Abstract
After encounter with antigen, the antibody repertoire is shaped by somatic hypermutation (SHM), which leads to an increase in the affinity of antibodies for the antigen, and class-switch recombination (CSR), which results in a change in the effector function of antibodies. Both SHM and CSR are initiated by activation-induced cytidine deaminase (AID), which deaminates deoxycytidine to deoxyuridine in single-stranded DNA (ssDNA). The precise mechanism responsible for the formation of ssDNA in V regions undergoing SHM has yet to be experimentally established. In this study, we searched for ssDNA in mutating V regions in which DNA–protein complexes were preserved in the context of chromatin in human B cell lines and in primary mouse B cells. We found that V regions that undergo SHM were enriched in short patches of ssDNA, rather than R loops, on both the coding and noncoding strands. Detection of these patches depended on the presence of DNA-associated proteins and required active transcription. Consistent with this, we found that both DNA strands in the V region were transcribed. We conclude that regions of DNA that are targets of SHM assemble protein–DNA complexes in which ssDNA is exposed, making it accessible to AID.
Ig genes encode antibodies whose V regions contain the antigen-binding site and whose C regions mediate effector functions. During the immune response, somatic hypermutation (SHM) generates point mutations at very high frequencies in Ig V regions of centroblasts (1) in germinal centers (GCs) (2). SHM is initiated by activation-induced cytidine deaminase (AID) (3), which deaminates deoxycytidines (dCs) in single-stranded DNA (ssDNA) and converts them to deoxyuridine (4–6). Deoxyuridine is subsequently processed by replication, uracil-N-glycosylase and base excision repair, error-prone polymerases, mismatch repair, and other proteins to produce mutations at a rate of ∼10−3 mutations per bp, per generation, i.e., 106 times higher than the rate of mutation of housekeeping genes (7–9). This extraordinary rate of mutation is focused on the expressed variable Ig regions and, to a lesser extent, to some non-Ig genes (10–13). Although the loss of AID results in immunodeficiency (14), the mistargeting of AID-induced mutation is thought to be a cause of many B cell lymphomas (10, 15). The mechanism responsible for targeting SHM to certain DNA regions and not to others is largely unknown (9, 16).
In addition to SHM, AID also mediates class-switch recombination (CSR) (3) by generating mutations and triggering recombination and deletion at switch regions (17). As a result of CSR, the V region rearranges to one of the downstream C regions that encode different effector functions. The transcribed strands of mammalian switch recombination sequences are C rich, so that their transcription results in the formation of R loops in which the nascent G-rich RNA transcript remains stably bound to the template strand, rendering the nontemplate strand single stranded (5, 18). AID deaminates the single-stranded, displaced nontemplate strand of R loops in vitro, and R loops may exist in vivo (5, 18). G quartets can form in switch recombination sequences (19) and bind mismatch-repair proteins that participate in the CSR reaction (20). R loops do not form in GC-poor switch recombination sequences, such as Xenopus laevis switch regions, where AID was found to be targeted to C's in AGCT motifs (21). Similarly, by virtue of their primary sequence, V regions are not predicted to form stable R loops. Experimental evidence that ssDNA is enriched in V regions in vivo has been lacking, and the mechanisms responsible for generating ssDNA and making it accessible to AID in some regions of highly transcribed genes but not in others have yet to be established.
In vitro, AID targets dCs on ssDNA (4, 5, 19, 22), whereas in Escherichia coli, AID acts on dCs in the nontemplate strand (6) or on both strands (23) of transcribed DNA. However, both DNA strands in a transcription bubble were shown to be deaminated by AID either in DNA transcribed by bacterial RNA polymerase (24) or in T7 polymerase–transcribed plasmids that are either supercoiled (25) or contain particular primary sequences (26). Replication protein A, an ssDNA binding protein, associates with AID and enhances its ability to deaminate dCs in T7-transcribed DNA in vitro (27). In vivo, transcription is necessary for SHM (28), and cis-acting elements that activate transcription of Ig loci also up-regulate SHM (29). Histone modifications such as acetylation and phosphorylation were shown to be enriched in DNA regions targeted by SHM and CSR (30–34), but it is not clear if changes in chromatin structure are responsible for selective targeting of AID (16).
As described above, the requirement of ssDNA for AID targeting has been established using in vitro assays and hypothesized to be made accessible in transcription bubbles in vivo based on the observation that SHM and CSR require the target genes to be highly transcribed (4–6). However, in B cells not all highly transcribed regions of DNA undergo AID-induced mutation (35). In this study, we sought to determine whether regions targeted for SHM are enriched in ssDNA. To do this, we established a strategy that enabled us to search for ssDNA in nuclei in the context of cross-linked chromatin. We did not find R loops in hypermutating V regions. However, we did find that ssDNA–protein complexes that depended on transcription were enriched in the V region compared with the C region, and these ssDNA–protein complexes were found in other regions that undergo SHM.
RESULTS
Detection of ssDNA in chromatinized substrates
In an effort to find out whether ssDNA is enriched in vivo in regions that undergo SHM in B cells, we used sodium bisulfite, which, like AID, deaminates dCs in ssDNA to form deoxyuridine (36). After PCR amplification and sequencing of bisulfite-treated DNA, clones derived from amplification of either the nontemplate (upper, nontranscribed) or the template (lower, transcribed) strand reveal the location and strand of single-stranded dCs. Thus, C to T conversions indicate single-stranded dCs on the upper strand, whereas G to A conversions on the lower strand indicate single-stranded dCs (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20062032/DC1). Bisulfite treatment of deproteinized genomic DNA has been used previously to detect the ssDNA in R loops in switch regions from mouse B lymphocytes undergoing CSR (18) and in ssDNA in plasmid substrates (26). A similar procedure as the one used to detect R loops (reference 18 and outlined in Fig. 1 A, left, Deproteinized DNA) was used to search for single-stranded dCs in the functional heavy chain V region of Ramos Burkitt's lymphoma cells that constitutively undergo SHM at a high frequency of ∼10−3 (Table I) (37, 38). As seen in Fig. 1 C, the V region in the bisulfite-treated, deproteinized genomic DNA had a similar low frequency of single-stranded dCs as the nonbisulfite-treated DNA (Fig. 1 B), in which C to T and G to A conversions were due to preexisting mutations or PCR errors.
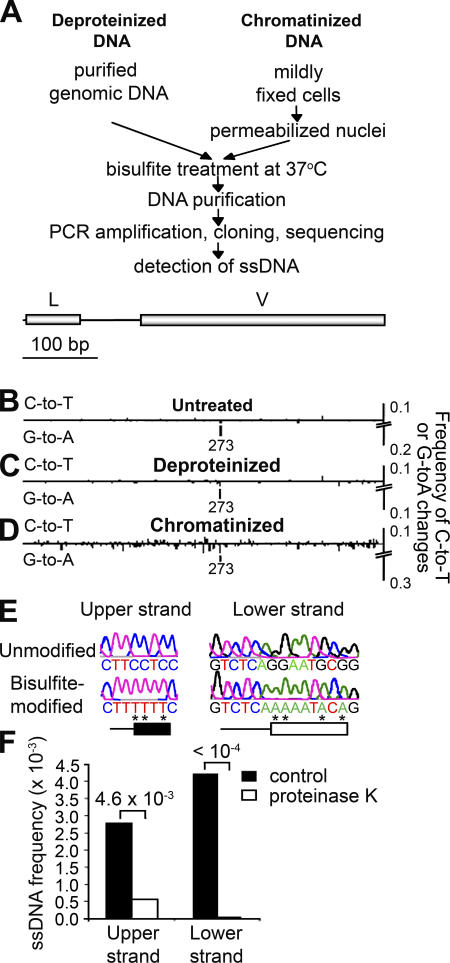
Figure 1.
A method to detect ssDNA in the context of chromatin. (A) Two assays were used to detect ssDNA, one using purified genomic DNA (Deproteinized DNA) (reference 18) and another using DNA in fixed nuclei (Chromatinized DNA). Both kinds of substrates were treated with bisulfite, amplified, cloned, and sequenced as described in Materials and methods. (B–D) Frequency of C to T conversions (indicative of ssDNA on the noncoding strand) and G to A conversions (indicating ssDNA on the template strand) was measured for untreated DNA (B), deproteinized DNA (C), and chromatinized DNA (D) treated with bisulfite in the IgH V region of Ramos cells (depicted above the diagrams). L indicates the leader sequence. Position 273 is a hotspot for G to A mutations. (E) Examples of ssDNA patches on the coding and noncoding strands. Asterisks indicate bases that have been converted by bisulfite. Because the length of these patches cannot be determined exactly, the minimum size is shown as boxes, whereas the maximum size is shown as a horizontal line. (F) The frequency of ssDNA patches was calculated for IgH V region in chromatinized DNA from Ramos 7 cells untreated (control) or pretreated with proteinase K. Numbers above the bar graphs indicate p-values calculated using the Chi-squared test. The results are representative of three experiments. Sample sizes are indicated in Table S2.
Table I.
Mutation frequencies of genes in Ramos cells
| Gene | Mutations | Bases sequenced | Mutation frequency |
|---|---|---|---|
| V | 58 | 55104 | 1.05 × 10−3 |
| C | 4 | 22349 | 1.79 × 10−4 |
| BCL6 | 1 | 15955 | 6.27 × 10−5 |
| RNAP II | 1 | 14792 | 6.76 × 10−5 |
| MDM2 | 1 | 11836 | 8.45 × 10−5 |
| CD27 | 2 | 12865 | 1.55 × 10−4 |
| CDKN2C | 0 | 10992 | <9.10 × 10−5 |
| CD4 | 1 | 10806 | 9.25 × 10−5 |
DNA was amplified from Ramos cells using the Pfu DNA polymerase, with the primers listed in Table S1.
Unlike mammalian switch regions, V and other regions that undergo SHM are not GC rich and thus are unlikely to form stable R loops. It is, however, possible that ssDNA forms in V regions as part of DNA–protein complexes in which the DNA strands are prevented from annealing. In this case, bisulfite would fail to detect ssDNA in assays in which DNA is deproteinized, as in the bisulfite assay that has been used to detect R loops in switch regions (18). To circumvent this potential problem, we devised an assay to detect ssDNA in the context of chromatin (Fig. 1 A, Chromatinized DNA). In this assay, cells were fixed with formaldehyde under conditions that preserved the binding of proteins to DNA, as in the chromatin immunoprecipitation assay (39), and nuclei were isolated and treated with bisulfite. The single-stranded status of a particular region was then assessed after PCR amplification, cloning, and sequencing. This assay is able to detect single-stranded regions at the single cell level and in a strand-specific manner (Fig. S1). We validated this assay by showing that it could detect R loops in situ in the GC-rich switch regions of mouse B lymphocytes undergoing CSR, and that these R loops were similar to the ones identified by others using deproteinized genomic DNA (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20062032/DC1) (18). This not only establishes the validity of the assay, but also shows that R loops do exist in the cross-linked chromatin in the nuclei of switching B cells.
Bisulfite was then used to examine the V regions of the cross-linked nuclei of constitutively mutating Ramos cells. As shown in Fig. 1 D, this revealed a considerable number of bisulfite-accessible dCs in chromatinized DNA in this V region. As a control for the overall efficiency of the bisulfite reaction, we used preboiled genomic DNA treated with bisulfite and found that typically ∼90% of dCs were converted to deoxyuridine (not depicted).
Unlike bisulfite-accessible dCs present in deproteinized DNA (Fig. 1 C), many of the single-stranded dCs found on chromatinized DNA were present in patches in which three or more single-stranded dCs were clustered together. As shown in the examples in Fig. 1 E, these patches often contained consecutive bisulfite-accessible dCs separated by other bases whose single-stranded status cannot be detected by this assay. Similar protein-associated patches of ssDNA were detected in the heavy and light chain V regions of the BL2 Burkitt's lymphoma cell line and in the V regions of light chain of Ramos, as well as in the V regions of other human cell lines (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20062032/DC1).
Our inability to detect ssDNA in deproteinized genomic DNA (Fig. 1 C) and the presence of ssDNA in chromatinized substrates (Fig. 1 D) implies that proteins might be needed for the formation of ssDNA. We therefore pretreated fixed Ramos cell nuclei with proteinase K and found that this treatment significantly diminished the frequency of ssDNA patches detected in the V region (Fig. 1 F). This finding indicates that proteins are needed to prevent the DNA strands from annealing and suggests that protein–DNA structures are required to maintain these patches of ssDNA.
ssDNA patches are enriched in regions that undergo SHM
To determine whether ssDNA detected on chromatinized substrates was more abundant in regions undergoing SHM, we compared the frequency of consecutive converted dCs in the hypermutating V and the nonmutating C regions of the Ramos Ig heavy chain (IgH) gene (Fig. 2 A) (37). To account for the fact that there is only one functional heavy chain V region allele, and there are two C region alleles of which only one is transcribed in Ramos cells, we divided the number of converted dCs in patches of different sizes in the V region by two and compared that to the frequency of such consecutive converted dCs in the C region. Even using this more stringent comparison, we found significantly more patches with three or more consecutive converted dCs in the V than in the C region (P = 0.0018 for the upper strand and P = 0.016 for the lower strand) (Fig. 2 A). Patches of three or more consecutive converted C's were present in 23% of the 286 Ramos V sequences analyzed, and most of these DNA molecules bore one ssDNA patch. Such patches were not detected in any of 98 deproteinized genomic DNA sequences, nor in 209 sequences derived from untreated nuclei. The presence of these ssDNA patches in chromatinized, but not deproteinized or untreated substrates suggests that patches of ssDNA form in the V region chromatin as part of higher order molecular complexes.
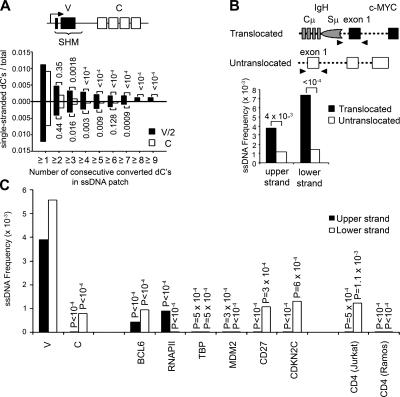
Figure 2.
Comparison of ssDNA frequencies in hypermutating and nonmutating regions. (A) The frequency of consecutive converted dCs was compared between the hypermutating VH (V) and the nonmutating Cμ (C) regions (depicted above the graph) in the IgH gene in Ramos cells. Single-stranded dCs were classified according to the number of consecutive converted dCs (for the upper strand) or dGs (for the lower strand). Values above or below pairs of bars indicate p-values. The V region frequencies were divided by two to account for the fact that there are half as many alleles of the functional V region compared with C, but one of the two C region alleles is not transcribed. (B) The frequency of ssDNA patches with three or more consecutive converted dCs was compared for the two c-MYC alleles. One allele is translocated in the proximity of the IgH locus, is expressed, and undergoes SHM, whereas the untranslocated allele is transcriptionally silent. Numbers indicate p-values. (C) The frequency of ssDNA patches was measured for nonmutating genes in Ramos cells and compared with the frequency of ssDNA in patches in the V and C region, and only patches with four or more consecutive converted Cs were taken into account for these calculations. Although BCL6 mutates in some human cells (references 12 and 72), it does not mutate in Ramos cells. No significant mutations were detected in any of these genes, except the V region (Table I). Sample sizes are indicated in Table S2. p-values indicate the significance of the difference in ssDNA frequency in the corresponding gene or region and the Ramos VH region.
Next, we determined the frequency of ssDNA patches in another hypermutating gene, c-MYC. In Burkitt's lymphoma cell lines, such as Ramos, one c-MYC allele is translocated to the IgH locus, and it is transcribed and undergoes SHM while the untranslocated allele is silent and does not mutate (40 and not depicted). We compared the same region of the two c-MYC alleles with respect to the presence of patches of ssDNA with three or more bisulfite-accessible dCs and found that the translocated allele had significantly more patches of ssDNA than the untranslocated allele (Fig. 2 B). The fact that these c-MYC regions have identical DNA sequences and significantly different ssDNA frequencies suggests that formation of ssDNA patches does not depend solely on the primary DNA sequence.
Several genes are highly transcribed but do not undergo SHM in Ramos (Table I) and other hypermutating cells (35). We tested whether patches of ssDNA were present in these genes in Ramos cells by analyzing regions >100 bp but <1 kb downstream of the promoter, which is the region that contains mutations in genes undergoing SHM (1). As shown in Fig. 2 C, BCL-6, RNAP II, TATA-binding protein (TBP), MDM2, CD27, and CDK2NC, as well as CD4, which is expressed in Jurkat, but not Ramos cells, had a significantly lower frequency of ssDNA patches than the Ig V region. These results support the idea that bisulfite-accessible patches of ssDNA are present at a higher abundance in genes that undergo SHM.
Characterization of ssDNA patches in the Ramos V region
We measured the average size of ssDNA patches in the Ramos V region and found their median length to be around 11 bases, although there was a wider distribution for upper strand patches (Fig. 3 A). We were able to detect more patches on the lower than on the upper DNA strand (Fig. 3 B). We were concerned that there might be even larger patches that would only have been detected if the dCs in the primers were converted to dTs (or dGs were converted to dAs) to allow the amplification of very long putative bisulfite-converted sequences (18). However, we did not detect patches of ssDNA when we used various combinations of mutated and wild-type primers in which all or only 50% of dCs and dGs were converted to dTs and dAs, respectively, to allow them to amplify such potential long stretches of bisulfite-converted DNA (18 and not depicted). These primers were efficient at amplifying bisulfite-converted genomic DNA, which was artificially rendered single stranded through boiling. Thus, most single-stranded patches in the Ramos V region are short (Fig. 3 B).
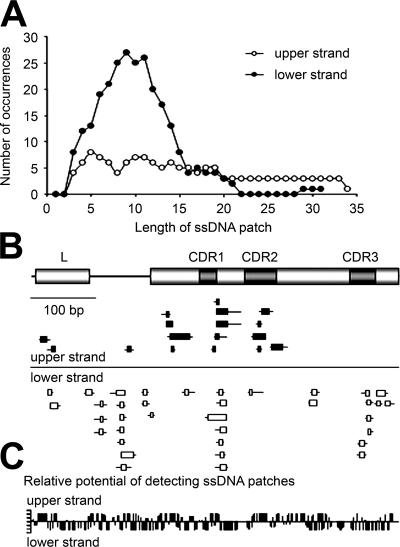
Figure 3.
Characterization of ssDNA patches in the Ramos V region. (A) The average length of all patches in A was calculated as described in Materials and methods. (B) ssDNA patches found in the Ramos V region (depicted at the top; L-leader exon, CDR1-three complementarity-determining regions in the V exon) were plotted for the upper (black) and lower (white) strands. Each box indicates a unique patch in individual Ramos cells. The TATA box is ∼60 bp upstream of the start of L. Because only single strandness of dCs could be determined using bisulfite, the minimum (boxes) and the maximum sizes (horizontal lines crossing boxes), as exemplified in Fig. 1 E, could be determined. (C) The graph shows, on the vertical axis, the relative potential to form ssDNA patches of three or more consecutive nucleotides for each dC residue in the analyzed V region on the horizontal axis. This potential was calculated as the ability of each dC to be found in a theoretical 11-base patch.
SHM-generated mutations occur in vivo on both the coding and noncoding strands (41, 42), and, likewise, ssDNA patches are located on both DNA strands (Fig. 3 B). Assuming that most clusters of dCs form within an ∼11-nucleotide interval (Fig. 3 A), we calculated that different subregions within V have different “relative potentials” to allow detection of bisulfite-accessible ssDNA patches of three or more consecutive nucleotides for each dC residue (Fig. 3 C). We found that ssDNA patches (Fig. 3 B) do not always form preferentially in regions that had a sufficient abundance of dCs (Fig. 3, B and C) and thus are not due solely to the primary nucleic acid sequence, in particular portions of the V region.
Transcription is required for the formation of ssDNA
Active transcription is necessary for SHM in vivo (11, 43), and the rate of transcription has been correlated with the extent of SHM (28, 44). It has been hypothesized that the source of ssDNA might be the nontemplate strand of transcription bubbles (4, 9) or supercoiled DNA at the end of transcription bubbles (25). We observed that the median length of ssDNA patches in the V regions analyzed was ∼11 bases (Fig. 3 A), a size similar to that of transcription bubbles (45). To determine whether transcription by RNA polymerase II (RNAP II) was coupled with the formation of ssDNA in regions subject to SHM, we incubated Ramos cells with α-amanitin, an inhibitor of RNAP II, and measured the frequency of ssDNA patches in treated and untreated cells. Incubation with α-amanitin significantly decreased the frequency of patches in the V region (Fig. 4 A), suggesting that RNAP II–mediated transcription is necessary for the formation of the ssDNA patches that we found.
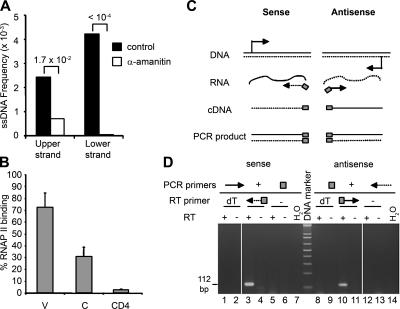
Figure 4.
The role of transcription in ssDNA formation. (A) Ramos cells were treated with α-amanitin as described in Materials and methods, and then used to determine the frequency of ssDNA patches in the IgH V region. Numbers indicate p-values. (B) Relative binding to RNAP II by pol II-ChIP (reference 71). Bars indicate the average of three experiments, and error bars indicate the standard deviation of these measurements. (C) Sense (filled line) or antisense (dotted line) RNA transcripts were reverse-transcribed using strand-specific primers (arrows) containing a Tag sequence (gray) on the 5′ end of the RT primer. Primer sequences are shown in Table S1. cDNA was amplified with the Tag PCR primer (gray box) in combination with either the forward primer (filled arrow) or the reverse (dotted arrow) VH-specific primer, and the products were run on an agarose gel and strained with ethydium bromide (D). RT-PCR products, expected to be 112 bp long, were sequenced and determined to contain the Ramos VH region and the Tag sequence on the 3′ end (lane 3) or on the 5′ end (lane 10).
If transcription is necessary for the formation of ssDNA, it is possible that the frequency of ssDNA patches is directly proportional to the rate of RNAP II–mediated transcription. Indeed, we observed patches on the transcribed, but not untranscribed c-MYC allele (Fig. 2 B). It is possible that the reason why there is more ssDNA in the V than in the C region is a higher level of RNAP II density in the V than in the C region. To test this possibility, we immunoprecipitated RNAP II–bound DNA and amplified it with primers specific for the V and C regions. (In Fig. 4 B, we normalized the amount of immunoprecipitated DNA by the amount of input DNA, and thus our analysis is independent of the efficiency of V- and C-specific PCR primers). The abundance of RNAP II in the C region was half that in the V region, but significantly more than in the silent CD4 gene. Although we observe a 50% difference in the density of RNAP II, if there is no RNAP II bound to the Cμ allele rearranged to c-MYC there would be no difference between the density of RNAP II in the V and C regions. Thus, although transcription is required for the formation of ssDNA, it is not sufficient to explain the increase in ssDNA patches, but passage of, or pausing of, RNAP II in a particular DNA region (11) could be a contributing factor for the formation of protein–DNA complexes containing ssDNA.
It has been assumed that only the template strand of protein-coding genes is transcribed. In vitro and in bacteria, the nontranscribed strand of various target genes accumulated more AID-induced mutations, perhaps because the transcribed strand is protected by RNA polymerase (6, 46). However, in vivo both strands are targeted for AID mutations (41). Both the coding and the noncoding strands are transcribed in a great number of genes in the genome (47), and antisense transcription in the IgH locus occurs in a regulated manner in early B cell development (48), although antisense transcription has not been reported in V regions or in centroblast cells. If the nontranscribed strand of RNAP II transcription bubbles is the source of ssDNA during SHM and ssDNA was found on both strands (Fig. 3 B), the V region should be transcribed in both orientations. To test this possibility, we used a method, outlined in Fig. 4 C, to specifically amplify RNA derived from the coding and noncoding strands. To ensure that the RT-PCR products are derived by amplifying cDNA and not DNA, we designed the RT primers to contain a unique sequence that is not present in the DNA (49). As shown in Fig. 4 D, this procedure allowed us to identify both sense and antisense RNA transcripts derived from the V region. Analysis of the relative abundance of sense and antisense RNAs revealed that the antisense transcript is present at approximately eightfold lower levels than the sense V region transcript in Ramos cells (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20062032/DC1). However, nonsense-mediated RNA decay or other mechanisms might render the antisense transcript more unstable than the sense transcript, so a quantitative comparison of the levels of the two transcripts may not reflect their rate of transcription.
Because replication forks have regions of ssDNA, we examined whether or not ssDNA patches are found exclusively in the S phase. It has been reported that mutations arise in the G1/S phase of the cell cycle (50), and double-strand breaks that are thought to be a byproduct of SHM accumulate in S/G2 (51). We fractionated cells by elutriation, treated individual fractions with bisulfite, and determined the frequency of ssDNA in both the upper and lower strands. As shown in Fig. S5 (available at http://www.jem.org/cgi/content/full/jem.20062032/DC1), patches could be detected throughout the cell cycle, and upper strand ssDNA patches accumulated in the G1/S, S/G2, and G2 phases, whereas lower strand patches were most abundant in G1/S. Thus, we found that, although ssDNA could be detected throughout the cell cycle, the extent of coding and noncoding strand ssDNA varied throughout the cell cycle and did not occur exclusively in conjunction with replication forks.
ssDNA patches can be detected in wild-type and AID−/− mice
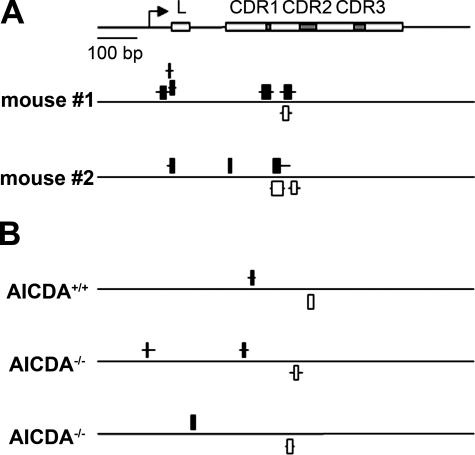
Because all of the studies described above were performed using B cell lines, we also examined the V regions in B cells from mice immunized with 4-hydroxy-3-nitrophenyl acetyl (NP) coupled to chicken γ globulin (CGG; Fig. 5 A) (52). As in human B cell lines, there were ssDNA patches on both strands of the rearranged V region of mouse splenocytes, but not in the Cγ1 region in splenocytes from immunized mice (not depicted). These patches were similar in size to the ones found in human cell lines (Fig. 3). Due to the need to analyze a very large number of cells for the bisulfite assay, we were unable to separate naive and GC lymphocytes, and were thus unable to determine the frequency of ssDNA patches in primary cells at different stages of differentiation.
Figure 5.
AID is dispensable for the formation of ssDNA in hypermutating regions. (A) Splenocytes from mice immunized with NP were isolated and fixed, and then treated with sodium bisulfite. DNA was purified, and the rearranged V186.2-D-JH region was amplified by PCR (references 52 and 73). ssDNA patches (black for the upper strand, white for the lower strand) are shown for each animal. Boxes indicate the minimum range of ssDNA patches, whereas lines indicate the maximum length. (B) ssDNA patches in the V regions of splenocytes from AID-deficient (AICDA−/−) and AICDA+/+ littermate control mice, analyzed as in A.
We then examined IgH V regions in AID-deficient (AICDA−/−) mice (3) and found ssDNA in both AICDA+/+ and AICDA−/− mice (Fig. 5 B). Likewise, the frequency of ssDNA patches was similar in two Ramos cell subclones, one of which had a fivefold decrease in AID mRNA expression relative to the other and did not undergo detectable levels of V region mutations (Fig. S6, available at http://www.jem.org/cgi/content/full/jem.20062032/DC1) (38). These findings suggest that ssDNA patches form independently of AID in V region genes.
DISCUSSION
During CSR, AID deaminates ssDNA in the context of DNA–RNA complexes called R loops, which form in switch recombination sequences. We sought to determine the origin of the ssDNA substrate of AID during SHM in V regions. Unlike switch recombination sequences, hypermutating V regions do not form R loops. By devising an assay to assess the presence of ssDNA in the context of chromatin, we have shown that V and some other hypermutating regions of cultured and primary B cell nuclei are enriched in chromatin-associated ssDNA. Unlike GC-rich switch regions that form R loops in which ssDNA is stabilized by DNA–RNA hybrids (18, 21), IgV regions do not form R loops. Indeed, we failed to identify ssDNA in deproteinized DNA from IgV regions using a protocol similar to that used to detect R loops in switch regions from B cell chromosomes (Fig. 1 C). Instead, we were able to detect ssDNA in the V region of B cell DNA when the binding of DNA-associated proteins was preserved (Fig. 1 D), whereas the identification of ssDNA was lost with pretreatment with proteinase K (Fig. 1 F). It is possible that unstable DNA–RNA hybrids also contribute to the formation of these ssDNA patches. To test this possibility, we pretreated fixed nuclei, as well as fixed chromatin fibers detached from the nuclear membrane, with RNaseH but were unable to detect a decrease in the frequency of ssDNA after treatment (not depicted). However, this negative result may not be informative because RNaseH might not have gained full access to DNA–RNA hybrids in complexes of fixed nucleic acids and proteins. In addition to bisulfite, other chemicals, such as bromoacetaldehyde (53), permanganate (54), or osmium tetroxide (55), could be used to detect ssDNA in cell nuclei. These assays nevertheless would have to be adapted to detect ssDNA in individual DNA molecules, as opposed to populations of molecules, because ssDNA patches appear to be relatively rare when detected using our bisulfite-based assay.
Mutations occur on both strands in Ig V regions, starting ∼100 bp after the start of transcription, with a peak around 300 bases 3′ of the promoter (56–58). To determine whether ssDNA patches are distributed in a similar manner to mutations in the Ramos V region, we plotted the location of ssDNA patches along the IgH V region (Fig. 3 B) and found that they were distributed unevenly along both strands of the V region. Moreover, lower strand ssDNA patches accumulated at higher levels starting ∼150 bases 3′ of the V promoter, whereas upper strand patches accumulated ∼240 bases 3′ of the V promoter, and both upper and lower strand patches peaked around 300 bases 3′ of the promoter. This distribution roughly corresponds to that of mutation accumulation in Ig V regions (58). Bisulfite detection of ssDNA is limited to determining whether dCs, but not other bases, are single stranded. Using the relatively small number of patches that we have identified, we were unable to determine rigorously whether hotspots of SHM (59) or other motifs were shared amongst various ssDNA regions. A program (60) previously used to detect potential secondary DNA structures in switch recombination sequences (61, 62) did not allow us to detect a correlation between predicted DNA secondary structures, such as loops in hairpin loops, and the location of ssDNA patches (not depicted).
Transcription is necessary for SHM, and the rate of transcription is proportional to the rate of SHM (28, 44). Several observations support the view that transcription is required to create the ssDNA patches that we have observed. We found that blocking RNAP II–mediated transcription ablated the formation of ssDNA patches in the Ig V regions (Fig. 4 A), and ssDNA could not be detected in the silent cMYC allele or CD4 (Fig. 2, B and C). Moreover, the modal size of ssDNA patches was similar to that of transcription bubbles (Fig. 3 A). Finally, both the density of single-stranded regions and the RNAP II density were higher in the V than in the C region, perhaps reflecting a higher rate of transcription in the 5′ end of genes (63) or pausing (11) (Fig. 4 B). Both strands of the Ig V region were actively transcribed (Fig. 4 D), and sense and antisense transcripts have been found in the c-MYC gene in Burkitt's lymphoma cells (64), providing a potential explanation for why ssDNA was found on both the template and the nontemplate strands. Our assay did not enable us to determine whether sense and antisense transcription occurred simultaneously in the same DNA molecule. If they did, it is possible that the sense and antisense transcription complexes may collide, as documented in budding yeast (65), thus stabilizing ssDNA on opposite strands. Alternatively, the source of stable ssDNA could be the collision of the RNAP II complex with other DNA-binding molecules such as topoisomerase I (66).
Evidence from other studies suggests that DNA in V regions that undergo SHM is subjected to single-strand and double-strand breaks (67–69), which could reflect labile DNA structures that are subject to cleavage. These breaks, however, could not be detected with the bisulfite assay we used in this study, which is based on PCR amplification of intact V region molecules, although the ssDNA patches we observe could be DNA molecules immediately after religation. Collectively, these results suggest that ssDNA patches form in conjunction with the transcription complex. However, transcription alone is not sufficient to generate SHM-specific ssDNA because the difference in the frequency of ssDNA patches between the V and C regions cannot be explained solely by the difference in RNAP II density between the two regions, and there are other transcribed genes in B cells that do not have ssDNA patches and do not undergo SHM (Fig. 2 C).
We also found ssDNA in primary cells that were undergoing SHM (Fig. 5) independently of the presence of AID (Fig. 5 B). Histone hyperacetylation in hypermutating regions and double-strand breaks that have been associated with SHM are also independent of the presence of AID (30, 70), whereas histone phosphorylation and histone hyperacetylation in switch regions occurred less efficiently or not at all in AID-deficient cells (33, 34). It is possible that ssDNA patches form before the expression of AID and thus represent an early step in targeting SHM to specific DNA regions.
We conclude that the ssDNA patches are generated as part of protein–DNA complexes whose formation requires transcription but not AID. We propose that chromatin-associated ssDNA serves as a molecular nidus to which AID as well as other proteins subsequently bind and mediate the SHM process. Bisulfite sequencing of chromatinized DNA could thus be used to search for other potential targets of SHM in the genome and to uncover the mechanism whereby SHM is targeted in healthy and cancerous B cells.
MATERIALS AND METHODS
Cell lines and mice.
The Ramos cell lines have been described (38). Jurkat cells were provided by T. Graf (Albert Einstein College of Medicine). AICDA knockout mice (3) were provided by T. Honjo (Kyoto University, Kyoto, Japan). Both wild-type and knockout mice were immunized intraperitoneally at 2–3 mo of age with 23NP-CGG (BioSearch Technologies) in alum (Pierce Chemical Co.). Mice were immunized on day zero with 100 mg NP-CGG, then at day 14 and again on day 15 with 100 mg NP-CGG in alum. Splenocytes were analyzed 1 wk after the secondary immunization. All of the mouse studies were approved by the Albert Einstein Animal Use Committee.
Bisulfite treatment of cells.
107 cells were fixed at room temperature with 1% formaldehyde for 5 min. The reaction was stopped with glycine to a final concentration of 125 mM. Nuclei were isolated and permeabilized by incubating cells in each of the following buffers for 20 min: buffer A (10 mM Tris-HCl, pH 8, 10 mM EDTA, pH 8, 0.5 mM EGTA, pH 8, 0.25% Triton X-100) and buffer B (0.2 M NaCl, 10 mM Tris-HCl, pH 8, 1 mM EDTA, 0.5 mM EGTA), and then resuspended in buffer C (0.3 M NaCl, 40 mM Tris-HCl, 4 mM EDTA, 1% Triton X-100). All buffers contained complete protease inhibitors (Roche). Nuclei or purified genomic DNA was incubated in a fresh solution containing 5 M sodium bisulfite and 20 mM hydroquinone and incubated at 37°C for 18 h. “Chromatinized” DNA was treated as described above, and “untreated” controls were handled similarly, except that no bisulfite was added during the final incubation. The cross-link was reversed, and DNA was purified with the addition of proteinase K (Roche) to a final concentration of 0.58 mg/ml by incubation at 55°C for 2 h, and then at 65°C overnight. DNA was purified using a Wizard genomic purification kit (Promega), and then disulphonated by incubation for 15 min with NaOH to a final concentration of 0.3 M, neutralized with ammonium acetate to a final concentration of 3 M, and purified by ethanol precipitation. In some cases, nuclei were pretreated overnight before incubation with bisulfite with 6 U proteinase K or 1 U RNaseH (Roche) per 106 cells. “Deproteinized” DNA was purified as genomic DNA, and then treated with bisulfite. Chromatinized, untreated, or deproteinized DNA was amplified by PCR using primers listed in the Table S1 (available at http://www.jem.org/cgi/content/full/jem.20062032/DC1) using Taq polymerase, which is able to copy deoxyuridine, cloned in TOPO TA vectors (Invitrogen), and sequenced with the M13 reverse primer. For mutation analysis, controls that were not treated with bisulfite were amplified with Pfu Turbo polymerase (Stratagene), cloned using the Zero Blunt PCR cloning system (Invitrogen), and sequenced. Mouse splenocytes were dislocated from spleens using the balloon method and immediately fixed, permeabilized, and treated with bisulfite, as described above.
Analysis of transcription.
Cells were treated with 0.25 μg/ml α-amanitin (Sigma-Aldrich) for 24 h. RNAP II chromatin immunoprecipitation was performed as described previously (71). In brief, 108 cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and the reaction was quenched by the addition of glycine to a final concentration of 0.125 M, and then washed three times with PBS. Cells were placed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0) and sonicated on ice in a Sonic Dismembrator (model 500; Fisher Scientific) with pulses of 30 s and a rest of 1 min in between pulses for a total time of 12 minutes at 55% amplitude to obtain an average DNA fragment length of 200 bp. The soluble chromatin fraction obtained after centrifugation was diluted 10 times and precleared with 30 μl/ml blocked protein A agarose beads (GE Healthcare). 100 μl of precleared lysate was saved to determine the chromatin input. Chromatin immunoprecipitation was achieved by overnight incubation at 4°C with 2 μg/ml anti–RNAP II (Santa Cruz Biotechnology, Inc.) or rabbit IgG anti-actin (Sigma-Aldrich). Controls immunoprecipitated with either anti-actin antibodies or no antibodies showed no detectable PCR product (unpublished data). Immunoprecipitated DNA was analyzed by real-time quantitative PCR using SYBRGreen (QIAGEN) and primers listed in Table S1. For strand-specific RT-PCR, RNA was prepared using the RNAWiz reagent (Ambion) according to the manufacturer's specifications. DNA was digested with DNaseI (Worthington) for 30 min at 37°C. Strand-specific RT was performed as described previously (48), except that RT primers contain the Tag sequence 5′CAGGTCATGGTGGCGA3′ on the 5′ end. PCR was performed using the Tag primer and gene-specific primers listed in Table S1.
Calculation of average length of ssDNA patches.
The minimum length of ssDNA patches that contained more than three consecutive converted dCs was determined by counting the distance, in nucleotides, between the furthest converted bases, including the intervening non-dC for the upper strand (or nondeoxyguanine for the lower strand). The maximum length of these patches was determined by counting the nucleotides between (and excluding) the nonconverted dCs to the 5′ and 3′ ends of the patch. The minimum and maximum lengths were determined separately for upper and lower strands, and the number of occurrences of each length was summed up and plotted.
Statistical analysis.
ssDNA patches are defined as stretches of DNA containing bisulfite-modified dCs on either strand without any intervening unmodified bases. Unless otherwise indicated, only patches longer than three consecutive C's were taken into account. The frequency of ssDNA was calculated as the ratio between the number of C's in ssDNA patches and the total number of C's sequenced. The number of modified and unmodified C's between different sequences was compared using the two-tailed Chi-squared test.
Online supplemental material.
Fig. S1 shows the detection of ssDNA using sodium bisulfite. Fig. S2 shows the detection of ssDNA in R loops using deproteinized and chromatinized DNA. Fig. S3 shows the detection of ssDNA in Ig-variable regions in human B cell lines. Fig. S4 depicts antisense transcripts in the Ramos V region, and Fig. S5 shows cell cycle distribution of ssDNA in Ramos cells. Fig. S6 is the ssDNA frequency in two Ramos cell subclones that differ in the amount of AID expression. Table S1 lists PCR and RT-PCR primers used in this study. Table S2 lists the sample size for bisulfite analysis. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20062032/DC1.
Supplemental Material
Acknowledgments
We are grateful to B.K. Birshtein, J. Warner, A. Skoultchi, D. Fyodorov, and M. Shulman for critical reading of the manuscript; to M. Sadofski, H. Ye, E. Bouhassira, and M. Brenowitz for helpful discussions; T. Honjo for AID-deficient mice; M. Kim for help with the statistical analysis; A. Melnick's lab for help with the ChIP assay; C. Schildkraut and E. Cook for help with cell elutriation; and S. Buhl, Z. Polonskaya, and C. Zhao for technical assistance.
This work was supported by National Institutes of Health grants CA72649 and CA102705 to M.D. Scharff and by a grant from the Canadian Cancer Society (16080) to A. Martin. M.D. Scharff is also supported by the Harry Eagle Chair provided by the National Women's Division of the Albert Einstein College of Medicine; D. Ronai is supported by a Cancer Research Institute Postdoctoral Fellowship and the Harry Eagle Fellowship; M.D. Iglesias-Ussel is supported by a Fellowship from the Northeast Biodefense Center (AI57158); Z. Li is a special fellow of the Leukemia and Lymphoma Society; and A. Martin is supported by a Canada Research Chair Award.
The authors have no conflicting financial interests.
Abbreviations used: AID, activation-induced cytidine deaminase; CSR, class-switch recombination; dC, deoxycytidine; GC, germinal center; NP, 4-hydroxy-3-nitrophenyl acetyl; RNAP II, RNA polymerase II; SHM, somatic hypermutation; ssDNA, single-stranded DNA.
D. Ronai's present address is Howard Hughes Medical Institute, Department of Molecular, Cellular and Developmental Biology, University of Colorado at Boulder, Boulder, CO 80309.
References
- 1.Maizels, N. 2005. Immunoglobulin gene diversification. Annu. Rev. Genet. 39:23–46. [DOI] [PubMed] [Google Scholar]
- 2.McHeyzer-Williams, L.J., D.J. Driver, and M.G. McHeyzer-Williams. 2001. Germinal center reaction. Curr. Opin. Hematol. 8:52–59. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 4.Bransteitter, R., P. Pham, M.D. Scharff, and M.F. Goodman. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA. 100:4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F.W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. [DOI] [PubMed] [Google Scholar]
- 6.Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. [DOI] [PubMed] [Google Scholar]
- 7.Di Noia, J., and M.S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48. [DOI] [PubMed] [Google Scholar]
- 8.Li, Z., C.J. Woo, M.D. Iglesias-Ussel, D. Ronai, and M.D. Scharff. 2004. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 18:1–11. [DOI] [PubMed] [Google Scholar]
- 9.Barreto, V.M., A.R. Ramiro, and M.C. Nussenzweig. 2005. Activation-induced deaminase: controversies and open questions. Trends Immunol. 26:90–96. [DOI] [PubMed] [Google Scholar]
- 10.Pasqualucci, L., P. Neumeister, T. Goossens, G. Nanjangud, R.S. Chaganti, R. Kuppers, and R. Dalla-Favera. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346. [DOI] [PubMed] [Google Scholar]
- 11.Storb, U., H.M. Shen, N. Michael, and N. Kim. 2001. Somatic hypermutation of immunoglobulin and non-immunoglobulin genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen, H.M., A. Peters, B. Baron, X. Zhu, and U. Storb. 1998. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 280:1750–1752. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, M.S., C.M. Kanegai, J.R. Doerr, and R. Wall. 2003. Somatic hypermutation of the B cell receptor genes B29 (Igbeta, CD79b) and mb1 (Igalpha, CD79a). Proc. Natl. Acad. Sci. USA. 100:4126–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 15.Ramiro, A.R., M.C. Nussenzweig, and A. Nussenzweig. 2006. Switching on chromosomal translocations. Cancer Res. 66:7837–7839. [DOI] [PubMed] [Google Scholar]
- 16.Odegard, V.H., and D.G. Schatz. 2006. Targeting of somatic hypermutation. Nat. Rev. Immunol. 6:573–583. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri, J., and F.W. Alt. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541–552. [DOI] [PubMed] [Google Scholar]
- 18.Yu, K., F. Chedin, C.L. Hsieh, T.E. Wilson, and M.R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442–451. [DOI] [PubMed] [Google Scholar]
- 19.Duquette, M.L., P. Pham, M.F. Goodman, and N. Maizels. 2005. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 24:5791–5798. [DOI] [PubMed] [Google Scholar]
- 20.Larson, E.D., M.L. Duquette, W.J. Cummings, R.J. Streiff, and N. Maizels. 2005. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 15:470–474. [DOI] [PubMed] [Google Scholar]
- 21.Zarrin, A.A., F.W. Alt, J. Chaudhuri, N. Stokes, D. Kaushal, L. Du Pasquier, and M. Tian. 2004. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 5:1275–1281. [DOI] [PubMed] [Google Scholar]
- 22.Dickerson, S.K., E. Market, E. Besmer, and F.N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beale, R.C., S.K. Petersen-Mahrt, I.N. Watt, R.S. Harris, C. Rada, and M.S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585–596. [DOI] [PubMed] [Google Scholar]
- 24.Besmer, E., E. Market, and F.N. Papavasiliou. 2006. The transcription elongation complex directs activation-induced cytidine deaminase-mediated DNA deamination. Mol. Cell. Biol. 26:4378–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen, H.M., and U. Storb. 2004. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc. Natl. Acad. Sci. USA. 101:12997–13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen, H.M., S. Ratnam, and U. Storb. 2005. Targeting of the activation-induced cytosine deaminase is strongly influenced by the sequence and structure of the targeted DNA. Mol. Cell. Biol. 25:10815–10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhuri, J., C. Khuong, and F.W. Alt. 2004. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 430:992–998. [DOI] [PubMed] [Google Scholar]
- 28.Fukita, Y., H. Jacobs, and K. Rajewsky. 1998. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 9:105–114. [DOI] [PubMed] [Google Scholar]
- 29.Storb, U., A. Peters, E. Klotz, N. Kim, H.M. Shen, J. Hackett, B. Rogerson, and T.E. Martin. 1998. Cis-acting sequences that affect somatic hypermutation of Ig genes. Immunol. Rev. 162:153–160. [DOI] [PubMed] [Google Scholar]
- 30.Woo, C.J., A. Martin, and M.D. Scharff. 2003. Induction of hypermutation is associated with modifications of variable region chromatin in BL2 cells. Immunity. 19:479–489. [DOI] [PubMed] [Google Scholar]
- 31.Nambu, Y., M. Sugai, H. Gonda, C.G. Lee, T. Katakai, Y. Agata, Y. Yokota, and A. Shimizu. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 302:2137–2140. [DOI] [PubMed] [Google Scholar]
- 32.Li, Z., Z. Luo, and M.D. Scharff. 2004. Differential regulation of histone acetylation and generation of mutations in switch regions is associated with Ig class switching. Proc. Natl. Acad. Sci. USA. 101:15428–15433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odegard, V.H., S.T. Kim, S.M. Anderson, M.J. Shlomchik, and D.G. Schatz. 2005. Histone modifications associated with somatic hypermutation. Immunity. 23:101–110. [DOI] [PubMed] [Google Scholar]
- 34.Wang, L., N. Whang, R. Wuerffel, and A.L. Kenter. 2006. AID-dependent histone acetylation is detected in immunoglobulin S regions. J. Exp. Med. 203:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen, H.M., N. Michael, N. Kim, and U. Storb. 2000. The TATA binding protein, c-Myc and survivin genes are not somatically hypermutated, while Ig and BCL6 genes are hypermutated in human memory B cells. Int. Immunol. 12:1085–1093. [DOI] [PubMed] [Google Scholar]
- 36.Hayatsu, H. 1976. Bisulfite modification of nucleic acids and their constituents. Prog. Nucleic Acid Res. Mol. Biol. 16:75–124. [DOI] [PubMed] [Google Scholar]
- 37.Sale, J.E., and M.S. Neuberger. 1998. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 9:859–869. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, W., P.D. Bardwell, C.J. Woo, V. Poltoratsky, M.D. Scharff, and A. Martin. 2001. Clonal instability of V region hypermutation in the Ramos Burkitt's lymphoma cell line. Int. Immunol. 13:1175–1184. [DOI] [PubMed] [Google Scholar]
- 39.Kuo, M.H., and C.D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 19:425–433. [DOI] [PubMed] [Google Scholar]
- 40.Bemark, M., and M.S. Neuberger. 2000. The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt's lymphoma line. Oncogene. 19:3404–3410. [DOI] [PubMed] [Google Scholar]
- 41.Milstein, C., M.S. Neuberger, and R. Staden. 1998. Both DNA strands of antibody genes are hypermutation targets. Proc. Natl. Acad. Sci. USA. 95:8791–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro, G.S., M.C. Ellison, and L.J. Wysocki. 2003. Sequence-specific targeting of two bases on both DNA strands by the somatic hypermutation mechanism. Mol. Immunol. 40:287–295. [DOI] [PubMed] [Google Scholar]
- 43.Peters, A., and U. Storb. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 4:57–65. [DOI] [PubMed] [Google Scholar]
- 44.Bachl, J., C. Carlson, V. Gray-Schopfer, M. Dessing, and C. Olsson. 2001. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 166:5051–5057. [DOI] [PubMed] [Google Scholar]
- 45.Gnatt, A. 2002. Elongation by RNA polymerase II: structure-function relationship. Biochim. Biophys. Acta. 1577:175–190. [DOI] [PubMed] [Google Scholar]
- 46.Bransteitter, R., P. Pham, P. Calabrese, and M.F. Goodman. 2004. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 279:51612–51621. [DOI] [PubMed] [Google Scholar]
- 47.Katayama, S., Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C.C. Yap, M. Suzuki, J. Kawai, et al. 2005. Antisense transcription in the mammalian transcriptome. Science. 309:1564–1566. [DOI] [PubMed] [Google Scholar]
- 48.Bolland, D.J., A.L. Wood, C.M. Johnston, S.F. Bunting, G. Morgan, L. Chakalova, P.J. Fraser, and A.E. Corcoran. 2004. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 5:630–637. [DOI] [PubMed] [Google Scholar]
- 49.Sangar, D.V., and A.R. Carroll. 1998. A tale of two strands: reverse-transcriptase polymerase chain reaction detection of hepatitis C virus replication. Hepatology. 28:1173–1176. [DOI] [PubMed] [Google Scholar]
- 50.Faili, A., S. Aoufouchi, Q. Gueranger, C. Zober, A. Leon, B. Bertocci, J.C. Weill, and C.A. Reynaud. 2002. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 3:815–821. [DOI] [PubMed] [Google Scholar]
- 51.Papavasiliou, F.N., and D.G. Schatz. 2000. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 408:216–221. [DOI] [PubMed] [Google Scholar]
- 52.Weiss, U., and K. Rajewsky. 1990. The repertoire of somatic antibody mutants accumulating in the memory compartment after primary immunization is restricted through affinity maturation and mirrors that expressed in the secondary response. J. Exp. Med. 172:1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohwi-Shigematsu, T., R. Gelinas, and H. Weintraub. 1983. Detection of an altered DNA conformation at specific sites in chromatin and supercoiled DNA. Proc. Natl. Acad. Sci. USA. 80:4389–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaret, K.S. 1999. In vivo analysis of chromatin structure. Methods Enzymol. 304:612–626. [DOI] [PubMed] [Google Scholar]
- 55.Palecek, E. 1992. Probing of DNA structure in cells with osmium tetroxide-2,2'-bipyridine. Methods Enzymol. 212:305–318. [DOI] [PubMed] [Google Scholar]
- 56.Lebecque, S.G., and P.J. Gearhart. 1990. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5′ boundary is near the promoter, and 3′ boundary is approximately 1 kb from V(D)J gene. J. Exp. Med. 172:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rada, C., J. Yelamos, W. Dean, and C. Milstein. 1997. The 5′ hypermutation boundary of kappa chains is independent of local and neighbouring sequences and related to the distance from the initiation of transcription. Eur. J. Immunol. 27:3115–3120. [DOI] [PubMed] [Google Scholar]
- 58.Rada, C., and C. Milstein. 2001. The intrinsic hypermutability of antibody heavy and light chain genes decays exponentially. EMBO J. 20:4570–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogozin, I.B., and M. Diaz. 2004. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172:3382–3384. [DOI] [PubMed] [Google Scholar]
- 60.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tashiro, J., K. Kinoshita, and T. Honjo. 2001. Palindromic but not G-rich sequences are targets of class switch recombination. Int. Immunol. 13:495–505. [DOI] [PubMed] [Google Scholar]
- 62.Min, I.M., C.E. Schrader, J. Vardo, T.M. Luby, N. D'Avirro, J. Stavnezer, and E. Selsing. 2003. The Smu tandem repeat region is critical for Ig isotype switching in the absence of Msh2. Immunity. 19:515–524. [DOI] [PubMed] [Google Scholar]
- 63.Mason, P.B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 17:831–840. [DOI] [PubMed] [Google Scholar]
- 64.Apel, T.W., J. Mautner, A. Polack, G.W. Bornkamm, and D. Eick. 1992. Two antisense promoters in the immunoglobulin mu-switch region drive expression of c-myc in the Burkitt's lymphoma cell line BL67. Oncogene. 7:1267–1271. [PubMed] [Google Scholar]
- 65.Prescott, E.M., and N.J. Proudfoot. 2002. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. USA. 99:8796–8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, J., and L.F. Liu. 1997. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 25:4181–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bross, L., Y. Fukita, F. McBlane, C. Demolliere, K. Rajewsky, and H. Jacobs. 2000. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 13:589–597. [DOI] [PubMed] [Google Scholar]
- 68.Zan, H., X. Wu, A. Komori, W.K. Holloman, and P. Casali. 2003. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 18:727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong, Q., and N. Maizels. 2001. DNA breaks in hypermutating immunoglobulin genes: evidence for a break and repair pathway of somatic mutation. Genetics. 158:369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papavasiliou, F.N., and D.G. Schatz. 2002. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med. 195:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sandoval, J., J.L. Rodriguez, G. Tur, G. Serviddio, J. Pereda, A. Boukaba, J. Sastre, L. Torres, L. Franco, and G. Lopez-Rodas. 2004. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 32:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pasqualucci, L., A. Migliazza, N. Fracchiolla, C. William, A. Neri, L. Baldini, R.S.K. Chaganti, U. Klein, R. Kuppers, K. Rajewsky, and R. Dalla-Favera. 1998. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA. 95:11816–11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiss, U., R. Zoebelein, and K. Rajewsky. 1992. Accumulation of somatic mutants in the B cell compartment after primary immunization with a T cell-dependent antigen. Eur. J. Immunol. 22:511–517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.