Abstract
Somatic hypermutation (SHM) introduces nucleotide substitutions into immunoglobulin variable (Ig V) region genes at all four bases, but the mutations at C/G and A/T pairs are achieved by distinct mechanisms. Mutations at C/G pairs are a direct consequence of the C→U deamination catalyzed by activation-induced deaminase (AID). Mutations at A/T pairs, however, require a second mutagenic process that occurs during patch repair of the AID-generated U/G mismatch. Several DNA polymerases have been proposed to play a role in SHM, but accumulating evidence indicates that the mutations at A/T are overwhelmingly achieved by recruitment of DNA polymerase η.
The initial response to antigen is provided by IgM antibodies whose binding sites usually exhibit a relatively low affinity for antigen. Over the subsequent days and weeks, the antibody response matures to yield antibodies (typically of the IgG class) that display greatly increased affinity for antigen. This affinity maturation is achieved during B cell expansion in germinal centers by an iterative alternation of SHM and antigen-mediated selection. SHM itself is characterized by the sequential introduction of (typically) single nucleotide substitutions over a region of DNA encompassing the expressed Ig VH and VL segments. The mutations themselves can occur at either C/G or A/T pairs and can be either transitions (purine–purine or pyrimidine–pyrimidine substitutions) or transversions (a swapping of purine and pyrimidine). Although the entire process of SHM is dependent on AID (1, 2), the mutations at A/T pairs are produced by a substantially different mechanism from those at C/G pairs (3). Thus, whereas AID-catalyzed deamination of C bases can directly explain the mutations at C/G pairs, those at A/T pairs require a second mutagenic process. Accumulating genetic evidence (4–9), including new data from Delbos et al. (on p. 17 of this issue [10]), increasingly points to a pivotal role for DNA polymerase η in this A/T-specific process.
The mechanics of SHM
AID triggers somatic hypermutation by attacking a small number of C residues within the Ig V domains, deaminating them to U and thereby transforming a few C/G pairs into U/G mispairs (for review see reference 11). The presence of this uracil in DNA triggers an ancient pathway of DNA repair (12, 13), in which the uracil is excised from the DNA deoxyribophosphate backbone by the UNG uracil-DNA glycosylase. This yields an abasic site, which when encountered on the DNA template strand, is likely to stall the progression of the DNA replication fork. Such stalling recruits specialized polymerases (14) that are able to insert a dNTP opposite the abasic site, despite the fact that the abasic site is “noninstructional.” Several translesion polymerases appear able to assist in this synthesis, although the evidence supporting a role for the REV1 polymerase (15–17) is especially clear, as its specificity of nucleotide insertion (almost exclusively dCTP) means that its absence alters the mutation spectrum. Mutations at C/G pairs can thus be envisaged as an inevitable consequence of replication over sites of AID-catalyzed C→U deamination and subsequent UNG-mediated uracil excision (Fig. 1 A).
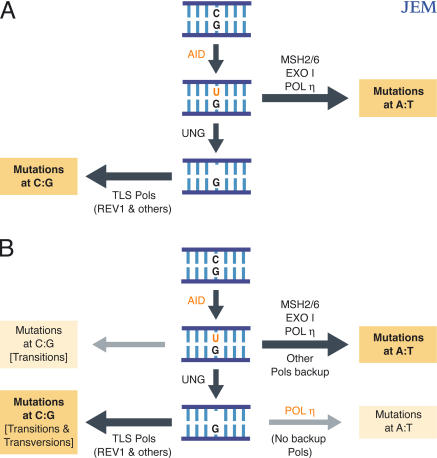
Figure 1.
(A) A model for the major pathways of somatic hypermutation in normal mice. After AID-catalyzed C→U deamination, the resulting U/G mispair is recognized by either UNG or MSH2–MSH6. (Mutations at C/G pairs) UNG-mediated recognition leads to the generation of an abasic site. Replication across this abasic site (by REV1 and other translesion polymerases) results in the generation of both transition and transversion substitutions at C/G pairs. (Mutations at A/T pairs) MSH2–MSH6-mediated recognition of the U/G mispair triggers an exonuclease I– and polymerase η–dependent patch repair process which results in mutations at A/T pairs. (B) Backup pathways that may operate in mutant mice. In the absence of UNG, replication across the U/G lesion leads solely to transition mutations at C/G pairs (the extent to which transitions at C/G are normally attributable to replication across the uracil as opposed to across the abasic site is unknown). In the absence of polymerase η, MSH2–MSH6 appears to recruit a backup polymerase that yields some residual mutations at A/T pairs. In the absence of MSH2, replication across the UNG-generated abasic site can also involve a mutagenic polymerase η–dependent patch repair process that generates mutations at A/T pairs. However, in the absence of polymerase η, no other polymerase is able to substitute for mutation creation at A/T pairs in this UNG-dependent pathway.
The mechanism by which mutations are generated at A/T pairs is less straightforward. These mutations depend on the original AID-catalyzed C-deamination but are clearly not a direct, inevitable consequence of that deamination. Genetic evidence suggests that a second mutagenic process takes place during patch repair of the original AID-generated U/G mispair (18). This mispair is a dual lesion in that it is both a mismatch and also contains a noncanonical DNA base. As a mismatch, it is recognized by the MSH2–MSH6 heterodimer, which conventionally functions to identify single-base mismatches for the purpose of initiating mismatch repair (Fig. 1 A). As a foreign base, it is recognized by the UNG uracil-DNA glycosylase. The patch repair process that generates the A/T mutations can be triggered by either MSH2–MSH6-mediated or UNG-mediated recognition of the initiating U/G lesion, although analysis of mutant mice suggests that MSH2–MSH6-mediated recognition is the major pathway (18–22).
Polymerase η: the prime suspect for mutations at A/T
The first breakthrough in identifying the major DNA polymerase involved in mutagenesis at A/T pairs came with the discovery by Zeng et al. that SHM at A/T (but not C/G) pairs is severely depressed in patients suffering from the variant form of Xeroderma pigmentosum (4). Xeroderma pigmentosum is a disease that renders patients highly susceptible to sun-induced skin cancers because of a deficiency in the cell's ability to repair ultraviolet-induced DNA damage. These patients carry inactivating mutations in the gene encoding DNA polymerase η, a translesion polymerase that is thought to play a role in allowing the replication fork to bypass cyclobutane pyrimidine dimers, which are major products of ultraviolet damage (23). Zeng et al. therefore proposed that mutations at A/T pairs were caused by errors of misincorporation by this low-fidelity polymerase during patch DNA synthesis (4). A similar depression of mutation accumulation at A/T pairs was later observed after disruption of the gene encoding DNA polymerase η in the mouse (6, 8).
Although these studies demonstrated a major role for DNA polymerase η in the generation of mutations at A/T pairs, it is clear that this polymerase is not the only DNA polymerase that can generate the A/T mutations. Although deficiency in polymerase η diminishes mutation accumulation at A/T pairs, it does not abolish it: the mutations at A/T are reduced from ∼50% to ∼20% of the total. The striking finding reported by Delbos et al. (10) is that mutations at A/T pairs are essentially totally abolished when deficiency in polymerase η is combined with deficiency in MSH2.
How should one interpret this result? Although genetics provides powerful insights into what happens in vivo, it is a dangerous practice to extrapolate from the phenotype of a mutant to deduce what goes on in the wild type. Simple epistasis analysis would suggest that, because mutations at A/T pairs are obliterated by simultaneous disruption of MSH2 and DNA polymerase η, but not by either disruption on its own, then the two proteins should lie on different pathways. The matter, however, is not quite so simple, as single disruptions in either MSH2 or polymerase η each yield a substantial, but not complete, depression in mutation accumulation at A/T pairs. The most likely explanation of the results is that the MSH2–MSH6-mediated recruitment of DNA polymerase η, which was previously thought to be a major pathway to mutations at A/T pairs, is in fact the overwhelming mechanism by which such mutations are generated. In the absence of DNA polymerase η, MSH2–MSH6 recruits a backup polymerase that provides a low background of mutations at A/T. In the absence of MSH2–MSH6, the need to replicate across the UNG-generated abasic site can still result in the recruitment of DNA polymerase η, generating a reduced load of A/T mutations (Fig. 1 B). However, unlike the MSH2-recruited mutagenic patch repair, mutagenesis at A/T pairs as a consequence of replicating across the abasic site is wholly dependent on polymerase η.
These observations result in a pleasing simplification of our view of SHM. Although multiple translesion polymerases have been implicated in antibody hypermutation (for review see references 24–26), DNA polymerase η appears to be the most dominant (if not the only) contributor to A/T mutations under normal conditions. That does not mean that there is no role in SHM for the other translesion polymerases. First, some other polymerase can, at least in the absence of polymerase η, provide some degree of backup mutations at A/T pairs in the MSH2-triggered pathway. Second, as discussed, the generation of transversion mutations at C/G pairs depends on replication across an abasic site and this process requires a translesion polymerase. Nevertheless, when considering mutation creation as opposed simply to the repair of AID-induced damage, it seems that the dominant polymerase is polymerase η.
Unanswered questions
The recruitment of DNA polymerase η for mutagenesis at A/T pairs raises many questions. For example, why (and how) does MSH2 recruit DNA polymerase η to the U/G lesion? The primary role of MSH2–MSH6 is to initiate postreplicative mismatch repair, correcting the occasional errors of misincorporation perpetrated by the replicative DNA polymerases (α, δ, and ɛ) (27). There is no evidence that polymerase η normally plays any role in mismatch repair, which is presumably usually a process of high fidelity. So, although the patch repair that generates mutations at A/T pairs is similar to conventional mismatch repair in that it is triggered by MSH2–MSH6 and likely involves strand degradation by exonuclease 1, it differs from conventional mismatch repair in that it is mutagenic and depends on polymerase η. Wilson et al. have provided evidence that MSH2 will associate with polymerase η and stimulate its activity (28). Is this association specific to B cells undergoing SHM? And what prevents polymerase η from being recruited during normal mismatch repair?
The mechanism by which polymerase η generates the mutations at A/T pairs also remains uncertain. The widely favored view is that these mutations are simply attributable to errors of base pairing by polymerase η during the patch DNA synthesis. This requires that the errors made by polymerase η occur largely opposite A or T on the template strand (and not opposite C or G)—a requirement that is well supported by the extensive in vitro analysis of the error spectrum of purified polymerase η from the Kunkel lab (9). This would predict that the nature of the nucleotide substitutions introduced during this second phase of SHM could be modified by altering the error spectrum of polymerase η.
Thus, antibody hypermutation involves two distinct mutagenic processes. Mutations at C/G pairs are a direct consequence of an active assault on the DNA molecule itself: these deviations from the parental DNA sequence can therefore be viewed as essentially a sin of commission. The mutations at A/T pairs, however, depend on a subsequent and distinct mutagenic process that involves polymerase η. If these mutations at A/T pairs are indeed simply errors resulting from a lack of fidelity in polymerase η, then these mutations can be considered a sin of omission. But the sin of omission would never have happened were it not for the original sin of commission (29).
M.S. Neuberger and C. Rada are at Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 2QH, UK
References
- 1.Muramatsu M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 2.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 3.Rada, C., M.R. Ehrenstein, M.S. Neuberger, and C. Milstein. 1998. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 9:135–141. [DOI] [PubMed] [Google Scholar]
- 4.Zeng, X., D.B. Winter, C. Kasmer, K.H. Kraemer, A.R. Lehmann, and P.J. Gearhart. 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2:537–541. [DOI] [PubMed] [Google Scholar]
- 5.Faili, A., S. Aoufouchi, S. Weller, F. Vuillier, A. Stary, A. Sarasin, C.A. Reynaud, and J.C. Weill. 2004. DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J. Exp. Med. 199:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delbos, F., A. De Smet, A. Faili, S. Aoufouchi, J.C. Weill, and C.A. Reynaud. 2005. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 201:1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayorov, V.I., I.B. Rogozin, L.R. Adkison, and P.J. Gearhart. 2005. DNA polymerase eta contributes to strand bias of mutations of A versus T in immunoglobulin genes. J. Immunol. 174:7781–7786. [DOI] [PubMed] [Google Scholar]
- 8.Martomo, S.A., W.W. Yang, R.P. Wersto, T. Ohkumo, Y. Kondo, M. Yokoi, C. Masutani, F. Hanaoka, and P.J. Gearhart. 2005. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl. Acad. Sci. USA. 102:8656–8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogozin, I.B., Y.I. Pavlov, K. Bebenek, T. Matsuda, and T.A. Kunkel. 2001. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2:530–536. [DOI] [PubMed] [Google Scholar]
- 10.F. Delbos, S. Aoufouchi, A. Faili, J.-C. Weill, and C.-A. Reynaud. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 203:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuberger, M.S., R.S. Harris, J. Di Noia, and S.K. Petersen-Mahrt. 2003. Immunity through DNA deamination. Trends Biochem. Sci. 6:305–312. [DOI] [PubMed] [Google Scholar]
- 12.Poole, A., D. Penny, and B.M. Sjoberg. 2001. Confounded cytosine! Tinkering and the evolution of DNA. Nat. Rev. Mol. Cell Biol. 2:147–151. [DOI] [PubMed] [Google Scholar]
- 13.Barnes, D.E., and T. Lindahl. 2004. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 38:445–476. [DOI] [PubMed] [Google Scholar]
- 14.Prakash, S., R.E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317–353. [DOI] [PubMed] [Google Scholar]
- 15.Simpson L.J., and J.E. Sale JE. 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 22:1654–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen, J.G., P. Langerak, A. Tsaalbi-Shtylik, P. van den Berk, H. Jacobs, and N. de Wind. 2006. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 203:319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross, A.L., and J.E. Sale. 2006. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol. Immunol. 43:1587–1594. [DOI] [PubMed] [Google Scholar]
- 18.Rada, C., J.M. Di Noia, and M.S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171. [DOI] [PubMed] [Google Scholar]
- 19.Martin, A., Z. Li, D.P. Lin, P.D. Bardwell, M.D. Iglesias-Ussel, W. Edelmann, and M.D. Scharff. 2003. Msh2 ATPase activity is essential for somatic hypermutation at a-T basepairs and for efficient class switch recombination. J. Exp. Med. 198:1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martomo, S.A., W.W. Yang, and P.J. Gearhart. 2004. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 200:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bardwell, P.D., C.J. Woo, K. Wei, Z. Li, A. Martin, S.Z. Sack, T. Parris, W. Edelmann, and M.D. Scharff. 2004. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 5:224–229. [DOI] [PubMed] [Google Scholar]
- 22.Shen, H.M., A. Tanaka, G. Bozek, D. Nicolae, and U. Storb. 2006. Somatic hypermutation and class switch recombination in Msh6(−/−)Ung(−/−) double-knockout mice. J. Immunol. 177:5386–5392. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, R.E., S. Prakash, and L. Prakash. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 283:1001–1004. [DOI] [PubMed] [Google Scholar]
- 24.Seki, M., P.J. Gearhart, and R.D. Wood. 2005. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 6:1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz, M., and C. Lawrence. 2005. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 26:215–220. [DOI] [PubMed] [Google Scholar]
- 26.Casali, P., Z. Pal, Z. Xu, and H. Zan. 2006. DNA repair in antibody somatic hypermutation. Trends Immunol. 27:313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiricny, J. 2006. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7:335–346. [DOI] [PubMed] [Google Scholar]
- 28.Wilson, T.M., A. Vaisman, S.A. Martomo, P. Sullivan, L. Lan, F. Hanaoka, A. Yasui, R. Woodgate, and P.J. Gearhart. 2005. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J. Exp. Med. 201:637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aquinas, T. 1947. Of the distinction of sins. In The Summa Theologica: Treatise on Habits (Question 72, Article 6). Benziger Bros. edition.