Abstract
The discovery of lymphocytes bearing two light chains in mice carrying self-reactive antibody transgenes has challenged the “one lymphocyte–one antibody” rule. However, the extent and nature of allelically included cells in normal mice is unknown. We show that 10% of mature B cells coexpress both Igκ alleles. These cells are not the result of failure in allelic exclusion per se, but arise through receptor editing. We find that under physiological conditions, editing occurs both by deletion and by inclusion with equal probability. In addition, we demonstrate that B lymphocytes carrying two B-cell receptors are recruited to germinal center reactions, and thus fully participate in humoral immune responses. Our data measure the scope of allelic inclusion and provide a mechanism whereby autoreactive B cells might “escape” central tolerance.
B-cell antigen receptor diversity is achieved by the assembly of antibody gene segments through V(D)J recombination (1). The random nature of the recombination process leads to the unavoidable production of self-reactive specificities, which are silenced both centrally and peripherally by clonal deletion, anergy, and receptor editing (2–6). Despite these mechanisms of tolerance, autoantibodies are frequently detected in normal mice (7, 8), and recent estimates suggest that up to 20% of long-lived B cells are self-reactive in humans (9). Although the exact mechanism by which these lymphocytes escape elimination is unknown, experiments with transgenic mice carrying prerecombined autoantibodies suggest that some self-reactive specificities may persist in the B-cell compartment by coexpression of a second “innocuous” light chain on the cell surface, a phenomenon referred to as allelic inclusion (10–12). Coexpression of a non–self-reactive light chain is thought to rescue the B cell from negative selection by diluting the self-reactive receptor (13). However, the presence of B cells bearing two receptors in transgenic mice poses a challenge to the mechanism of allelic exclusion (14–17), as well as to the “one lymphocyte–one antibody” theory (18). Thus, the extent and function of light chain allelic inclusion under physiological conditions is unknown.
Until recently, the study of light chain gene recombination at the κ loci was hampered by a lack of natural κ allelic polymorphisms in humans or mice. To overcome this difficulty, we used gene targeting to replace the mouse Igκ constant region (mCκ) with its human counterpart (hCκ; references 19, 20). Using mice heterozygous for the hCκ allele (Igκm/h), we showed that ∼3–5% of B lymphocytes express equal amounts of cell surface mCκ and hCκ light chains, as measured by flow cytometry (reference 19; Fig. 1 A, mCκ+hCκ+ population). This analysis, however, failed to consider allelic inclusion within mCκ−hCk+ or mCk+hCk− B-cell populations (Fig. 1 A), which might conceal Igκ double producers expressing predominantly one of two light chains on the cell surface. We determined the full extent of allelic inclusion in the B-cell compartment of Igκm/h mice by several independent assays. We show that ∼10% of B lymphocytes express two cell surface receptors. Moreover, we demonstrate that these double producers arise from light chain editing, and not as a result of defects in the mechanism of allelic exclusion.
Figure 1.
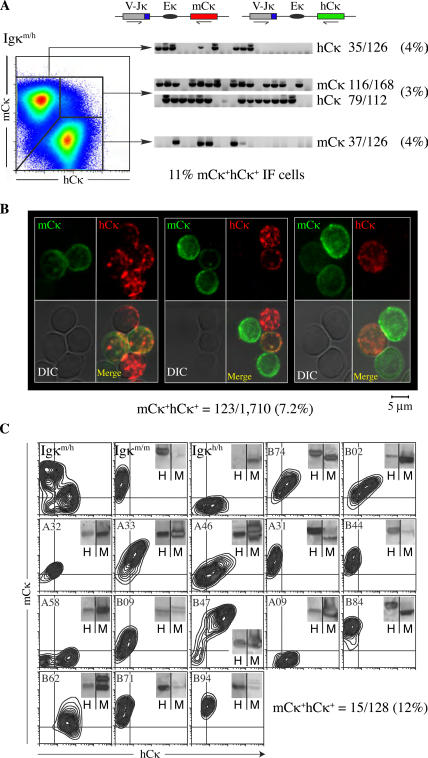
Allelic inclusion in Igκm/h mice. (A) Left pseudocolor plot shows analysis of mouse and human κ expression in Igκm/h splenocytes gated on B220+Igλ− and stained with rat monoclonal antibodies against mCκ and hCκ. Numbers indicate percentages of gated lymphocytes. VJκ-mCκ and -hCκ transcripts (top schematics) were amplified by RT-PCR from single cells sorted from mCκ+, hCκ+, and mCκ+hCκ+ fractions. Numbers in parentheses represent percentage of in-frame transcripts amplified from the various fractions of total B220+ B cells (a more detailed analysis is given in Table S1). TO-PRO3 was used to exclude dead cells from analysis. (B) Igκ protein expression in Igκm/h lymphocytes. Total splenic B cells were enriched by magnetic bead depletion of nonB cells and stained with anti-hCκ (red, Alexa Fluor 546) and anti-mCκ (green, Alexa Fluor 488). Cells were cytospun and analyzed by confocal microscopy. Values were summed from two independent experiments (1,192 and 518 cells scored). This analysis was also reproduced using a colorimetric assay on additional mice (Fig. S1). (C) mCκ and hCκ expression in 15 allelically included Igκm/h hybridomas (from a total of 128), as determined by flow cytometry (contour plots) and Western blot (insets). Control staining included splenocytes from Igκm/m and Igκh/h mice (first two plots). Fig. S1 and Table S1 are available at http://www.jem.org/cgi/content/full/jem.20061918/DC1).
RESULTS
B lymphocytes frequently express two cell surface receptors
To measure the scope of allelic inclusion, we first stained Igκm/h splenic B cells with anti-hCκ and -mCκ antibodies and characterized light chain transcripts in single cells by RT-PCR and sequencing (9). In agreement with our previous observations (19), VJκ-mCκ and -hCκ transcripts were readily detected in mCκ+hCκ+ lymphocytes. These cells make up 3–5% of the total B-cell pool, and most express in-frame transcripts from both alleles (Fig. 1 A and Table S1, available at http://www.jem.org/cgi/content/full/jem.20061918/DC1; 183/195 in-frame). VJκ-mCκ mRNAs were detected in ∼30% (37/126) of apparently single-positive mCκ−hCκ+ B cells, but only one third of these transcripts (10/37) were in-frame (Fig. 1 A). Likewise, hCκ+ transcripts were present at a similar frequency in mCκ+hCκ− B cells (Fig. 1 A, 35/126; 10 in-frame). Analysis of a control mixture of Igκm/m and Igκh/h lymphocytes demonstrated that <0.5% of the analyzed cells might be improperly scored as dual κ producers by this method. Taking into account the efficiency of the PCR reaction, we conclude that 11% of all B lymphocytes in Igκm/h mice express in-frame light chain transcripts from both κ alleles (4% of the hCκ+mCκ− cells + 4% of the hCκ−mCκ+ cells + 3% of the hCκ+mCκ+ cells; Table S1).
To measure the extent of allelic inclusion at the protein level, isolated Igκm/h splenic B cells were stained with anti-hCκ (Fig. 1 B, red, Alexa Fluor 546) and anti-mCκ (green, Alexa Fluor 488) antibodies and characterized by confocal microscopy. The specificity of this staining was assessed in B cells isolated from Igκm/m and Igκh/h mice, from which ∼1 in 250 cells were nonspecifically recognized by both antibodies. Of 1,710 individual B cells analyzed, we found that 123 (7.2%) coexpressed detectable levels of mouse and human Cκ light chains on the cell surface (Fig. 1 B). However, the intensity of staining varied between alleles, with some cells staining predominantly with anti-mCκ and others with anti-hCκ (Fig. 1 B, middle and right, and Fig. S1 available at http://www.jem.org/cgi/content/full/jem.20061918/DC1). Finally, we produced hybridomas from Igκm/h splenic lymphocytes and measured mCκ and hCκ light chain protein production. Of 128 hybridomas screened by ELISA and Western blotting, 12% were found to be allelically included, with 4% (5/128) secreting high levels of both mCκ and hCκ light chains (Fig. 1 C, B47), whereas the remaining 8% (10/128) predominantly expressed one of two alleles (Fig. 1 C, sample A32). The divergent pattern of cell surface mCκ/hCκ expression observed in the latter population was a direct correlation of Igκ transcription levels (Fig. S2), and may reflect inherent differences in Vκ promoter strength (20). Therefore, based on single-cell PCR, confocal microscopy, and hybridoma analyses, we conclude that ∼10% of peripheral B lymphocytes are allelically included, indicating that B cells frequently develop expressing two B-cell surface receptors.
Allelic inclusion results from light chain receptor editing
At least two mechanisms could account for dual κ light chain expression. Secondary VJκ rearrangements might be specifically induced to abolish self-reactivity or to rescue cells carrying less than optimal combinations of Ig heavy and light chains. Alternatively, inefficient allelic exclusion could lead to primary V–J recombination on both κ alleles. To discriminate between these two possibilities, we compared the developmental kinetics of bone marrow B cells carrying one or two κ light chains by BrdU labeling in vivo. The thymidine analogue BrdU is incorporated in the genome of cycling proB cells that give rise to the small resting preB cells in which light chains are actively rearranged (21). Edited B lymphocytes are developmentally delayed compared with unedited cells in this assay (19). In agreement with published observations (19, 22, 23), BrdU+B220low B cells expressing only mCκ or hCκ emerged first into the immature B-cell compartment, ∼4 h after BrdU injection (Fig. 2 A). In contrast, allelically included lymphocytes were delayed at the preB-cell stage for at least an additional 4 h, appearing in the IgM+ immature B-cell compartment 8 h after BrdU injection (Fig. 2 A). This developmental delay suggests that dual receptor expression is not the result of simultaneous biallelic Igκ recombination or failure in allelic exclusion, but instead, results from secondary light chain gene rearrangements in cells arrested at the early preB-cell stage of development.
Figure 2.
Allelically included B cells result from receptor editing. (A) Kinetics of bone marrow development of included and excluded B cells. Linear regression analysis shows the percentage of B220lowBrdU+ B cells plotted against time. Percentage values of excluded mCκ+ (blue ovals) and hCκ+ (green squares) cells are represented in the left y axis, and the percentage of mCκ+hCκ+-included cells (red circles) is depicted in the right y axis. Igκm/h mice were injected with 0.5 mg of BrdU intraperitoneally and killed after 6, 12, 18, 24, and 48 h (three mice per time point). Cells were permeabilized and stained with anti–BrdU-APC, mCκ-PE, hCκ-FITC, and B220-PerCP antibodies. (B) Comparative analysis of Jκ usage (percentage) in allelically included (red bars; n = 168 transcripts) and excluded (blue bars; n = 233 transcripts) B cells. (C) Antibodies purified from the supernatants of 15 mCκ+hCκ+ hybridoma clones were compared with 38 antibodies from hybridomas expressing only one allele for binding to HEp-2 cells and double-stranded DNA. HEp-2 binding was compared with positive and negative control sera provided by the manufacturer. To ensure HEp-2 binding was not caused by xenoreactivity, self-specificities were verified by a commercial ANA assay designed for mice (not depicted).
Increased downstream Jκ usage has been associated with receptor editing, as it suggests continuing V–Jκ rearrangements (24). Our analysis of 401 antibody sequences obtained from Igκm/h primary cells and hybridomas showed that Jκ1 usage is predominant in allelically excluded cells (35% vs. 20%; χ2 test, P = 0.002; Fig. 2 B). In contrast, Jκ5 was significantly enriched in cells carrying dual receptors (21% vs. 34%; χ2 test, P = 0.005; Fig. 2 B). This observation further reinforces the conclusion that allelic inclusion occurs in cells undergoing extensive light chain editing, presumably because of self-reactivity. To examine whether allelically included lymphocytes carry autoreactive receptors, we tested individual antibodies from Igκ-included (15 clones) and -excluded (38 clones) hybridomas for binding to double-stranded DNA or HEp-2 cells, which are commonly used to diagnose autoimmune diseases such as lupus (25). We found that >30% of mCκ+hCκ+ cells carried self-reactivity conferred predominantly by either the human or the mouse light chain (Fig. 2 C). Autoreactivity in these clones was confirmed using a test for mouse antinuclear antigen (ANA) reactivity (see Materials and methods). Self-reactivity in allelically excluded cells was nearly threefold less frequent (13%) than in hybridomas expressing dual receptors, although the low number of included hybridomas precluded a sufficient n-value to reach statistical significance (χ2 test, P = 0.09). Collectively, our data indicate that receptor editing leads to light chain allelic inclusion and retention of self-specificities in the B-cell compartment.
Allelically included B cells preferentially home to MZs and can contribute to immune responses
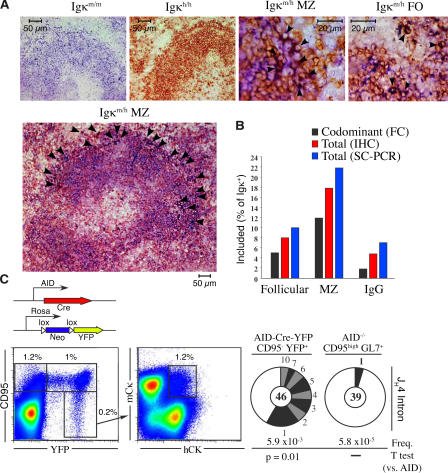
B cells carrying natural autoantibodies have been shown to accumulate preferentially in the marginal zone (MZ) of mice (26), but not of humans (27). To investigate the distribution of Igκ-included cells in the long-lived B-cell compartment, we assessed mCκ and hCκ expression in Igκm/h spleens by immunohistochemistry, flow cytometry, and single-cell PCR. We found allelically included cells both within the follicles and MZs (Fig. 3 A). However, whereas between 8–11% of follicular B cells (B220+CD23highCD21low) were mCκ+hCκ+, ∼20% of MZ lymphocytes (B220+CD23lowCD21high) expressed light chains from both alleles (Fig. 3 B; χ2 test, P < 0.0001). Preferential homing of Igκ double producers to MZs is consistent with the reported accumulation of edited Igκ+Igλ+ lymphocytes in MZs of anti-DNA transgenic mice (10). These results further suggest that included B cells often retain self-specificities in Igκm/h mice, as reported in transgenic mice carrying autoantibodies (10–13).
Figure 3.
Allelically included B cells home preferentially to MZs and participate in the germinal center reaction. (A) Control spleen sections from Igκm/m and Igκh/h mice (first and second images, respectively) were stained with antibodies against mCκ (blue) and hCκ (brown). Allelically included cells (dark brown) are identified with arrowheads. (B) Frequency of included lymphocytes in follicular, MZ, and IgG+ B cell populations, as determined by flow cytometry (FC), immunohistochemistry (IHC), and single-cell PCR (SC-PCR). (C) To permanently tag the progeny of germinal center cells, mice expressing the Cre recombinase gene under the AID promoter were crossed to Rosa-neo-YFP transgenic mice (schematic). In AID-Cre-YFP mice B220+CD95high, germinal center B cells are permanently labeled with YFP (1% of total B220+), and recirculating postgerminal center cells are identified as B220+CD95−YFP+ (0.2% of B220+, left pseudocolor plot). Allelic inclusion is assessed in postgerminal center Igκm/h cells (1.2%, right pseudocolor plot). Mutation analysis of JH4 intron from B220+CD95−YFP+ postgerminal center and B220+CD95highAID−/− germinal center cells is shown with pie charts. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated in the periphery of the charts. The frequency of mutations per base pair sequenced and the total number of independent sequences analyzed is indicated underneath and in the center of each chart, respectively. Statistical significance was determined by a two-tailed Student's t test assuming unequal variance and comparing to AID−/−.
To investigate whether Igκ double producers participate in the humoral immune response, we analyzed IgG+ B cells from Igκm/h mice. We readily found IgG+ lymphocytes that were mCκ+hCκ+ (2–5%; Fig. 3 B). To confirm the presence of allelically included cells in the postgerminal center B-cell compartment, we analyzed memory B cells in Igκm/hAID-Cre-yellow fluorescent protein (YFP) mice. In these mice, germinal center and memory B cells are indelibly labeled with YFP (Fig. 3 C, left pseudocolor plot; unpublished data). Flow cytometry analysis showed the presence of allelically included cells within postgerminal center CD95−YFP+ cells, which carry high levels of somatic hypermutation (Fig. 3 C). These results demonstrate that B cells expressing two cell surface receptors are not excluded from the germinal center reaction and fully participate in the humoral immune response.
Editing occurs by deletion and inclusion with similar probability
Using mice carrying prerecombined light chains, we established that 25% of all developing B cells undergo light chain editing (19). To measure the extent of allelic inclusion within the edited compartment, we analyzed mice carrying a VκαHEL-Jκ2–targeted transgene and the hCκ allele (IgκαHEL/h mice). In these mice, the prerearranged light chain is normally paired to the physiological heavy chain repertoire, and V–Jκ replacements at the hCκ allele are readily monitored by flow cytometry or PCR analysis (19). Single B cells were sorted from IgκαHEL/h mice and mCκ+ and hCκ+ transcripts were analyzed by single-cell PCR. We found that 61% of all developing B cells in these mice express the prerecombined αHEL light chain (Fig. 4 A). However, in ∼18% of all IgκαHEL/h B lymphocytes, the VκαHEL was edited by deletion, either through nested recombination events at the mCκ allele alone (αHEL−mCκ+, 6%; Fig. 4 A), or together with recombination at the hCκ allele (αHEL−hCκ+, 12%; Fig. 4 A). In close agreement with results obtained with Igκm/h mice (Figs. 1 and 2), editing by inclusion occurred in 14% of IgκαHEL/h B cells. The majority (13%) of these lymphocytes retained the VκαHEL light chain and carried in-frame V–J rearrangements at the hCκ allele (αHEL+hCκ+; Fig. 4 A), whereas a small number (1%) edited the VκαHEL by functional rearrangements in both alleles (αHEL−hCκ+mCκ+; Fig. 4 A). In addition, we noted that nearly all rearrangements at the hCk allele were in-frame (Fig. 4 B; 95%), which contrasts to the 2:1 OOF/in-frame ratio expected from random recombination. This observation indicates that cells carrying successful V–Jκ rearrangements on the human allele were selected during development. Because hCκ+ B cells are developmentally delayed by editing in these mice (19), these results argue that allelic inclusion in IgκαHEL/h mice originates from secondary gene rearrangements and not as a result of defective exclusion of light chain gene recombination. We conclude that receptor editing occurs either by deletion or by allelic inclusion with roughly equal probability (Fig. 4).
Figure 4.
Extent of allelic inclusion in the edited B-cell compartment. (A) IgκαHEL/h splenic B cells were isolated by single-cell sorting, and 387 Igκ transcripts were amplified by RT-PCR and analyzed by sequencing (top). From this analysis, six populations were characterized as shown (bottom). The VκαHEL prerecombined light chain is depicted by a yellow rectangle, and secondary recombination events at the mCκ and hCκ alleles are depicted with red or blue rectangles, respectively. OOF rearrangements are depicted by rectangles with a slash, and use of the lambda locus is represented with a green rectangle. (B) Analysis of in-frame and OOF status of Igκ transcripts in IgκαHEL/h or Igκm/h B cells. IgκαHEL/h cells were the same as those shown in A.
DISCUSSION
Studies with immunoglobulin knock-in mice suggested that autoreactive B cells are edited primarily by deletional recombination (28, 29). However, the preference for deletion in autoimmune mouse models might reflect the difficulty of vetoing somatically hypermutated receptors, which recognize autoantigens with high affinity and are not normally found in developing B cells (2, 4). Indeed, Ig light chain editing of naturally occurring antiself antibodies is less stringent (30). We show that under physiological conditions, editing occurs both by deletion and by allelic inclusion with nearly equal probability. Mechanistically, an equivalent deletion/inclusion ratio implies that although V–Jκ rearrangements initially target a single allele, ensuring allelic exclusion in most cells (Fig. 4; references 14–16, 31), autoreactivity and/or a 4-h developmental delay at the preB-cell stage promotes RAG accessibility to both alleles. This idea is consistent with recent findings showing that activation of both κ alleles is favored as development progresses from the preB to the immature B-cell stages (32).
Although natural autoantibodies were discovered >100 yr ago (33), their origin has remained obscure. In normal individuals, large numbers of self-specificities are believed to escape or bypass imperfect B-cell tolerance checkpoints (10), and this phenomenon appears to be exacerbated in patients with autoimmune diseases such as systemic lupus erythematosus (8, 9, 34). Our studies suggest that light chain receptor editing, which normally accounts for 25–50% of all antibodies (19, 35), also retains autoantibodies in the wild-type repertoire by allelic inclusion. As previously proposed (10, 11, 13), coexpression of an innocuous receptor might dampen signaling from self-reactive receptors, and thus promote development. Alternatively, light chain editors may tolerize autoreactive B cells by outcompeting self-reactive light chains for pairing with the IgH chain. This feature may, in fact, explain the higher frequency of allelic inclusion as measured by single-cell RT-PCR analysis (11%; Fig. 1 A), which detects Igκ transcription, versus confocal microscopy (7.2%; Fig. 1 B), which monitors light chain cell surface expression. Another possibility is that autoreactive receptors are continuously internalized by autoantigen binding, as shown in B lymphocytes expressing anti-MHC antibodies (11) or in T cells expressing two α chains (36), although whether double-producer T cells arise from a lack of α chain allelic exclusion or through receptor editing is still controversial (37).
The accumulation of allelically included B cells in MZs is intriguing. MZ B cells are commonly hyperreactive to T cell–independent type II antigens, such as phosphorylcholine, and represent the first line of defense against blood-borne pathogens (38). Conceivably, self-reactive allelically included cells are preferentially selected into the MZ because of their chronic activation by self-antigens. Diversion to this compartment might help prevent these cells from undergoing full cell differentiation (10). If recruited to germinal centers, allelically included cells could, in fact, function as Trojan horses; by introducing stop codons or otherwise disabling the innocuous editor light chain, hypermutation might unveil autoantibodies during immune responses.
MATERIALS AND METHODS
Flow cytometry.
Mouse B cells were purified from suspensions of spleen cells using a RosetteSep murine B cell isolation kit (Stemcell Technologies). Antibodies included rat monoclonal antibodies against mCκ (PE; BD Biosciences) and hCκ (FITC; previously available from SouthernBiotech). Cells were sorted using either a MoFlo (Dako Cytomation) or a FACS-Aria (Becton Dickinson) cytometer. Cytometric analyses were done using a LSR II analyzer (Becton Dickinson). Analyses and figures were done with FlowJo software.
Immunohistochemistry.
For analysis of the total frequency of allelic inclusion by immunofluorescence and confocal microscopy, total splenic B cells from Igκh/m mice were enriched by magnetic bead depletion of non–B cells (Stemcell Technologies) and stained with biotin-conjugated goat anti- hCk that had been absorbed with mCκ protein (SouthernBiotech), followed by streptavidin-conjugated Alexa Fluor 546 (Invitrogen) and rat anti-mCk (SouthernBiotech) directly conjugated in-house to Alexa Fluor 488 (Invitrogen). The cells were adhered to microscope slides and scored using a confocal microscope (LSM-510 META; Carl Zeiss MicroImaging, Inc.). Analyses for sorted fractions of hCκ+ cells for surface expression of mCκ+ were similar, except that the cells were stained with anti-mCκ conjugated to alkaline phosphatase (SouthernBiotech), adhered to slides, and developed with Alkaline Phosphatase Substrate kit III (Vector Laboratories). For analysis of splenic tissue, sections from snap-frozen Igκm/h, Igκm/m, and Igκh/h mouse spleens were stained with rat anti-mCκ (SouthernBiotech) and developed as per the sorted fractions (above). Sections were then stained with mouse anti–hCκ-HRP (SouthernBiotech) and developed with Vector NovaRED Substrate kit (Vector Laboratories). Analyses were done with a fluorescent microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.).
Single-cell RT-PCR.
Cells were presorted in bulk using a FACS-Aria cytometer, and single cells were resorted into 96-well plates containing 10 mM Tris-HCl with 40 U/μl of RNase inhibitor (Promega) with a MoFlo cytometer fitted with a cell dispenser. The plates were immediately frozen on dry ice and stored at −80°C until analysis. Each cell was amplified in a one-step RT-PCR reaction (OneStep RT-PCR kit; QIAGEN) using a cocktail of sense primers specific for the leader region (primers used were as follows: 5′-ATGGAATCACAGRCYCWGGT-3′; 5′-TTTTGCTTTTCTGGATTYCAG-3′; 5′-TCTTGTTGCTCTGGTTYCCAG-3′; 5′-ATTWTCAGCTTCCTGCTAATC-3′; 5′-TGCTGCTGCTCTGGGTTCCAG-3′; and 5′-TCGTGTTKCTSTGGTTGTCTG-3′), and antisense primers specific for either mCκ (5′-AGCTCTTGACGATGGGTGAAGTTG-3′) or hCκ (5′-GTTTCTCGTAGTCTGCTTTGCTCA-3′). RT-PCR conditions were as follows: 50°C for 30 min, 95°C for 15 min, followed by 39 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 1 min, and, finally, 72°C for 10 min. 1 μl from each RT-PCR reaction was reamplified using nested PCR primers specific for FW1 regions of the various Vκ genes (5′-GCGAAGCTTCCCTGATCGCTTCACAGGCAGTGG-3′, 5′-GCGAAGCTTCCCTGCTCGCTTCAGTGGCAGTGG-3′, 5′-GCGAAGCTTCCCAKCCAGGTTCAGTGGCAGTGG-3′, and 5′-GCGAAGCTTSCCATCRAGGTTCAGTGGCAGTGG-3′), and antisense primers specific to either mCκ (5′-ATCTGGAGGTGCCTCAGTC-3′) or hCκ (5′-GTGCTGTCCTTGCTGTCCTGCTC-3′). Nested PCR reaction conditions were as follows: 95°C for 15 min, followed by 39 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 1 min, and a final 72°C for 10 min. PCR products were resolved using 1% Agarose gels, and products were purified (Gel Extraction kit; QIAGEN) and sequenced on an ABI 3730 capillary sequencer by the Oklahoma Medical Research Foundation DNA sequencing core facility.
Analysis of Igκ proteins from hybridomas (ELISA, Western blot, and ANA).
Hybridomas were produced and subcloned by the Monoclonal Antibody Core Facility at the Memorial Sloan-Kettering Cancer Center. Igκ splenic B cells from 4-wk-old mice were stimulated for 3 d in fetal calf serum–supplemented RPMI with 25 μg/ml of lipopolysacharide (Sigma-Aldrich) and fused to the SP2 cell line. Two separate fusions with different mice produced similar results and were combined herein (58 and 70 clones for 128 total Igκm/h hybridomas). Each clone expressing protein or transcripts from both Ig alleles was subcloned in triplicate by limiting dilution to verify monoclonality. Clones from which no Igκ expression was detectable were discarded. Antibodies were expressed in serum-free RPMI media supplemented with Nutridoma (Roche) and purified from supernatant using Agarose-bead–conjugated L-protein (Pierce Chemical Co.). Capture ELISA assays were performed using purified anti-hCκ or -mCκ (Jackson ImmunoResearch Laboratories) and horse-radish peroxidase (HRP)–conjugated anti-hCκ or anti-mCκ (Jackson ImmunoResearch Laboratories). For the anti-DNA ELISAs, purified antibodies were captured on microtiter plates coated with 10 g/ml of salmon-sperm double-stranded DNA (Roche). The HRP was developed using a HRP detection kit (Bio-Rad Laboratories). All ELISA assays were analyzed using a SpectraMax plus microtiter plate reader (Invitrogen). Western blots were performed using HRP-conjugated antibodies (Jackson ImmunoResearch Laboratories). Purified antibodies were screened for ANA reactivity using commercially prepared HEp-2 slides (BION Enterprises, Ltd.) and the aforementioned FITC-conjugated anti-mCκ or -hCκ antibodies. The HEp-2–reactive clones were verified using a commercial ANA assay kit designed to test mouse autoantibodies (The Binding Site).
Ribonuclease-protection assays.
RNA was isolated from 1 million cells for each hybridoma clone using the RNAwiz reagent (Ambion) and treated with RNase-free DNase for 15 min at 37°C. The reaction was stopped with addition of one twentieth volume of 0.5 M EDTA, followed by heat inactivation of the DNase I for 10 min at 90°C, phenol/chloroform extraction, and ETOH precipitation. 5 μg of RNA were used in each of three replicate RPA reactions for each hybridoma analyzed. RPA probes were designed as described in Fig. S2 and cloned in the PcR2.1 vector containing both T3 and T7 RNA polymerase promoters. The [32P]UTP-labeled RNA probes were generated using the MAXIscript In Vitro Transcription kit (Ambion). To ensure full-length probes, each probe was purified from a 5% polyacrylamide/8 M urea gel. RNA probes were eluted overnight at room temperature from excised bands of the appropriate length with NH4-acetate/1 mM EDTA/0.2% SDS, followed by ETOH precipitation and resuspension in diethylpyrocarbonate-treated water at a concentration of 5 × 104 cpm in 10 μl. RPA reactions were performed using the RPA III kit (Ambion). All antisense probes were hybridized simultaneously to each sample. Protected fragments were resolved using a 5% denaturing polyacrylamide DNA sequencing gel and quantified using a phosphorimager (Storm 840; Molecular Dynamics). Protected bands were normalized relative to the Sp2 cell line fusion partner OOF mCκ transcript that is produced by each hybridoma to account for variations in RNA sample quality.
Online supplemental material.
Fig. S1 provides an analysis of the abundance of either allele expressed as measured by immunohistochemistry. Fig. S2 is an analysis of transcript expression levels from either Igκ allele. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20061918/DC1.
Supplemental Material
Acknowledgments
We thank Martin Weigert, Michael Lenardo, Paul Kincade, J. Donald Capra, Paul Plotz, Richard Siegel, John Knight, and Elizabeth Crouch for comments, and Francis Weiss-Garcia and Kenneth Wilson for technical assistance. Microscopy and DNA sequencing were performed at the Oklahoma Medical Research Foundation core facilities.
This research was supported in part by National Institutes of Health (NIH) grants P20RR018758-01 and P20RR15577-02 (to P.C. Wilson), and by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH (to R. Casellas).
The authors have no conflicting financial interests.
Abbreviations used: ANA, antinuclear antigen; hCκ, human κ constant region; HRP, horse-radish peroxidase; mCκ, mouse κ constant region; MZ, marginal zone; OOF, out-of-frame; YFP, yellow fluorescent protein.
R. Casellas and Q. Zhang contributed equally to this paper.
References
- 1.Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature. 302:575–581. [DOI] [PubMed] [Google Scholar]
- 2.Gay, D., T. Saunders, S. Camper, and M. Weigert. 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 177:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodnow, C.C., J. Crosbie, S. Adelstein, T.B. Lavoie, S.J. Smith-Gill, R.A. Brink, H. Pritchard-Briscoe, J.S. Wotherspoon, R.H. Loblay, K. Raphael, et al. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 334:676–682. [DOI] [PubMed] [Google Scholar]
- 4.Tiegs, S.L., D.M. Russell, and D. Nemazee. 1993. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177:1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamoto, M., M. Murakami, A. Shimizu, S. Ozaki, T. Tsubata, S. Kumagai, and T. Honjo. 1992. A transgenic model of autoimmune hemolytic anemia. J. Exp. Med. 175:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemazee, D.A., and K. Burki. 1989. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 337:562–566. [DOI] [PubMed] [Google Scholar]
- 7.Imai, H., S. Suzuki, K. Uchida, K. Kikuchi, H. Sugiyama, H. Kohno, M. Umeda, and K. Inoue. 1994. Natural autoantibody against apolipoprotein A-I. Detection and characterization of the monoclonal antibody established from normal unimmunized BALB/c mice. J. Immunol. 153:2290–2301. [PubMed] [Google Scholar]
- 8.Lacroix-Desmazes, S., S.V. Kaveri, L. Mouthon, A. Ayouba, E. Malanchere, A. Coutinho, and M.D. Kazatchkine. 1998. Self-reactive antibodies (natural autoantibodies) in healthy individuals. J. Immunol. Methods. 216:117–137. [DOI] [PubMed] [Google Scholar]
- 9.Wardemann, H., S. Yurasov, A. Schaefer, J.W. Young, E. Meffre, and M.C. Nussenzweig. 2003. Predominant autoantibody production by early human B cell precursors. Science. 301:1374–1377. [DOI] [PubMed] [Google Scholar]
- 10.Li, Y., H. Li, and M. Weigert. 2002. Autoreactive B cells in the marginal zone that express dual receptors. J. Exp. Med. 195:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, S., M.G. Velez, J. Humann, S. Rowland, F.J. Conrad, R. Halverson, R.M. Torres, and R. Pelanda. 2005. Receptor editing can lead to allelic inclusion and development of B cells that retain antibodies reacting with high avidity autoantigens. J. Immunol. 175:5067–5076. [DOI] [PubMed] [Google Scholar]
- 12.Huang, H., J.F. Kearney, M.J. Grusby, C. Benoist, and D. Mathis. 2006. Induction of tolerance in arthritogenic B cells with receptors of differing affinity for self-antigen. Proc. Natl. Acad. Sci. USA. 103:3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes, T., and M. Wabl. 2004. Autoreactivity and allelic inclusion in a B cell nuclear transfer mouse. Nat. Immunol. 5:1282–1287. [DOI] [PubMed] [Google Scholar]
- 14.Schlissel, M.S. 2004. Regulation of activation and recombination of the murine Igkappa locus. Immunol. Rev. 200:215–223. [DOI] [PubMed] [Google Scholar]
- 15.Bergman, Y., and H. Cedar. 2004. A stepwise epigenetic process controls immunoglobulin allelic exclusion. Nat. Rev. Immunol. 4:753–761. [DOI] [PubMed] [Google Scholar]
- 16.Mostoslavsky, R., F.W. Alt, and K. Rajewsky. 2004. The lingering enigma of the allelic exclusion mechanism. Cell. 118:539–544. [DOI] [PubMed] [Google Scholar]
- 17.Pernis, B., G. Torrigiani, L. Amante, A.S. Kelus, and J.J. Cebra. 1968. Identical allotypic markers of heavy polypeptide chains present in different immunoglobulin classes. Immunology. 14:445–451. [PMC free article] [PubMed] [Google Scholar]
- 18.Nossal, G.J.V., and J. Lederberg. 1958. Antibody production by single cells. Nature. 181:1419–1420. [PubMed] [Google Scholar]
- 19.Casellas, R., T.A. Shih, M. Kleinewietfeld, J. Rakonjac, D. Nemazee, K. Rajewsky, and M.C. Nussenzweig. 2001. Contribution of receptor editing to the antibody repertoire. Science. 291:1541–1544. [DOI] [PubMed] [Google Scholar]
- 20.Casellas, R., M. Jankovic, G. Meyer, A. Gazumyan, Y. Luo, R. Roeder, and M. Nussenzweig. 2002. OcaB is required for normal transcription and V(D)J recombination of a subset of immunoglobulin kappa genes. Cell. 110:575–585. [DOI] [PubMed] [Google Scholar]
- 21.Opstelten, D., and D.G. Osmond. 1983. Pre-B cells in mouse bone marrow: immunofluorescence stathmokinetic studies of the proliferation of cytoplasmic mu-chain-bearing cells in normal mice. J. Immunol. 131:2635–2640. [PubMed] [Google Scholar]
- 22.Allman, D.M., S.E. Ferguson, V.M. Lentz, and M.P. Cancro. 1993. Peripheral B cell maturation. II. Heat-stable antigen(hi) splenic B cells are an immature developmental intermediate in the production of long-lived marrow-derived B cells. J. Immunol. 151:4431–4444. [PubMed] [Google Scholar]
- 23.Rocha, B., C. Penit, C. Baron, F. Vasseur, N. Dautigny, and A.A. Freitas. 1990. Accumulation of bromodeoxyuridine-labeled cells in central and peripheral lymphoid organs: minimal estimates of production and turnover rates of mature lymphocytes. Eur. J. Immunol. 20:1697–1708. [DOI] [PubMed] [Google Scholar]
- 24.Nemazee, D., and M. Weigert. 2000. Revising B cell receptors. J. Exp. Med. 191:1813–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradwell, A.R., R.P. Stokes, and G.D. Johnson. 1995. Atlas of HEp-2 Patterns. Univerity of Birmingham, Birmingham, England. 129 pp.
- 26.Kearney, J.F. 2005. Innate-like B cells. Springer Semin. Immunopathol. 26:377–383. [DOI] [PubMed] [Google Scholar]
- 27.Tsuiji, M., S. Yurasov, K. Velinzon, S. Thomas, M.C. Nussenzweig, and H. Wardemann. 2006. A checkpoint for autoreactivity in human IgM+ memory B cell development. J. Exp. Med. 203:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, C., E.L. Prak, and M. Weigert. 1997. Editing disease-associated autoantibodies. Immunity. 6:97–105. [DOI] [PubMed] [Google Scholar]
- 29.Pelanda, R., S. Schwers, E. Sonoda, R.M. Torres, D. Nemazee, and K. Rajewsky. 1997. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 7:765–775. [DOI] [PubMed] [Google Scholar]
- 30.Wardemann, H., J. Hammersen, and M.C. Nussenzweig. 2004. Human autoantibody silencing by immunoglobulin light chains. J. Exp. Med. 200:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamagami, T., E. ten Boekel, J. Andersson, A. Rolink, and F. Melchers. 1999. Frequencies of multiple IgL chain gene rearrangements in single normal or kappaL chain-deficient B lineage cells. Immunity. 11:317–327. [DOI] [PubMed] [Google Scholar]
- 32.Liang, H.E., L.Y. Hsu, D. Cado, and M.S. Schlissel. 2004. Variegated transcriptional activation of the immunoglobulin kappa locus in pre-b cells contributes to the allelic exclusion of light-chain expression. Cell. 118:19–29. [DOI] [PubMed] [Google Scholar]
- 33.Besredka, M. 1901. Les antihemolysines naturelles. Ann. Inst. Pasteur (Paris). 15:785–807. [Google Scholar]
- 34.Yurasov, S., H. Wardemann, J. Hammersen, M. Tsuiji, E. Meffre, V. Pascual, and M.C. Nussenzweig. 2005. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 201:703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Retter, M.W., and D. Nemazee. 1998. Receptor editing occurs frequently during normal B cell development. J. Exp. Med. 188:1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacorazza, H.D., and J. Nikolich-Zugich. 2004. Exclusion and inclusion of TCR alpha proteins during T cell development in TCR-transgenic and normal mice. J. Immunol. 173:5591–5600. [DOI] [PubMed] [Google Scholar]
- 37.Sarukhan, A., C. Garcia, A. Lanoue, and H. von Boehmer. 1998. Allelic inclusion of T cell receptor alpha genes poses an autoimmune hazard due to low-level expression of autospecific receptors. Immunity. 8:563–570. [DOI] [PubMed] [Google Scholar]
- 38.Fagarasan, S., and T. Honjo. 2000. T-independent immune response: new aspects of B cell biology. Science. 290:89–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.