Abstract
Macroautophagy (hereafter referred to as autophagy) is a well-conserved intracellular degradation process. Recent studies examining cells lacking the autophagy genes Atg5 and Atg7 have demonstrated that autophagy plays essential roles in cell survival during starvation, in innate cell clearance of microbial pathogens, and in neural cell maintenance. However, the role of autophagy in T lymphocyte development and survival is not known. Here, we demonstrate that autophagosomes form in primary mouse T lymphocytes. By generating Atg5−/− chimeric mice, we found that Atg5-deficient T lymphocytes underwent full maturation. However, the numbers of total thymocytes and peripheral T and B lymphocytes were reduced in Atg5 chimeras. In the periphery, Atg5−/− CD8+ T lymphocytes displayed dramatically increased cell death. Furthermore, Atg5−/− CD4+ and CD8+ T cells failed to undergo efficient proliferation after TCR stimulation. These results demonstrate a critical role for Atg5 in multiple aspects of lymphocyte development and function and suggest that autophagy may be essential for both T lymphocyte survival and proliferation.
Macroautophagy, microautophagy, and chaperone-mediated autophagy are conserved catabolic processes in eukaryotic cells that deliver cytoplasmic material to lysosomes (1). Macroautophagy (hereafter referred to as autophagy) is uniquely defined by the formation of autophagosomes, double membrane vesicles 0.5–1.5 μm in diameter arising through the elongation of a cup-shaped isolation membrane around cellular cargo (1). A screen of yeast mutants incapable of surviving nitrogen starvation has identified a network of Atg genes important for the formation of autophagosomes (2). Homologues of these genes have been identified and characterized in the mammalian system. Although autophagy has been long recognized, only recently have its essential roles in various cellular functions been established (3, 4). For example, mice lacking the autophagy gene Atg5 or Atg7 cannot withstand short periods of starvation and display severe neurodegenerative disease (5–7). Autophagy is also essential for maintaining cell survival after growth factor withdrawal (8). Furthermore, autophagy contributes to the clearance of intracellular pathogens including group A Streptococcus and Mycobacterium tuberculosis (9, 10).
Autophagy is initiated through a well-conserved molecular pathway. Class III phosphatidylinoside 3-kinase in complex with Beclin-1 (yeast Atg6) induces autophagosome formation (11, 12). Two ubiquitin-like conjugation systems downstream of Beclin-1–phosphatidylinoside 3-kinase are important for the induction of autophagy. The terminal products of one of these two pathways, an Atg5–Atg12 covalent complex, is required for the elongation of the isolation membrane (13). The second pathway results in the conjugation of microtubule-associated protein 1 light chain 3 (LC3; yeast Atg8) to phospholipids for incorporation into the autophagosome membrane (14).
Although autophagy is important for survival during periods of nutrient deprivation (5), its role in regulating cellular life and death decisions remains largely unknown (for review see reference 15). The induction of autophagy may either program or prevent cell death (3, 4, 16). Regardless, the regulation of autophagy and apoptosis is likely to be linked at the molecular level. The autophagy protein Beclin-1 binds to antiapoptotic Bcl-2 family proteins, including Bcl-xL and Bcl-2 (17). Atg5 interacts with Fas-associated via death domain (FADD), a death receptor adaptor protein (18). Autophagy-dependent death occurs in cells whose apoptotic machinery has been disabled (19, 20).
Although innate immune cells, such as macrophages, use autophagy to clear microbial pathogens, it is not known whether autophagy has a role in the adaptive immune system. Only recently has evidence emerged that T cells may be capable of inducing autophagy. Human T lymphocytes stimulated and cultured in vitro form autophagosomes as identified by transmission electron microscopy (21). CD4+ human umbilical cord T cells also can be induced to form autophagosomes by the HIV-1 envelope glycoprotein (22). Yet a role for autophagy in T cells remains undefined.
To study the role of autophagy in T lymphocyte development and function, we first assessed the induction of autophagy in primary mouse T lymphocytes. We then generated autophagy-deficient lymphocytes by transferring Atg5−/− fetal hematopoietic progenitor cells into lethally irradiated wild-type congenic hosts. We found that Atg5−/− lymphocytes exhibit multiple defects, including a reduced lymphocyte compartment, enhanced CD8+ T cell apoptosis, and an inability to undergo TCR-induced proliferation. Our results suggest that autophagy may play an essential role in T lymphocyte survival and function.
RESULTS AND DISCUSSION
Autophagy in mouse T lymphocytes
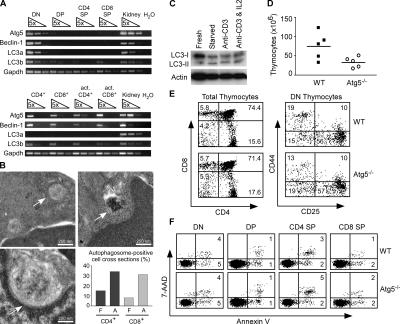
Recent reports suggest that thymocytes (19) and cultured human lymphocytes (21, 22) are capable of inducing autophagy. To determine whether primary mouse T cells can form autophagosomes, we took three approaches. First, to determine if T lymphocytes express autophagy machinery, thymocytes and peripheral T cells were double sorted (>99% pure) and total RNA was isolated. Semiquantitative RT-PCR was performed to detect the expression of Atg5, Beclin-1 (yeast Atg6), and LC3 (yeast Atg8) (Fig. 1 A). Expression of these essential autophagy genes could be detected in both thymocytes and mature T cells. In the thymus, Atg5, Beclin-1, and LC3 were most highly expressed in the double negative (DN) compartment. In the periphery, both CD4+ and CD8+ T cells expressed autophagy machinery. This expression was maintained after 2 d of in vitro stimulation with anti-CD3.
Figure 1.
Atg5−/− thymocytes are reduced in number but develop normally. (A) Rag-2−/− DN thymocytes, sorted DP, CD4+ SP, CD8+ SP, and peripheral T cell subsets from C57BL/6 mice were analyzed for the expression of autophagy genes by semiquantitative RT-PCR. Both sorted CD4+ and CD8+ peripheral T cells were activated in vitro for 2 d with anti-CD3 and subjected to RT-PCR analysis. (B) Transmission electron microscopy of peripheral T cells. Freshly isolated (F) and anti-CD3–activated (A) CD4+ and CD8+ T cells (n = 50) were cross sectioned and analyzed for the presence of autophagosomes (arrows). Activated cells were cultured in vitro for 2 d with anti-CD3. (C) T cell immunoblot for LC3. Purified T cells from C57BL/6 mice were either immediately lysed or amino acid starved in vitro for 4 h in a balanced salt solution, or stimulated with plate-bound anti-CD3 with or without hIL-2 (100 U/ml) for 16 h. The lysates were probed for LC3 processing. Actin serves as a loading control. (D) Total thymocyte number in Atg5−/− and wild-type chimeras. Circles and squares represent individual mice. P = 0.04. (E) FACS profile of Atg5−/− thymocytes. Total thymocytes were stained with anti-CD4 and anti-CD8. DN thymocytes were pregated on CD3−CD4−CD8− cells. (F) FACS analysis of thymocyte apoptosis. Thymocytes were stained with CD4, CD8, 7-AAD, and annexin V. Numbers indicate the percentage of cells in each region.
Next, to determine if autophagosomes are present in T lymphocytes, we purified CD4+ and CD8+ T cells from the spleen and lymph nodes of naive mice (≥96% pure). Using transmission electron microscopy, we could identify characteristic double membrane autophagosomes in both T cell populations (Fig. 1 B). Quantification revealed that ∼10% of CD4+ and CD8+ T cell cross sections contained autophagosomes (Fig. 1 B). We also analyzed autophagosome formation in T cells purified after in vitro stimulation for 2 d with anti-CD3. Approximately 30% of activated cell cross sections contained autophagosomes (Fig. 1 B). However, the narrow depth of the section does not allow us to distinguish between an increase in the number of autophagosome per cell and an increase in the number of autophagosome-positive cells.
Finally, we used immunoblotting for LC3 to detect autophagy in T cells. LC3 undergoes regulated modifications during the induction of autophagy, delivering a faster migrating processed band (termed LC3-II) to autophagosomes (14). Although a small amount of LC3-II was present in freshly purified peripheral T cells, the ratio of LC3-II to LC3-I was increased after 4 h of amino acid starvation in a balanced salt solution and after anti-CD3–induced T cell activation with or without exogenous IL-2 (Fig. 1 C). Collectively, these three results suggest that primary T lymphocytes form autophagosome and can regulate the induction of autophagy.
Thymocyte development in Atg5−/− chimeras
To determine the role of autophagy-related genes in T cell development and function, we reconstituted the immune system of adult mice with autophagy-deficient hematopoietic precursors from mice lacking Atg5 (5). Atg5-deficient mice die within 24 h of birth because of their inability to survive brief periods of nutrient deprivation. We generated chimeric mice by transferring day 14 fetal liver cells from Atg5−/− and wild-type mice into lethally irradiated (950 rad) CD45.1+ congenic hosts. We analyzed the thymus of Atg5−/− and wild-type chimeras by flow cytometry 6–10 wk after reconstitution. In all chimeric mice, >99% of thymocytes recovered were CD45.2+ (unpublished data), indicating a complete reconstitution by donor-derived cells. When compared with wild-type controls, Atg5−/− chimeras demonstrated a 40–60% reduction in their total thymic cellularity (Fig. 1 D). Although the number of Atg5−/− thymocytes was reduced, the developmental profile of cells within the thymus appeared normal (Fig. 1 E). Atg5−/− chimeras had normal percentages of CD4−CD8− DN, CD8+CD4+ double positive (DP), CD4+ single positive (SP), and CD8+ SP cells. Furthermore, the progressive subsets within the DN compartment as defined by the expression of CD25 and CD44 were comparable between Atg5−/− and wild-type control thymus. Atg5−/− thymocytes did not exhibit enhanced apoptosis as detected by annexin V and 7–aminoactinomycin D (7-AAD) staining (Fig. 1 F). Collectively, these results demonstrate that Atg5-deficient thymocytes underwent full maturation without any obvious developmental blockade, suggesting that Atg5 is dispensable in thymocyte differentiation. The reduced thymic cellularity may be caused by reduced lymphoid precursor activity in the absence of Atg5 (see following paragraph).
Reduced peripheral lymphocyte compartment in Atg5−/− chimeras
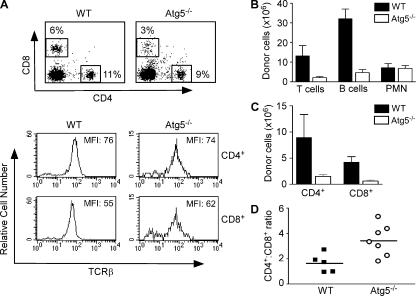
We examined the peripheral lymphocyte compartment of chimeric mice 6–10 wk after fetal liver reconstitution. CD45.2+ donor-derived CD4+ and CD8+ T lymphocytes were detected in the spleen of both Atg5−/− and wild-type chimeras (Fig. 2 A). Atg5−/− T cells expressed similar levels of TCR on their surface compared with control cells (Fig. 2 A). However, the total numbers of Atg5−/− T and B lymphocytes in the spleen were only ∼10% of control chimeras (Fig. 2 B). In addition, the total number of cells obtained from the LNs of Atg5−/− chimeras was 10% of that in control mice (Atg5−/−: 0.83 + 0.8 × 106, n = 4; control: 8.2 + 6 × 106, n = 3). In contrast, the number of Atg5−/− Gr-1+CD11b+ neutrophils in the spleen was comparable to that of controls (Fig. 2 B), suggesting that Atg5−/− lymphoid progenitors have a defective capacity to reconstitute the lymphoid compartment.
Figure 2.
Defective peripheral lymphocyte compartment in Atg5−/− chimeras. (A) FACS profiles of donor-derived CD4+ and CD8+ T cells in the spleens of Atg5−/− and wild-type chimeric mice 6–10 wk after reconstitution. All cells are pregated on CD45.2+ donor-derived cells. (B) The numbers of T cells, B cells, and neutrophils in the spleen of Atg5−/− and wild-type chimeras. The numbers are derived from multiplying the percentage of CD4+ and CD8+ T cells, B220+, or Gr-1+CD11b+ CD45.2+ cells by the total numbers of splenocytes. p values for comparison between Atg5−/− and wild-type chimeras are: T cells, P = 0.0267; B cells, P = 0.0001; PMN, P = 0.8785. (C) The numbers of CD4+ and CD8+ T lymphocytes in the spleen of Atg5−/− and wild-type chimeras. CD4+ T cells, P = 0.0678; CD8+ T cells, P = 0.002. (D) The CD4+ to CD8+ T cell ratio in Atg5−/− and wild-type chimeras. Shown are data from individual mice. P = 0.0163. All p values are derived from unpaired, two-tailed Student's t test.
Interestingly, although the numbers of both Atg5−/− CD4+ and CD8+ T lymphocytes were dramatically reduced (Fig. 2 C), the reduction of Atg5−/− CD8+ T lymphocytes (5% of control cells) was more severe than that of Atg5−/− CD4+ T lymphocytes (10% of control cells) (Fig. 2 A and C). This was further indicated by the elevated CD4+ to CD8+ ratio of Atg5−/− T lymphocytes compared with that of control T cells (Fig. 2 D). Although the number of thymocytes in Atg5−/− chimeras is reduced, this dramatic and selective reduction in peripheral T lymphocytes suggests a role for autophagy in mature T cells.
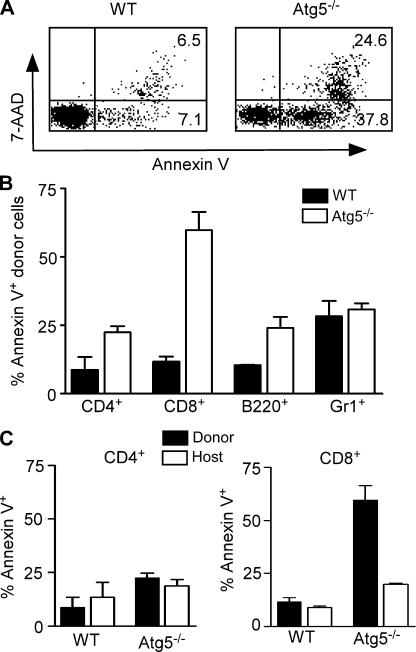
Selective survival defect in Atg5−/− CD8+ T cells
The process of autophagy is intimately linked to survival. Atg5 was first identified in a screen for yeast incapable of surviving nitrogen starvation (2). Given that CD8+ T cells exhibited a more severe reduction than CD4+ T cells in Atg5−/− chimeras, we examined whether Atg5 was required for mature CD8+ T cell survival in vivo. In freshly isolated splenocytes, <10% of wild-type but nearly 40% of Atg5−/− CD8+ T cells were annexin V+7-AAD− (Fig. 3 A), indicating that these cells had initiated apoptosis in vivo. Furthermore, the total apoptotic rate of Atg5−/− CD8+ T cells as indicated by annexin V+7-AAD− and annexin V+7-AAD+ staining was five to six times higher than that of control CD8+ T cells (Fig. 3, A and B). In contrast, the apoptotic rates of Atg5−/− CD4+ T lymphocytes and B220+ B lymphocytes were only slightly higher than those of control cells, whereas the apoptotic rate of Atg5−/− Gr-1+ neutrophils was similar to that of control cells (Fig. 3 B). Because a small fraction of host peripheral CD4+ and CD8+ T lymphocytes survived the lethal irradiation, we examined the apoptosis in these cells as an internal control. The apoptotic rates of CD4+ T lymphocytes from either wild-type and Atg5−/− donors or C57BL/6 congenic hosts were comparable (Fig. 3 C). However, the apoptotic rates of Atg5−/− CD8+ T cells were dramatically higher than those of either wild-type CD8+ or host long-lived CD8+ T cells (Fig. 3 C). This result suggests that Atg5, and perhaps the ability to induce autophagy, is required for the survival of CD8+ T cells. Interestingly, we noticed that the apoptotic rates of CD4+ and CD8+ host T cells in Atg5−/− chimeras were also slightly higher than those of their counterparts in wild-type chimeras (Fig. 3 C). Whether this elevation is indirectly caused by the massive apoptosis of Atg5−/− CD8+ T cells remains to be determined.
Figure 3.
Selective survival defect in Atg5−/− CD8+ T cells. (A) FACS analysis of CD8+ T cell apoptosis. Donor-derived CD8+ T cells in Atg5−/− and wild-type chimeras were stained with 7-AAD and PE–annexin V. Numbers indicate the percentage of cells in each region. (B). Apoptotic rates of various cell populations in Atg5−/− and wild-type chimeras. Donor-derived cells (CD45.2+) were stained using the indicated lineage markers plus PE–annexin V and analyzed by FACS. CD8+ T cells, P = 0.0118; CD4+, P = 0.0544; B220+, P = 0.0826; Gr1+ cells, P = 0.6605. n = 3. (C) Apoptotic rates of donor-derived and host long-term surviving CD4+ and CD8+ T cells. p values for donor versus host T cells in Atg5−/− chimeras: CD4+ T cells, P = 0.72; CD8+ T cells, P = 0.004.
Our results suggest that the survival defect seems to be specific to peripheral T cells. Not only do Atg5−/− CD8+ SP thymocytes develop normally (Fig. 1 E), but Atg5−/− CD8+ SP thymocytes do not display an enhanced rate of apoptosis in vivo compared with wild-type controls (Fig. 1 F). How does Atg5 contribute to the survival of CD8+ T cells? One possibility is that peripheral CD8+ T cells depend on autophagy for their survival. This is consistent with the expression of autophagy genes and presence of autophagosomes in these cells. This possibility implies that CD8+ T cells encounter either a nutrient shortage or cytokine deprivation or both after they exit the thymus. Growth factor deprivation–induced autophagy has been shown to be critical for cell survival (8). Given that CD4+ T cells also express autophagy machinery and form autophagosomes, it is interesting to see that CD4+ T cell survival is less dependent on Atg5 than CD8+ T cells.
Alternatively, Atg5 may regulate CD8+ T cell survival by interacting with antiapoptotic molecules. Published data have demonstrated a pattern of reciprocal relationships between autophagy and apoptosis. Both the inhibition of apoptosis can induce autophagy (19, 20, 23), and the inhibition of autophagy can induce apoptosis (24). Molecular and functional studies have only begun to explore the complex relationship between autophagy and apoptosis. The mammalian Atg6 homologue, Beclin-1, was first identified as a Bcl-2–Bcl-xL interacting protein (17); however, the role of this interaction remains controversial. Bcl-2 has been shown to inhibit autophagosome formation in nutrient-starved yeast and human cancer cell lines (25), yet Bcl-2 expression in Bax−/− Bak−/− mouse embryonic fibroblasts enhances autophagy after etoposide treatment (19). In each of these in vitro studies, autophagy was required for the observed cellular death. However, the massive apoptosis we observe in Atg5−/− CD8+ T cells suggests that the capacity to induce autophagy in these cells is essential for their survival in vivo. Whether direct interactions between autophagy and apoptotic machinery contribute to the enhanced apoptosis in Atg5−/− CD8+ T cells will be an important area of future study.
Impaired proliferation of Atg5−/− T lymphocytes to TCR stimulation
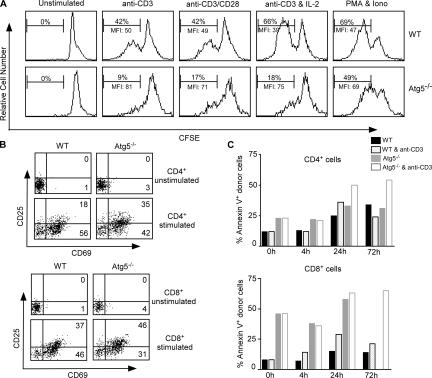
Next, we examined whether Atg5−/− T cells respond normally to TCR stimulation. Total splenocytes from Atg5−/− and wild-type chimeras were labeled with the fluorescent dye CFSE. After 3 d of anti-CD3 stimulation, 7-AAD− CD45.2+ donor-derived T cells were analyzed by flow cytometry for proliferation as measured by CFSE dilution. Although 42% of wild-type CD4+ T cells proliferated in response to plate-bound anti-CD3, only 9% of Atg5−/− CD4+ T cells proliferated (Fig. 4 A). In addition, the number of divisions within the dividing population was reduced in Atg5−/− T cells. Measured by the mean fluorescence intensity, the dilution of CFSE was greater in dividing wild-type T cells than in Atg5−/− cells. These proliferation defects could be rescued by neither the addition of an anti-CD28 costimulatory signal nor the proproliferative cytokine IL-2. When we bypassed membrane-proximal TCR/CD3 signaling by the use of PMA and ionomycin, 49% of Atg5−/− CD4+ T cells proliferated compared with 69% of wild-type controls; however, the dilution of CFSE in dividing cells remained reduced in Atg5−/− T cells (Fig. 4 A). These results suggest that downstream mitogen-activated protein kinase activation and calcium mobilization can partially rescue the proliferation defect in Atg5−/− CD4+ T lymphocytes. Similar results were obtained for Atg5−/− CD8+ T cells (unpublished data).
Figure 4.
Impaired proliferation of Atg5−/− T cells upon anti-CD3 stimulation. (A) Proliferation of Atg5−/− and wild-type CD4+ T cells upon TCR stimulation. Splenocytes from control and Atg5−/− chimeras were labeled with CFSE and stimulated under the following conditions for 3 d: 5 μg/ml of plate-bound anti-CD3, or plus 1 μg/ml plate-bound anti-CD28, or plus 100 U/ml of recombinant IL-2, or 10 ng/ml PMA plus 300 ng/ml of ionomycin. Shown are the percentage of cells with diluted CFSE staining and the mean fluorescence intensity of the proliferating population. (B) Up-regulation of CD25 and CD69 on Atg5−/− and wild-type T cells after overnight stimulation with plate-bound anti-CD3. (C) Apoptosis rates of Atg5−/− and wild-type T cells after TCR stimulation. Total splenocytes were cultured either in media alone or with 5 μg/ml of plate bound anti-CD3 for the indicated times. Apoptosis was measured by annexin V staining. Data are representative of two experiments.
Although Atg5−/− T cells did not proliferate in response to CD3 stimulation, they did express normal levels of TCR (Fig. 2 A). After overnight stimulation, both wild-type and Atg5−/− T cells also expressed similar levels of the activation markers CD69 and CD25 (Fig. 4 B). This result suggests that Atg5−/− T cells can be activated through the TCR complex. Given that Atg5−/− CD8+ T cells displayed a survival defect, we determined whether enhanced apoptosis in activated Atg5−/− T cells might account for the reduced number of proliferating cells. At 4 and 24 h after stimulation, Atg5−/− CD4+ T cells did not display a dramatically enhanced rate of apoptosis compared with wild-type controls (Fig. 4 C). However, at 72 h we did observe an increased death in anti-CD3–stimulated Atg5−/− CD4+ T cells. We observed the same result for Atg5−/− CD8+ T cells, although the baseline rate of cell death was dramatically enhanced in these cells. The absence of early death after TCR stimulation in T cells lacking the autophagy gene Atg5 suggests that the enhanced death observed at 72 h may result from the impaired activation of these cells. Although it is also possible that proliferating Atg5−/− T cells rely on autophagy to maintain their metabolism late in culture, the addition of the cell-permeable metabolic substrate methylpyruvate did not enhance our recovery of proliferating Atg5−/− T lymphocytes (unpublished data). It should be noted that at 72 h very few Atg5−/− CD8+ T cells were detected in the culture without anti-CD3 stimulation and the apoptotic rate cannot be assessed.
Although autophagy has emerging roles in cellular survival and death, no specific function in proliferation has been identified. Our results clearly demonstrate that in activated CD4+ and CD8+ T cells Atg5 is essential for T cell proliferation but not activation. It will be important to determine how the induction of autophagy is regulated during TCR stimulation. Although Atg5 is required for the induction of autophagy in mouse cells (1), it is also possible that Atg5 may serve an autophagy-independent role in T cell proliferation. Atg5 interacts with the death domain of the adaptor protein FADD (18). In HeLa cells, this interaction is required for IFN-γ–mediated autophagic cell death. FADD-deficient T cells display a defect in proliferation, with normal up-regulation of activation markers (26, 27). This similarity suggests that Atg5 and FADD may act together, either in the autophagic process or independent of autophagy, to transduce signals important for T cell proliferation.
In summary, our results demonstrate that the autophagy gene Atg5 plays multiple roles in lymphocyte development and function. The reduced number of thymocytes and B lymphocytes suggests that Atg5 regulates lymphoid precursor activity. The reduced CD4+ and CD8+ T cell compartment in Atg5−/− chimeras is likely caused by both a lowered lymphoid precursor activity and an inability of these cells to undergo homeostatic proliferation because during fetal liver reconstitutions, there is a transient period of lymphopenia in the host after irradiation. In addition, the selective apoptosis of Atg5−/− CD8+ T cells further contribute to the dramatically reduced T cell compartment. Our results suggest that autophagy may play an essential role in T lymphocyte survival and proliferation.
MATERIALS AND METHODS
Animals.
Atg5-deficient mice were generated and characterized previously (5). The mice were backcrossed for seven generations to C57BL/6. 6–8-wk-old CD45.1 C57BL/6 mice were used as hosts for fetal liver transfers (The Jackson Laboratory). All mice were housed in a specific pathogen-free facility and used according to protocols approved by the Duke University Institutional Animal Care and Use Committee.
RT-PCR.
Thymocytes and peripheral T cells were isolated from C57BL/6 mice by fluorescence-activated cell sorting (99% pure), and total RNA from 106 cells was extracted with an RNeasy Mini kit (Qiagen). DN thymocytes were isolated from Rag-2−/− mice, and activated T cells were generated by culturing sorted peripheral T cells for 2 d with 5 μg/ml plate-bound anti-CD3. First-strand cDNA was reverse transcribed with the iScript Reverse Transcriptase kit (Bio-Rad Laboratories). The expression of autophagy genes was determined by RT-PCR using the following primers: Atg5, 5′-GACAAAGATGTGCTTCGAGATGTG-3′ and 5′-GTAGCTCAGATGCTCGCTCAG-3′; Beclin-1, 5′-CTGAAACTGGACACGAGCTTCAAG-3′ and 5′-CCAGAACAGTATAACGGCAACTCC-3′; LC3a,5′-AGCTTCGCCGACCGCTGTAAG-3′ and 5′-CTTCTCCTGTTCATAGATGTCAGC-3′; LC3b, 5′-CGGAGCTTTGAACAAAGAGTG-3′ and 5′-TCTCTCACTCTCGTACACTTC-3′; and Gapdh, 5′-CCTGGAGAAACCTGCCAAGTAT-3′ and 5′-AGAGTGGGAGTTGCTGTTGAAG-3′.
Transmission electron microscopy.
CD4+ and CD8+ T cells were purified from the spleen and lymph node of C57BL6 mice (≥96% pure) using EasySep negative bead selection (StemCell Technologies). Activated cells were stimulated for 2 d in vitro with 5 μg/ml soluble anti-CD3 before purification. Cell pellets were fixed in a 4% gluteraldehyde in 0.1 M sodium cacodylate buffer overnight. The samples were rinsed in 0.1 M cacodylate buffer containing 7.5% sucrose three times for 15 min each and fixed in 1% osmium in cacodylate buffer for 1 h. After being washed three times in 0.11 M veronal acetate buffer for 15 min each, the samples were incubated with 0.5% uranyl acetate in veronal acetate buffer for 1 h at RT. Specimens were then dehydrated in an ascending series of ethanol (35%, 70%, 95%, and 2 changes of 100%) for 10 min each, followed by two changes of propylene oxide for 5 min each. The samples were incubated with a 1:1 mixture of 100% resin and propylene oxide for 1 h, followed by two changes of 100% resin, each for 30 min. Finally, the samples were embedded in resin and polymerized at 60°C overnight. Thick sections (0.5 μm) were cut and stained with toluidine blue for light microscopy selection of the appropriate area for ultrathin sections. Thin sections (60–90 nm) were cut, mounted on copper grids, and then stained with uranyl acetate and lead citrate. Micrographs were taken with a Philips LS 410 electron microscope.
Western blot.
Total T cells were purified from the spleen and lymph node of C57BL6 mice (≥96% pure) using EasySep negative bead selection (StemCell Technologies). The cells were lysed in 50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, and 100 mM dithioreitol. LC3 was visualized using a rabbit polyclonal antibody (from Ron Kopito, Stanford University, Stanford, CA), HRP-conjugated donkey anti–rabbit (Jackson ImmunoResearch Laboratories), and Femto chemiluminescent substrate (Pierce Chemical Co.).
Generation of chimeric mice.
Fetuses were harvested from day 14 Atg5 heterozygous pregnant female mice. DNA was isolated from 106 fetal liver cells using a DNeasy Tissue kit (Qiagen) for genotyping. The primers for the wild-type and deletion allele of the Atg5 gene were 5′-GAATATGAAGGCACACCCCTGAAATG-3′, 5′-ACAACGTCGAGCACAGCTGCGCAAGG-3′, and 5′-GTACTGCATAATGGTTTAACTCTTGC-3′, and the PCRs were performed at 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min for 30 cycles. 1–3 × 106 liver cells from wild-type and Atg5−/− fetuses were injected into lethally irradiated (950 rad) CD45.1 congenic hosts. Chimeric mice were analyzed 6–10 wk after reconstitution.
Flow cytometry analysis.
Single cell suspensions of spleen and thymus were lysed of RBCs and incubated with FcR blocker (2.4G2; eBioscience). Cells were incubated with FITC, PE, PE/Cy5, APC, and biotinylated antibodies followed by streptavidin-Alexa 594 for 30 min on ice. Antibodies used included anti-CD4, -CD8, -TCR-β, -B220, -Gr1, -Mac1, -CD25, -CD69, and -CD45.2 (eBioscience, BioLegend, and BD Biosciences). Dead cells were excluded by 7-AAD or propidium iodide staining (BD Biosciences). Cell events were collected on a FACS STAR and analyzed using Cell Quest software (Becton Dickinson). Apoptotic cells were assayed by annexin V–PE kit and 7-AAD staining (BD Biosciences).
Cell proliferation.
Splenocytes were labeled with CFSE (Molecular Probes) and stimulated with 5 μg/ml plate-bound anti-CD3 (2C11). In some cultures, plate-bound anti-CD28 (clone 37.51; BioLegend) or human recombinant IL-2 was added. Proliferation was assayed by flow cytometry after 3 d.
Acknowledgments
We thank Dr. Montonari Kondo (Duke University, Durham, NC) for CD45.1 congenic mice and Dr. Ron Kopito (Stanford University, Stanford, CA) for the anti-LC3 antibody. We thank Mr. Phillip Christopher, Ms. Susan Hester, and Dr. Sara E Miller at the EM lab in the Department of Pathology at Duke University Medical Center for their assistance with the EM sample preparation and photography. We also thank Dr. Jeff Rathmell for useful discussion and Dr. Nu Zhang for critical review of this manuscript.
This work was supported by National Institutes of Health grant CA92123 (to Y.-W. He).
The authors have no conflicting financial interests.
References
- 1.Mizushima, N., Y. Ohsumi, and T. Yoshimori. 2002. Autophagosome formation in mammalian cells. Cell Struct. Funct. 27:421–429. [DOI] [PubMed] [Google Scholar]
- 2.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169–174. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima, N. 2005. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 12(Suppl 2):1535–1541. [DOI] [PubMed] [Google Scholar]
- 4.Lum, J.J., R.J. DeBerardinis, and C.B. Thompson. 2005. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 6:439–448. [DOI] [PubMed] [Google Scholar]
- 5.Kuma, A., M. Hatano, M. Matsui, A. Yamamoto, H. Nakaya, T. Yoshimori, Y. Ohsumi, T. Tokuhisa, and N. Mizushima. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu, M., S. Waguri, T. Chiba, S. Murata, J.I. Iwata, I. Tanida, T. Ueno, M. Koike, Y. Uchiyama, E. Kominami, and K. Tanaka. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884. [DOI] [PubMed] [Google Scholar]
- 7.Hara, T., K. Nakamura, M. Matsui, A. Yamamoto, Y. Nakahara, R. Suzuki-Migishima, M. Yokoyama, K. Mishima, I. Saito, H. Okano, and N. Mizushima. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. [DOI] [PubMed] [Google Scholar]
- 8.Lum, J.J., D.E. Bauer, M. Kong, M.H. Harris, C. Li, T. Lindsten, and C.B. Thompson. 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 120:237–248. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa, I., A. Amano, N. Mizushima, A. Yamamoto, H. Yamaguchi, T. Kamimoto, A. Nara, J. Funao, M. Nakata, K. Tsuda, et al. 2004. Autophagy defends cells against invading group A Streptococcus. Science. 306:1037–1040. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez, M.G., S.S. Master, S.B. Singh, G.A. Taylor, M.I. Colombo, and V. Deretic. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 119:753–766. [DOI] [PubMed] [Google Scholar]
- 11.Petiot, A., E. Ogier-Denis, E.F. Blommaart, A.J. Meijer, and P. Codogno. 2000. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275:992–998. [DOI] [PubMed] [Google Scholar]
- 12.Tassa, A., M.P. Roux, D. Attaix, and D.M. Bechet. 2003. Class III phosphoinositide 3-kinase–Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem. J. 376:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baehrecke, E.H. 2005. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 6:505–510. [DOI] [PubMed] [Google Scholar]
- 16.Yu, L., M.J. Lenardo, and E.H. Baehrecke. 2004. Autophagy and caspases: a new cell death program. Cell Cycle. 3:1124–1126. [PubMed] [Google Scholar]
- 17.Liang, X.H., L.K. Kleeman, H.H. Jiang, G. Gordon, J.E. Goldman, G. Berry, B. Herman, and B. Levine. 1998. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72:8586–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyo, J.O., M.H. Jang, Y.K. Kwon, H.J. Lee, J.I. Jun, H.N. Woo, D.H. Cho, B. Choi, H. Lee, J.H. Kim, et al. 2005. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 280:20722–20729. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu, S., T. Kanaseki, N. Mizushima, T. Mizuta, S. Arakawa-Kobayashi, C.B. Thompson, and Y. Tsujimoto. 2004. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 6:1221–1228. [DOI] [PubMed] [Google Scholar]
- 20.Yu, L., A. Alva, H. Su, P. Dutt, E. Freundt, S. Welsh, E.H. Baehrecke, and M.J. Lenardo. 2004. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 304:1500–1502. [DOI] [PubMed] [Google Scholar]
- 21.Gerland, L.M., L. Genestier, S. Peyrol, M.C. Michallet, S. Hayette, I. Urbanowicz, P. Ffrench, J.P. Magaud, and M. Ffrench. 2004. Autolysosomes accumulate during in vitro CD8+ T-lymphocyte aging and may participate in induced death sensitization of senescent cells. Exp. Gerontol. 39:789–800. [DOI] [PubMed] [Google Scholar]
- 22.Espert, L., M. Denizot, M. Grimaldi, V. Robert-Hebmann, B. Gay, M. Varbanov, P. Codogno, and M. Biard-Piechaczyk. 2006. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Invest. 116:2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, L., F. Wan, S. Dutta, S. Welsh, Z. Liu, E. Freundt, E.H. Baehrecke, and M. Lenardo. 2006. Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. USA. 103:4952–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boya, P., R.A. Gonzalez-Polo, N. Casares, J.L. Perfettini, P. Dessen, N. Larochette, D. Metivier, D. Meley, S. Souquere, T. Yoshimori, et al. 2005. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 25:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattingre, S., A. Tassa, X. Qu, R. Garuti, X.H. Liang, N. Mizushima, M. Packer, M.D. Schneider, and B. Levine. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 122:927–939. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, J., D. Cado, A. Chen, N.H. Kabra, and A. Winoto. 1998. Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature. 392:296–300. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, Y., S. Rosenberg, H. Wang, H.Z. Imtiyaz, Y.J. Hou, and J. Zhang. 2005. Conditional Fas-associated death domain protein (FADD): GFP knockout mice reveal FADD is dispensable in thymic development but essential in peripheral T cell homeostasis. J. Immunol. 175:3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]