the effect of our CXCR7 antagonist in animal models of lung cancer, because we observed in immunohistochemical stains that many biopsy samples of primary human lung tumors expressed CXCR7 (one example of many shown in Fig. 9 B). To evaluate potential efficacy of CCX754 in a xenograft lung carcinoma model, immunodeficient mice were implanted subcutaneously with fragments of A549 human lung tumor. Animals were randomized and treated with either CCX754, vehicle alone, or Melphalan as a positive control. At 49 d, cohorts receiving either CCX754 or Melphalan exhibited reduced tumor volume as compared with the vehicle control group (P < 0.005; Fig. 9 C). In a separate syngeneic tumor model, immunocompetent mice were inoculated subcutaneously with LL/2 Lewis lung carcinoma cells. Cohorts of mice were randomized and treated with either CCX754, vehicle alone, or mitomycin C as a positive control. Mice receiving the CXCR7 antagonist developed markedly smaller tumors than those found in animals receiving vehicle only (P < 0.004; Fig. 9 D). Indeed, some tumors were similar in size to those in animals receiving the known potent antitumor agent mitomycin C (Fig. 9 D).
DISCUSSION
This study provides extensive characterization of the novel high affinity SDF-1 (CXCL12)–binding receptor that we reported in an earlier publication (14). The novel receptor, initially designated CCX CKR2 (14) but renamed CXCR7 in this paper, is a 7-transmembrane receptor encoded by RDC1 (21, 22), a gene that, before our initial report (14), belonged to the family of orphan receptors with unknown ligands (20). In addition to binding SDF-1, CXCR7 is also a high affinity receptor for I-TAC (CXCL11) that, before our investigations, was regarded as a ligand for CXCR3 only. Our data show that CXCR7 regulates several important biological processes including cell survival, cell adhesion, and tumor development in animal models. CXCR7 is expressed on many tumor cells but not on most nontransformed cells. Although not expressed on unstimulated endothelial cells, CXCR7 can be induced, in vitro, on cells that form the neovasculature, a finding that is consistent with the independent studies of Madden et al., who have observed up-regulation of RDC1 expression in neovasculature associated with malignant gliomas (23). Some of the effects observed have been previously associated with SDF-1 activity but have been thought, perhaps erroneously, to be mediated entirely through CXCR4. The elucidation of CXCR7 may require a reexamination of much of the previous body of work that presumed a mutually exclusive biological interaction between SDF-1 and CXCR4. Indeed, suggestions of discrepant CXCR4 expression and SDF-1 responsiveness already exist in the literature. For example, previous studies demonstrate that adhesion of E11 fetal liver cells to bone marrow endothelium can be neutralized by anti–SDF-1 antibodies despite the fact that the E11 fetal liver cells do not migrate in response to SDF-1 (24).
The gene that encodes CXCR7, RDC1, was initially cloned from a dog thyroid cDNA library using degenerate PCR primers corresponding to consensus sequences in the transmembrane domains of known GPCR (21, 25). Subsequent isolation of human and mouse RDC1 homologues (22) revealed >90% identity of both nucleotide and protein sequences of all three species, indicating an extremely high level of evolutionary conservation. RDC1 protein has been reported to be a vasoactive intestinal peptide receptor (26) and an adrenomedulin receptor (27), but these designations have fallen from general acceptance (28–30). RDC1- encoded CXCR7 is structurally similar to CXCR2, and the CXCR7 gene maps between those of CXCR2 and CXCR4 on mouse chromosome 1 (22). It is likely that the RDC1/CXCR7 escaped earlier deorphanization and identification as a CXCR because it lacks certain typical and easily accessible functional properties of CXCRs, namely the ability to mediate chemotaxis and calcium mobilization after ligand binding. In this context, previous reports of CXCR4 −/− mice demonstrated the absence of SDF-1–induced functional responses such as chemotaxis (1, 7) but lacked binding experiments with radiolabeled SDF-1 that would have revealed the existence of the second SDF-1 receptor (i.e., CXCR7). Although numerous database search engines suggest that CXCR7/RDC1 is broadly expressed at the mRNA level (e.g., http://www.sagenet.org and http://www.symatlas.org; see also references 22, 25), our study demonstrates this is not the case at the level of membrane-associated CXCR7. This seeming discrepancy is explained by our direct demonstration of certain nontransformed cells that express CXCR7-specific mRNA but lack surface CXCR7 protein as measured by ligand binding assays or anti-CXCR7 antibody staining (Fig. 4). This observation potentially reflects some type of posttranslational regulation in CXCR7 expression. Although we have observed many examples in which nontransformed cells express CXCR7 mRNA but lack surface CXCR7, we have seen complete concordance to date in CXCR7 mRNA and surface CXCR7 expression in tumor cells, E13 mouse fetal liver cells, and activated endothelial cells (Figs. 3, 5, and 7).
The absence of ligand-induced CXCR7-mediated calcium mobilization or cell migration suggests that the CXCR7 signaling pathway is distinct from the typical GPCR mechanism of other CXCRs. Although an alternative CXCR7-linked signal transduction pathway has not been identified in this manuscript, receptor-mediated signaling is implied by our observations that CXCR7 provides a growth/survival advantage and increased adhesiveness of cells (Figs. 6 and 7). Indeed, preliminary evidence from microarray analyses suggests that CXCR7 may be constitutively active in tumor cell lines, thereby resembling numerous constitutively active non-CXCR GPCRs (31–33), as well as some virally encoded CXCRs; e.g., CMV-encoded US28 (34, 35). The same studies (31–35) indicate that constitutively active 7TM-GPCRs can nevertheless be regulated by receptor-binding ligands via a process of inverse agonism rather than by acting as traditional agonists or antagonists.
Our experiments suggest a potential role for CXCR7 in tumor development. In separate experiments, we have surveyed a broad panel of primary human tumors for CXCR7 expression by immunohistochemistry and found that many human tumors, as well as the neovasculature feeding these tumors (but not normal blood vessels), express CXCR7 (unpublished data). It is not yet clear whether the suppressed tumor growth observed in the presence of CXCR7-binding small molecules in vivo (Fig. 9) reflects action on tumor CXCR7, vasculature CXCR7, or both. In support of a direct role for CXCR7 on the tumor cells, we have done an extensive series of RNAi experiments showing that 90% reduction of CXCR7 expression on two separate tumor lines leads to dramatic reduction of in vivo growth of these tumors (unpublished data). We have also observed that introduction of CXCR7 into MDA MB435s cells transforms their in vivo growth from exceedingly slow to dramatically faster (unpublished data). Although these data implicate the importance of tumor-associated CXCR7, they do not exclude an additional role of CXCR7 expressed by neovasculature in tumor development.
Like CXCR4 and CCR5, CXCR7/RDC1 has been shown to be a coreceptor for HIV and SIV, in this case strains that are neither M- nor T-tropic (36). However, the role of CXCR7/RDC1 in the transmission and pathogenesis of HIV and SIV remains to be elucidated. A study published in late 2005 by Balabanian et al. (37) evaluated the shared HIV coreceptor function of CXCR4 and RDC1 and found that SDF-1–induced T cell chemotaxis could be blocked by specific antibodies to either CXCR4 or RDC1. Balabanian's work reproduced our finding (14) that both receptors used SDF-1 ligand; however, a major distinction between their study and our own is that SDF-1 binding to CXCR7 caused T cell chemotaxis in their hands, whereas we have not observed RDC1-mediated chemotaxis of any cell tested to date, including primary T cells (unpublished data). Furthermore, we have not detected surface CXCR7 on mouse or human T cells, either by radiolabeled SDF-1 binding analyses or CXCR7-specific mAb binding (unpublished data). The basis of the discrepant observations of Balabanian's study and our own is currently unknown and warrants further investigation.
In conclusion, CXCR7 is a new CXCR with properties that affect a spectrum of important biological and pathological processes, including cell growth/survival and adhesion, as well as the promotion of tumor growth. Our data suggest that the homeostatic and inflammatory events regulated by SDF-1 and I-TAC are much more complex than previously thought and that reinterpretation of earlier findings related to these chemokines in light of finding an additional high affinity receptor for them may be warranted. CXCR7 may provide a new molecular link in the chain of connections between inflammation and cancer, and in this context the interrelationships between CXCR7, CXCR4, CXCR3, and their shared chemokines, SDF-1 and I-TAC, will be of considerable interest. Finally, the elucidation of this new receptor may introduce new avenues of potential therapeutic intervention in important clinical indications, including oncology.
MATERIALS AND METHODS
Reagents and cells.
Chemokines were obtained from R&D Systems and PeproTech. 125I SDF-1 and 125I I-TAC were purchased from PerkinElmer and GE Healthcare, respectively. mAbs used in flow cytometry, anti-CXCR4 (clone 12G5) and normal mouse IgG2a, were obtained from R&D Systems. Goat anti–mouse IgG PE conjugate (Coulter Immunotech) was used to detect antibody binding. Unless otherwise indicated, cell lines were obtained from the American Type Culture Collection. CEM-NKr cells were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. HUVECs were purchased from Clonetics. Human PBMCs were collected from buffy coats of healthy donors (Stanford Blood Center), as described previously (38). Primary mouse cells were prepared from harvested organs by mechanical dispersion through a 70-μm nylon strainer.
Mice.
8-wk-old female C57BL/6 mice, SCID mice, and timed-pregnant C57BL/6 mice were purchased from Charles River Laboratories. All animal procedures and studies were performed in strict accordance with protocols approved by the ChemoCentryx, Inc. institutional animal care and use committee.
Radioligand binding assays.
Assays to assess radioligand binding to CXCR7 were performed as previously described (16). Radioligand binding was quantitated by analyzing the cells in a γ counter (PerkinElmer). Data were analyzed and plotted using software (Prism; GraphPad).
Calcium mobilization.
Calcium mobilization responses were performed as described previously using Indo-1, an intracellular ratiometric fluorescent dye (38).
Cell migration.
Cell migration assays were performed using a 96-well microchamber (Chemo Tix; NeuroProbes, Inc.). Cells were resuspended in chemotaxis buffer (HBSS with Ca2+, Mg2+, and 0.1% BSA) and placed on top of the filter (pore size of 3 or 5 μm, depending on cell size). Chemokines were placed in chamber below the filter. After 90 min at 37°C in a humidified incubator, filters were removed, 5 μl of CyQuant (Invitrogen) was added to the lower chamber, and the microplate was analyzed on a plate reader (Molecular Devices) at 540-nm wavelength.
Flow cytometry.
Cells were labeled using standard procedures and analyzed on a FACScan (Becton Dickinson). The data presented are gated for viable cells using light scatter.
CXCR7 transfectants.
The complete coding sequence of the gene encoding human CXCR7, RDC1, was isolated from MCF-7 cells using an mRNA isolation kit (μMACs; Miltenyi Biotec). DNA contamination was removed by DNase digestion on RNeasy columns (QIAGEN), and cDNA was generated using an RNA PCR core kit (GeneAmp; Applied Biosystems). PCR of cDNA samples was performed with Taq PCR Master Mix kit (QIAGEN) using RDC1 primers harboring 5′ and 3′ NotI sites (hRDC1F, 5′-GAATGCGGCCGCTATGGATCTGCATCTCTTCGACT-3′; hRDC1R, 5′-GAATGCGGCCGCTCATTTGGTGCTCTGCTCCAAG-3′). NotI-digested PCR products were ligated into NotI-digested pcDNA3.1(+) (Invitrogen) and screened for orientation, then the cDNA sequence was confirmed. Plasmid DNA was isolated from overnight bacterial cultures by Maxiprep (QIAGEN). 10 μg Plasmid DNA was added to MDA MB 435s cells via electroporation (0.22 kV, 960 uF) using a Gene Pulser (Bio-Rad Laboratories). 48 h after electroporation, cells were transferred to selection medium (1,000 μg/ml G418).
mAb production.
mAbs to human CXCR7 were obtained by DNA vaccination using an RDC1-containing plasmid. CXCR7-specific antibodies were identified by binding to human RDC1 transfectants versus transfectants expressing an irrelevant protein. Two CXCR7 specific antibodies designated 11G8 and 6E10 were generated. Specificity was confirmed by a lack of reactivity to a panel of chemokine receptors.
Antibody purification.
Antibodies were purified from harvested supernatants using protein G columns with subsequent acidic elution (glycin, pH 3) and dialysis against PBS. Antibody concentration was determined by Lowry assay against BSA.
Northern blot analyses.
10 μg total RNA was subjected to electrophoresis, transferred to nitrocellulose filters, and probed using reagents (NorthernMax; Ambion). Specific antisense riboprobes were generated from cloned mouse or human CXCR4 and CXCR7 open reading frames using an RNA kit (Strip-EZ; Ambion). The FirstChoice Mouse Blot 1 (Ambion) was purchased and probed with mouse CXCR4 and CXCR7 riboprobes.
Cell apoptosis assay.
Annexin V–FITC (Invitrogen) and propidium iodide (Invitrogen) were added to washed cells (106 cells/ml in FACS buffer) for 15 min at room temperature in the dark. FACS buffer was added, and cells were analyzed immediately by flow cytometry.
Adhesion assay.
HUVECs were allowed to adhere to 24-well plastic tissue culture plates overnight. The monolayer was treated with medium containing 10 ng/ml TNF-α and 10 ng/ml IL-1β for 5 h. Cells were loaded with calcein AM (Invitrogen) in PBS for 30 min at room temperature, washed, and added to the HUVEC monolayers for 15 min at 37°C. Adherent cells were quantitated by microscopy and by fluorescence intensity.
In vivo tumor models.
In the syngeneic model, C57BL/6 mice were inoculated with the mouse Lewis lung carcinoma line LL/2. In the xenograft model, CB17 SCID mice were inoculated with IM9 human lymphoma or transplanted with A549 human lung carcinoma fragments. In all experiments, s.c. treatment with CXCR7 antagonist (100 mpk qd) or vehicle (equivalent volume) commenced at the time of tumor transplantation. Positive control groups received known chemotherapeutic agents (either mitomycin or melphalan) administered at 2 mg/kg intraperitoneally every 2 d.
Immunohistochemistry.
5-μm sections of formalin-fixed, paraffin-embedded human or mouse spleen were deparaffinized, hydrated, and exposed to anti-CXCR7 mAb clone 11G8 at 10 μg/ml for 1 h. Rinsed slides were exposed to biotin-conjugated Fab′2 fragments of goat anti–mouse IgG for 30 min. The slides were rinsed and exposed to avidin-biotinylated alkaline phosphatase complex for 20 min. The slides were rinsed, exposed to fuchsin+ substrate for 5–20 min, and rinsed with deionized water. The slides were counterstained in Mayer's hematoxylin for 3 min, rinsed with tap water, and mounted with coverslips.
Acknowledgments
We thank Drs. K. Moore, C. Gerard, H. Showell, B. Premack, M. Stein, and R. Ransohoff for insightful comments. We gratefully acknowledge R&D Systems for providing many reagents and materials used in these studies. D. Littman is affiliated with Howard Hughes Medical Institute.
This work was funded entirely by ChemoCentryx, Inc.
The small molecule CXCR7 antagonists used in some experiments are the proprietary property of ChemoCentryx, Inc., and they may be developed for clinical application and, if successful, marketed as novel therapeutics. The compounds are used as research tools only in this study. The authors have no additional conflicting financial interests to report.
Abbreviations used: CXCL, chemokine ligand; CXCR, chemokine receptor; GPCR, G protein–coupled receptor; HUVEC, human umbilical vein endothelial cell; I-TAC, interferon-inducible T cell α chemoattractant; SDF-1, stromal cell–derived factor.
B.E. McMaster died on 17 May 2006.
References
- 1.Nagasawa, T., S. Hirota, K. Tachibana, N. Takakura, S. Nishikawa, Y. Kitamura, N. Yoshida, H. Kikutani, and T. Kishimoto. 1996. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 382:635–638. [DOI] [PubMed] [Google Scholar]
- 2.Ma, Q., D. Jones, P.R. Borghesani, R.A. Segal, T. Nagasawa, T. Kishimoto, R.T. Bronson, and T.A. Springer. 1998. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc. Natl. Acad. Sci. USA. 95:9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiuti, A., I.J. Webb, C. Bleul, T. Springer, and J.C. Gutierrez-Ramos. 1997. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J. Exp. Med. 185:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tachibana, K., S. Hirota, H. Iizasa, H. Yoshida, K. Kawabata, Y. Kataoka, Y. Kitamura, K. Matsushima, N. Yoshida, S. Nishikawa, et al. 1998. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 393:591–594. [DOI] [PubMed] [Google Scholar]
- 5.Salcedo, R., K. Wasserman, H.A. Young, M.C. Grimm, O.M. Howard, M.R. Anver, H.K. Kleinman, W.J. Murphy, and J.J. Oppenheim. 1999. Vascular endothelial growth factor and basic fibroblast growth factor induce expression of CXCR4 on human endothelial cells: in vivo neovascularization induced by stromal-derived factor-1alpha. Am. J. Pathol. 154:1125–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleul, C.C., M. Farzan, H. Choe, C. Parolin, I. Clark-Lewis, J. Sodroski, and T.A. Springer. 1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 382:829–832. [DOI] [PubMed] [Google Scholar]
- 7.Zou, Y.R., A.H. Kottmann, M. Kuroda, I. Taniuchi, and D.R. Littman. 1998. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 393:595–599. [DOI] [PubMed] [Google Scholar]
- 8.Feng, Y., C.C. Broder, P.E. Kennedy, and E.A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 272:872–877. [DOI] [PubMed] [Google Scholar]
- 9.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J.L. Virelizier, F. Arenzana-Seisdedos, O. Schwartz, J.M. Heard, I. Clark-Lewis, D.F. Legler, et al. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 382:833–835. [DOI] [PubMed] [Google Scholar]
- 10.Muller, A., B. Homey, H. Soto, N. Ge, D. Catron, M.E. Buchanan, T. McClanahan, E. Murphy, W. Yuan, S.N. Wagner, et al. 2001. Involvement of chemokine receptors in breast cancer metastasis. Nature. 410:50–56. [DOI] [PubMed] [Google Scholar]
- 11.Kijima, T., G. Maulik, P.C. Ma, E.V. Tibaldi, R.E. Turner, B. Rollins, M. Sattler, B.E. Johnson, and R. Salgia. 2002. Regulation of cellular proliferation, cytoskeletal function, and signal transduction through CXCR4 and c-Kit in small cell lung cancer cells. Cancer Res. 62:6304–6311. [PubMed] [Google Scholar]
- 12.Bertolini, F., C. Dell'Agnola, P. Mancuso, C. Rabascio, A. Burlini, S. Monestiroli, A. Gobbi, G. Pruneri, and G. Martinelli. 2002. CXCR4 neutralization, a novel therapeutic approach for non-Hodgkin's lymphoma. Cancer Res. 62:3106–3112. [PubMed] [Google Scholar]
- 13.Rubin, J.B., A.L. Kung, R.S. Klein, J.A. Chan, Y. Sun, K. Schmidt, M.W. Kieran, A.D. Luster, and R.A. Segal. 2003. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc. Natl. Acad. Sci. USA. 100:13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melikian, A., J. Burns, B.E. McMaster, T. Schall, and J.J. Wright. 2004. Inhibitors of human tumor-expressed CCX CKR2. Patent Cooperation Treaty application WO04058705 (15 July 2004) and USA patent publication US 20040170634 (2 September 2004).
- 15.Hatse, S., K. Princen, G. Bridger, E. De Clercq, and D. Schols. 2002. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 527:255–262. [DOI] [PubMed] [Google Scholar]
- 16.Dairaghi, D.J., R.A. Fan, B.E. McMaster, M.R. Hanley, and T.J. Schall. 1999. HHV8-encoded vMIP-I selectively engages chemokine receptor CCR8. Agonist and antagonist profiles of viral chemokines. J. Biol. Chem. 274:21569–21574. [DOI] [PubMed] [Google Scholar]
- 17.Kledal, T.N., M.M. Rosenkilde, F. Coulin, G. Simmons, A.H. Johnsen, S. Alouani, C.A. Power, H.R. Luttichau, J. Gerstoft, P.R. Clapham, et al. 1997. A broad-spectrum chemokine antagonist encoded by Kaposi's sarcoma-associated herpesvirus. Science. 277:1656–1659. [DOI] [PubMed] [Google Scholar]
- 18.Cole, K.E., C.A. Strick, T.J. Paradis, K.T. Ogborne, M. Loetscher, R.P. Gladue, W. Lin, J.G. Boyd, B. Moser, D.E. Wood, et al. 1998. Interferon-inducible T cell α chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J. Exp. Med. 187:2009–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loetscher, M., B. Gerber, P. Loetscher, S.A. Jones, L. Piali, I. Clark-Lewis, M. Baggiolini, and B. Moser. 1996. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T lymphocytes. J. Exp. Med. 184:963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joost, P., and A. Methner. 2002. Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands. Genome Biol. 3:RESEARCH0063. [DOI] [PMC free article] [PubMed]
- 21.Libert, F., M. Parmentier, A. Lefort, J.E. Dumont, and G. Vassart. 1990. Complete nucleotide sequence of a putative G protein coupled receptor: RDC1. Nucleic Acids Res. 18:1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heesen, M., M.A. Berman, A. Charest, D. Housman, C. Gerard, and M.E. Dorf. 1998. Cloning and chromosomal mapping of an orphan chemokine receptor: mouse RDC1. Immunogenetics. 47:364–370. [DOI] [PubMed] [Google Scholar]
- 23.Madden, S.L., B.P. Cook, M. Nacht, W.D. Weber, M.R. Callahan, Y. Jiang, M.R. Dufault, X. Zhang, W. Zhang, J. Walter-Yohrling, et al. 2004. Vascular gene expression in nonneoplastic and malignant brain. Am. J. Pathol. 165:601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazo, I.B., E.J. Quackenbush, J.B. Lowe, and U.H. von Andrian. 2002. Total body irradiation causes profound changes in endothelial traffic molecules for hematopoietic progenitor cell recruitment to bone marrow. Blood. 99:4182–4191. [DOI] [PubMed] [Google Scholar]
- 25.Libert, F., M. Parmentier, A. Lefort, C. Dinsart, J. Van Sande, C. Maenhaut, M.J. Simons, J.E. Dumont, and G. Vassart. 1989. Selective amplification and cloning of four new members of the G protein-coupled receptor family. Science. 244:569–572. [DOI] [PubMed] [Google Scholar]
- 26.Sreedharan, S.P., A. Robichon, K.E. Peterson, and E.J. Goetzl. 1991. Cloning and expression of the human vasoactive intestinal peptide receptor. Proc. Natl. Acad. Sci. USA. 88:4986–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libert, F., E. Passage, M. Parmentier, M.J. Simons, G. Vassart, and M.G. Mattei. 1991. Chromosomal mapping of A1 and A2 adenosine receptors, VIP receptor, and a new subtype of serotonin receptor. Genomics. 11:225–227. [DOI] [PubMed] [Google Scholar]
- 28.Nagata, S., T. Ishihara, P. Robberecht, F. Libert, M. Parmentier, J. Christophe, and G. Vassart. 1992. RDC1 may not be VIP receptor. Trends Pharmacol. Sci. 13:102–103. [DOI] [PubMed] [Google Scholar]
- 29.Kapas, S., and A.J. Clark. 1995. Identification of an orphan receptor gene as a type 1 calcitonin gene-related peptide receptor. Biochem. Biophys. Res. Commun. 217:832–838. [DOI] [PubMed] [Google Scholar]
- 30.McLatchie, L.M., N.J. Fraser, M.J. Main, A. Wise, J. Brown, N. Thompson, R. Solari, M.G. Lee, and S.M. Foord. 1998. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 393:333–339. [DOI] [PubMed] [Google Scholar]
- 31.Dupre, D.J., M. Rola-Pleszczynski, and J. Stankova. 2004. Inverse agonism: more than reverting constitutively active receptor signaling. Biochem. Cell Biol. 82:676–680. [DOI] [PubMed] [Google Scholar]
- 32.Soudijn, W., I. van Wijngaarden, and A.P. Ijzerman. 2005. Structure-activity relationships of inverse agonists for G-protein-coupled receptors. Med. Res. Rev. 25:398–426. [DOI] [PubMed] [Google Scholar]
- 33.Vauquelin, G., and I. Van Liefde. 2005. G protein-coupled receptors: a count of 1001 conformations. Fundam. Clin. Pharmacol. 19:45–56. [DOI] [PubMed] [Google Scholar]
- 34.Bakker, R.A., P. Casarosa, H. Timmerman, M.J. Smit, and R. Leurs. 2004. Constitutively active Gq/11-coupled receptors enable signaling by co-expressed G(i/o)-coupled receptors. J. Biol. Chem. 279:5152–5161. [DOI] [PubMed] [Google Scholar]
- 35.Hulshof, J.W., P. Casarosa, W.M. Menge, L.M. Kuusisto, H. van der Goot, M.J. Smit, I.J. de Esch, and R. Leurs. 2005. Synthesis and structure-activity relationship of the first nonpeptidergic inverse agonists for the human cytomegalovirus encoded chemokine receptor US28. J. Med. Chem. 48:6461–6471. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu, N., Y. Soda, K. Kanbe, H.Y. Liu, R. Mukai, T. Kitamura, and H. Hoshino. 2000. A putative G protein-coupled receptor, RDC1, is a novel coreceptor for human and simian immunodeficiency viruses. J. Virol. 74:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balabanian, K., B. Lagane, S. Infantino, K.Y. Chow, J. Harriague, B. Moepps, F. Arenzana-Seisdedos, M. Thelen, and F. Bachelerie. 2005. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280:35760–35766. [DOI] [PubMed] [Google Scholar]
- 38.Burns, J.M., R.C. Gallo, A.L. DeVico, and G.K. Lewis. 1998. A new monoclonal antibody, mAb 4A12, identifies a role for the glycosaminoglycan (GAG) binding domain of RANTES in blocking the antiviral effect against HIV-1 and intracellular Ca++ signaling. J. Exp. Med. 188:1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]