Abstract
To combat the human immune response, bacteria should be able to divert the effectiveness of the complement system. We identify four potent complement inhibitors in Staphylococcus aureus that are part of a new immune evasion cluster. Two are homologues of the C3 convertase modulator staphylococcal complement inhibitor (SCIN) and function in a similar way as SCIN. Extracellular fibrinogen-binding protein (Efb) and its homologue extracellular complement-binding protein (Ecb) are identified as potent complement evasion molecules, and their inhibitory mechanism was pinpointed to blocking C3b-containing convertases: the alternative pathway C3 convertase C3bBb and the C5 convertases C4b2aC3b and C3b2Bb. The potency of Efb and Ecb to block C5 convertase activity was demonstrated by their ability to block C5a generation and C5a-mediated neutrophil activation in vitro. Further, Ecb blocks C5a-dependent neutrophil recruitment into the peritoneal cavity in a mouse model of immune complex peritonitis. The strong antiinflammatory properties of these novel S. aureus–derived convertase inhibitors make these compounds interesting drug candidates for complement-mediated diseases.
The complement system is essential for an effective immune response against invading pathogens. Conserved microbial sugars or microbe-bound antibodies initiate a cascade of protein–protein interactions and proteolytic steps, which result in the formation of target-bound convertases that cleave C3 and C5 into biologically important complement fragments (1, 2). The small fragments C3a and C5a serve as potent proinflammatory mediators that attract phagocytes to the site of infection and activate them. Deposition of large C3 cleavage products (C3b and iC3b) on microbial surfaces greatly enhances bacterial uptake by phagocytic cells (3, 4). Furthermore, complement can directly kill gram-negative bacteria via formation of a membrane attack complex (MAC), composed of C5b, C6, C7, C8, and multiple C9, that inserts into target membranes and promotes osmotic lysis (5). Next to its beneficial role in host defense, complement also promotes many unwanted inflammatory reactions in pathological conditions such as autoimmune diseases (6), allergies (7), allograft/xenograft rejection (8), and systemic inflammation during sepsis (9).
Complement activation occurs via three different recognition pathways that converge at the level of C3 by forming C3-cleaving proteases, the C3 convertases (10). Activation of the classical pathway (CP) and lectin pathway (LP) results in the cleavage of C4 and C2 by C1s or mannose-binding lectin–associated serine protease 2, generating a C4b2a complex, the C3 convertase of the CP/LP (11, 12). The alternative pathway (AP) is initiated by formation of either spontaneously hydrolyzed C3 (C3H2O) in fluid-phase or surface-bound C3b deposited by the CP/LP. Generated C3b can form a complex with factor B (fB) that is activated by factor D (fD) to generate the C3bBb complex, the C3 convertase of the AP (13).
The C3 convertases (C4b2a and C3bBb) are of major biological importance, as they mediate the release of C3a and the deposition of large numbers of C3b molecules onto bacterial surfaces. C3 convertases consist of a covalently surface-bound noncatalytic subunit (C4b or C3b), which is in complex with the catalytic subunit (C2a or Bb). Because of the rapid and irreversible dissociation of the catalytic subunits, convertases only have a short lifespan of 1–2 min. C3 convertases also function as precursors of C5 convertases (11, 14, 15). In response to the deposition of high C3b concentrations on target surfaces, C3 convertases switch to C5 convertases to cleave C5, initiating the release of C5a and the formation of the cytolytic MAC (16–18). During this process, assembly of one C3b molecule on the C3 convertase results in the formation of C4b-C3b or C3b-C3b dimers that have high affinities for C5.
In recent years, it has become evident that bacteria have evolved sophisticated strategies to escape complement-mediated immune responses. Staphylococcus aureus secretes several complement modulators to target different steps in the complement cascade (19, 20). Phagocyte activation by C5a is effectively blocked by the chemotaxis inhibitory protein of S. aureus (CHIPS) that binds the C5a receptor (C5aR), as well as the formylated peptide receptor (FPR) (21). The recently described staphylococcal complement inhibitor (SCIN) interferes with all complement activation pathways by blocking C3 convertases (22–24). The CHIPS and SCIN genes are part of the first immune evasion cluster (IEC-1) in S. aureus, a bacteriophage-localized element that also encodes staphylokinase and staphylococcal enterotoxin A (25). Although CHIPS and SCIN are promising molecules to target complement diseases, their strict specificity for human complement proteins has limited activity studies in vivo. Nevertheless, their importance as staphylococcal immune modulators prompted us to seek homologous proteins in S. aureus. This led to the identification of a CHIPS homologue (28% amino acid homology) that blocks FPR-like 1 receptor signaling: FPR-like 1 inhibitory protein (FLIPr) (26). A 73% homologue of FLIPr (FLIPr-like) is also involved in modulation of FPRs (unpublished data).
In this study, we identify two SCIN homologues that have complement inhibitory properties similar to SCIN. Intriguingly, the genomic location of both CHIPS and SCIN homologues reveals a novel second IEC in S. aureus, IEC-2. Extracellular fibrinogen-binding protein (Efb) and its homologue, extracellular complement-binding protein (Ecb), both located on IEC-2, are now identified as potent complement inhibitors acting via a novel convertase-inhibitory mechanism. Because complement inhibition by Efb and Ecb is not human specific, we could for the first time demonstrate the in vivo potency of an S. aureus–derived complement inhibitor.
RESULTS
CHIPS and SCIN homologues are part of a new IEC
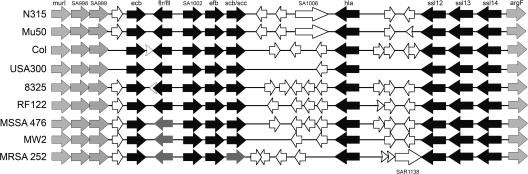
Database analyses revealed that S. aureus contains three open reading frames (ORFs) with a high homology to SCIN, whereas no noteworthy homologues were found in other microorganisms. We identified SCIN-B, SCIN-C, and ORF-D as sharing 48, 46, and 33% homology with SCIN, respectively (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20070818/DC1). Surprisingly, we found that CHIPS and SCIN homologues are clustered on the genome of all sequenced S. aureus strains (Fig. 1). The cluster contains the gene for FLIPr (flr) or FLIPr-like (fll) in combination with the gene for SCIN-B (scb) or SCIN-C (scc). The ORF-D gene is outside this cluster, directly upstream of the staphylocoagulase precursor gene. Next to CHIPS and SCIN homologues, we found this cluster to harbor the genes for Efb (efb) and a 33% homologue of Efb, which we termed Ecb (ecb). Efb has dual functions because its N terminus binds fibrinogen, whereas its C-terminal part (C3-binding domain of Efb [Efb-C]) binds different C3 molecules (27–30). The homology between Efb and Ecb is mainly located in Efb-C (Fig. S1 B). Both the C3-binding properties of Efb-C and the high homology with Ecb suggest that these molecules may play an important role in staphylococcal complement evasion. Furthermore, we found this cluster to encode a putative outer membrane protein (SA1002), α-hemolysin, and three exotoxin-like molecules. Because the exotoxins are structural homologues of staphylococcal superantigens, they are referred to as staphylococcal superantigen–like 12 (SSL12), SSL13, and SSL14 (Fraser, J.D., personal communication) (31). The borders of IEC-2 are represented by the housekeeping genes murI (glutamate racemase) and argF (ornithine carbamoyltransferase), as well as SA0998 and SA0999 (homologues of the housekeeping genes). Of note, the presence of transposases and bacteriophage remnants suggests that this cluster has evolved through horizontal gene transfer (32). In summary, the clustering of several known and potential immune evasion molecules indicates this region represents a novel second IEC in S. aureus (IEC-2). In this paper, we will investigate the immune evasive properties of four molecules on IEC-2: SCIN-B, SCIN-C, Efb, and Ecb.
Figure 1.
A new IEC in S. aureus. Graphic representation of the novel IEC-2 in sequenced S. aureus strains. Black arrows indicate known or putative immune evasion molecules: Ecb (ecb), FLIPr (flr), FLIPr-like (fll), Efb (efb), SCIN-B (scb), SCIN-C (scc), α-hemolysin (hla), SSL12 (ssl12), SSL13 (ssl13), and SSL14 (ssl14). Genes with unknown functions are named according to their locus number in S. aureus N315. All strains carry either flr (black) or fll (dark gray) and scb (black) or scc (dark gray). The household genes (light gray) murI (glutamate racemase), SA998, SA999, and argF (ornithine carbamoyltransferase) form the borders of IEC-2. White arrows delineate ORFs similar to bacteriophage proteins. Three transposases for insertion sequences were found: SA1006 in N315 and Mu50, and SAR1138 in MRSA252. S. aureus strain RF122 represents a bovine isolate.
Prevalence of SCIN and Efb homologues in S. aureus strains
PCR analyses of 84 clinical S. aureus strains and 6 classical lab strains revealed that the genes encoding SCIN-B (scb), SCIN-C (scc), and ORF-D (orf-d) are carried by 47, 32, and 98% of S. aureus strains, respectively. As observed for sequenced strains, clinical S. aureus strains also carry either scb or scc. The genes encoding Efb (efb) and Ecb (ecb) were found in 85 and 98% of S. aureus strains, respectively. All are secreted proteins because they contain a signal peptide and a signal peptide cleavage site.
Innate immune evasion by the SCIN homologues, Efb-C and Ecb
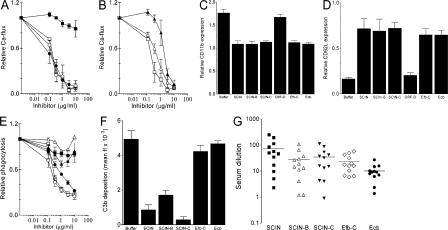
The recombinant SCIN homologues, Efb-C and Ecb were tested in several in vitro assays that mimic critical steps of complement-mediated immune responses against S. aureus. To analyze complement activation at the level of C5a, S. aureus was initially incubated with human serum in the presence or absence of inhibitors. Collected supernatants were subsequently tested for their capacity to activate neutrophils, a response that is completely C5a dependent (23). We observed that SCIN-B and SCIN-C effectively blocked C5a-mediated calcium mobilization, whereas ORF-D had no effect (Fig. 2 A). C5a responses were also inhibited by Efb-C and Ecb (Fig. 2 B). C5a responses were not affected when proteins were added to supernatants after opsonization. As a different readout for neutrophil activation, we analyzed expression of CD11b and CD62L. In concordance with calcium mobilization assays, supernatants generated in the presence of SCIN-B, SCIN-C, Efb-C, or Ecb contained less C5a because they could not up- or down-regulate CD11b and CD62L, respectively (Fig. 2, C and D) (33). Next to C5a formation, we studied the effect of SCIN-B, SCIN-C, ORF-D, Efb-C, and Ecb on phagocytosis. Neutrophil uptake of FITC-labeled S. aureus in the presence of human serum was strongly inhibited by SCIN-B and SCIN-C (Fig. 2 E). In contrast, Efb-C, Ecb, and ORF-D did not affect phagocytosis (Fig. 2 E), even at different serum concentrations (not depicted). Because effective phagocytosis of bacteria depends on the presence of opsonic C3 fragments (34, 35), it was not surprising to find that Efb-C and Ecb also did not inhibit deposition of C3b/iC3b on S. aureus (Fig. 2 F). Only SCIN, SCIN-B, and SCIN-C strongly prevented deposition of C3b/iC3b in normal human serum. To study whether SCIN-B, SCIN-C, Efb, and Ecb are produced in vivo, we analyzed the presence of antibodies against SCIN-B, SCIN-C, Efb-C, and Ecb in humans. We found that antibodies capable of recognizing each of the recombinant molecules are present in humans (Fig. 2G).
Figure 2.
Innate immune evasion by four putative complement inhibitors on IEC-2. (A) SCIN-B and SCIN-C inhibit C5a production. S. aureus was incubated with 10% human serum in the presence of 10 μg/ml SCIN (□), SCIN-B (•), SCIN-C (○), or ORF-D (▪). C5a formation was measured by using supernatants as stimuli for calcium mobilization of human neutrophils. SCIN-B and SCIN-C show a dose-dependent inhibition of C5a formation, whereas ORF-D had no effect. (B) Efb-C and Ecb block C5a production during opsonization. Dose-dependent inhibition of C5a-mediated neutrophil activation by Efb-C (▴), Ecb (Δ), and SCIN (□). (C) Supernatants that were activated in the presence of 10 μg/ml SCIN, SCIN-B, SCIN-C, Efb-C, or Ecb were less potent in up-regulating CD11b expression on neutrophils. (D) Supernatants that were activated in the presence of 10 μg/ml SCIN, SCIN-B, SCIN-C, Efb-C, or Ecb were less potent in down-regulation of CD62L expression on neutrophils. (E) SCIN (□), SCIN-B (•), and SCIN-C (○) inhibit phagocytosis of S. aureus by human neutrophils in 10% human serum. ORF-D (▪), Efb-C (▴), and Ecb (Δ) did not affect phagocytosis. (F) SCIN, SCIN-B, and SCIN-C inhibit C3b/iC3b deposition on the bacterial surface in 10% human serum, whereas Efb-C and Ecb do not influence opsonization. (G) Antibody titers against SCIN, SCIN-B, SCIN-C, Efb, and Ecb in sera of 12 healthy lab volunteers. Horizontal lines represent the mean. All data represent the mean ± SE of three separate experiments. Mean fl, mean fluorescence.
In conclusion, SCIN-B and SCIN-C are efficient complement inhibitors that prevent C5a generation, C3b/iC3b deposition, and phagocytosis. ORF-D is not a complement inhibitor, and its function remains unknown. In contrast to SCIN molecules, Efb-C and Ecb do not inhibit phagocytosis but strongly interfere with C5a production.
SCIN-B and SCIN-C function similar to SCIN
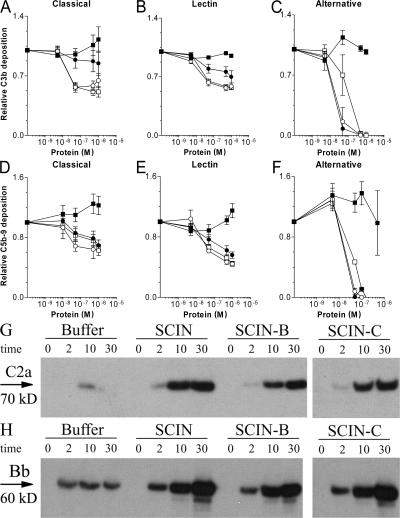
To gain more insight into the complement inhibitory mechanisms of SCIN-B and SCIN-C, we determined their regulatory impact on the different pathways individually (36, 37). ELISA experiments showed that SCIN-B and SCIN-C share pathway specificity with SCIN (Fig. 3). Both at the level of C3b and C5b-9 formation, SCIN-B and SCIN-C inhibit the CP and LP by 50% (Fig. 3, A, B, D, and E), and the AP by 100% (Fig. 3, C and F). Of note, SCIN-B appears less potent in blocking CP and LP activation at the level of C3b (Fig. 3, A and B). Like SCIN, SCIN-B and SCIN-C did not block C4b deposition during CP or LP activation (24). Because stabilization of inherently labile C3 convertases is a hallmark of SCIN activity, we examined the convertase-stabilizing capacities of SCIN-B and SCIN-C. Opsonization of S. aureus in the presence of SCIN-B or SCIN-C resulted in increased amounts of bacterium-bound C2a and Bb, indicating that these molecules stabilize C3 convertases (Fig. 3, G and H). Collectively, these data show that SCIN-B and SCIN-C inhibit complement by stabilization of C3 convertases, which is similar to the inhibitory mechanism of SCIN.
Figure 3.
SCIN-B and SCIN-C function similar to SCIN. ELISA experiments showing that SCIN (□), SCIN-B (•), and SCIN-C (○) inhibit C3b deposition after CP (5% serum; A), LP (5% serum; B), and AP activation (30% serum; C). SCIN, SCIN-B, and SCIN-C also prevent C5b-9 deposition during CP (5% serum; D), LP (5% serum; E), and AP activation (30% serum; F). ORF-D (▪) cannot inhibit C3b or C5b-9 deposition in ELISA. Data shown in A–F represent the mean ± SE of three separate experiments. (G and H) SCIN, SCIN-B, and SCIN-C enhance convertase stability during opsonization of S. aureus in 20% human serum. Opsonization was performed for 0, 2, 10, and 30 min. Surface-bound convertases were detected by immunoblotting using anti-C2 (G) or anti-fB (H) antibodies. Blots represent three separate experiments.
Complement inhibitory properties of Efb-C and Ecb
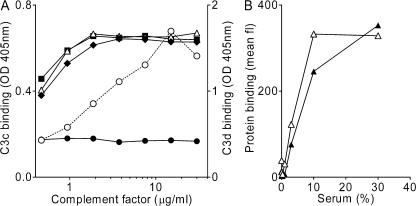
The pathway specificity of Efb-C and Ecb was also analyzed by ELISA. At the level of C3b deposition, we found that Efb-C and Ecb block the AP but not the CP and LP (Fig. 4, A–C). However, C5b-9 formation was effectively blocked by Efb-C and Ecb in all pathways (Fig. 4, D–F). In contrast to SCIN, Efb-C and Ecb did not enhance convertase stability during opsonization of S. aureus (Fig. 4, G and H). Because Efb-C binds different C3 molecules via the C3d domain (29), we also studied the C3-binding properties of Ecb. Therefore, Ecb was coated to microtiter wells and incubated with C3 or its fragments. Similar to Efb-C, we found that Ecb specifically binds all C3d-containing C3 molecules (C3, C3b, iC3b, and C3d), whereas no binding was observed to C3c, which is devoid of C3d (Fig. 5 A). Binding specificity was demonstrated by a lack of binding to C4, C5, fB, and fibrinogen.
Figure 4.
Efb-C and Ecb act differently than SCIN. ELISA experiments showing that Efb-C (▴) and Ecb (Δ) do not inhibit C3b deposition after CP (5% serum; A) or LP (5% serum; B) activation but strongly block C3b deposition after AP activation (30% serum; C). Efb-C and Ecb strongly prevent C5b-9 deposition in response to activation of all pathways: CP (5% serum; D), LP (5% serum; E), and AP (30% serum; F). Data shown in A–F are the mean ± SE of three separate experiments. (G and H) Efb-C and Ecb do not enhance convertase stability during opsonization of S. aureus in 20% human serum. Opsonization was performed for 0, 2, 10, and 30 min. Surface-bound convertases were detected by immunoblotting using anti-C2 (G) or anti-fB (H) antibodies. Blots represent three separate experiments.
Figure 5.
Complement-binding properties of Efb-C and Ecb. (A) Ecb binds the C3d domain of C3 (fragments). 5 μg/ml Ecb was coated to ELISA plates, and subsequent binding of C3 (▪), iC3b (Δ), C3b (♦), C3c (•), and C3d (○) was determined. Ecb exclusively binds C3d-containing C3 molecules. (B) Efb-C (▴) and Ecb (Δ) bind to S. aureus in a serum-dependent manner. 10 μg/ml of His-tagged Efb-C and Ecb were incubated with S. aureus and human serum for 30 min at 37°C, and binding was detected using anti-His antibodies and flow cytometry. His-tagged SSL7 served as a negative control. Graphs show one representative figure out of three separate experiments. Mean fl, mean fluorescence.
Our findings provide clear evidence that Efb-C and Ecb are potent complement inhibitors that act differently than SCIN. By binding to C3d-containing molecules, Efb-C and Ecb can block C3b deposition in the AP but not in the CP/LP. However, Efb-C and Ecb are potent inhibitors of C5b-9 formation in all complement pathways.
Efb-C and Ecb act on the bacterial surface
To assess whether Efb-C and Ecb act on the bacterial surface, we studied the binding of histidine (His)-tagged Efb-C and Ecb to S. aureus. The His tag did not influence complement inhibitory activities, as was shown by ELISA (unpublished data). We clearly observed a serum-dependent binding of His–Efb-C and His-Ecb to S. aureus (Fig. 5 B). The C5-binding protein of S. aureus, His-SSL7 (19), was used as a negative control and did not bind to the surface. His-tagged Efb-C and Ecb did not bind to bacteria in the presence of heat-inactivated serum, demonstrating a complement-dependent interaction.
Efb-C and Ecb specifically block C3b-containing convertases
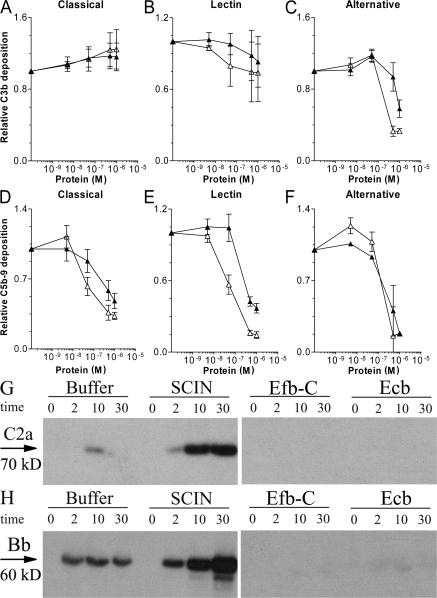
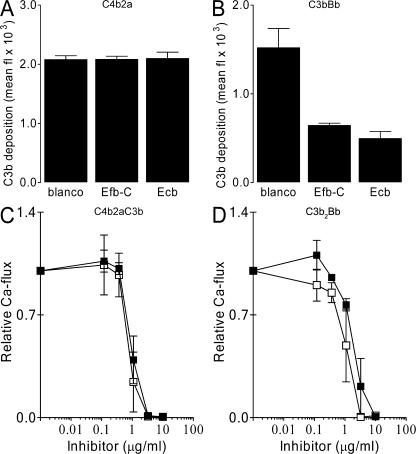
Our finding that Efb and Ecb act on the bacterial surface prompted us to study their regulatory impact on C3 and C5 convertases. Therefore, S. aureus was incubated with human serum and the proteolytic activity of C3 convertases was tested by measuring C3b deposition, whereas C5 convertase activity was monitored by testing C5a release in supernatants using neutrophil calcium mobilization assays (Fig. 6). To exclusively assess CP and LP convertases, incubations were performed in fD-deficient serum. AP convertases were analyzed in the presence of Mg-EGTA to chelate calcium ions needed for the CP and LP (22). We found no inhibition of C3b deposition in the presence of Efb-C and Ecb after CP or LP activation (Fig. 6 A). In contrast, C5a production was strongly inhibited in response to CP or LP activation (Fig. 6 C). Importantly, Efb-C and Ecb blocked C3b deposition and C5a-mediated calcium mobilization after AP activation (Fig. 6, B and D). Collectively, we demonstrate that Efb-C and Ecb do not affect the CP/LP convertase C4b2a but specifically block C3b-containing convertases, i.e., the AP C3 convertase (C3bBb) and the C5 convertases of CP/LP (C4b2aC3b) and AP (C3b2Bb).
Figure 6.
Efb-C and Ecb act on C3b-containing convertases. Efb-C and Ecb specifically block C3b-containing convertases. S. aureus was incubated with 10% fD-depleted serum (to measure CP and LP activation) or 10% human serum in the presence of Mg-EGTA (to measure AP activation). Complement activation was measured at the level of C3b deposition by flow cytometry using anti-C3 antibodies (A and B) or at the level of C5a production by calcium mobilization (C and D). Efb-C (▪) and Ecb (□) do not prevent C3b deposition by the CP/LP (A) but inhibit C3b deposition by the AP (B). Formation of C5a was blocked in response to CP and LP activation (C), as well as AP activation (D). Data shown represent the mean ± SE of three separate experiments. Mean fl, mean fluorescence.
Efb-C and Ecb block convertase activity
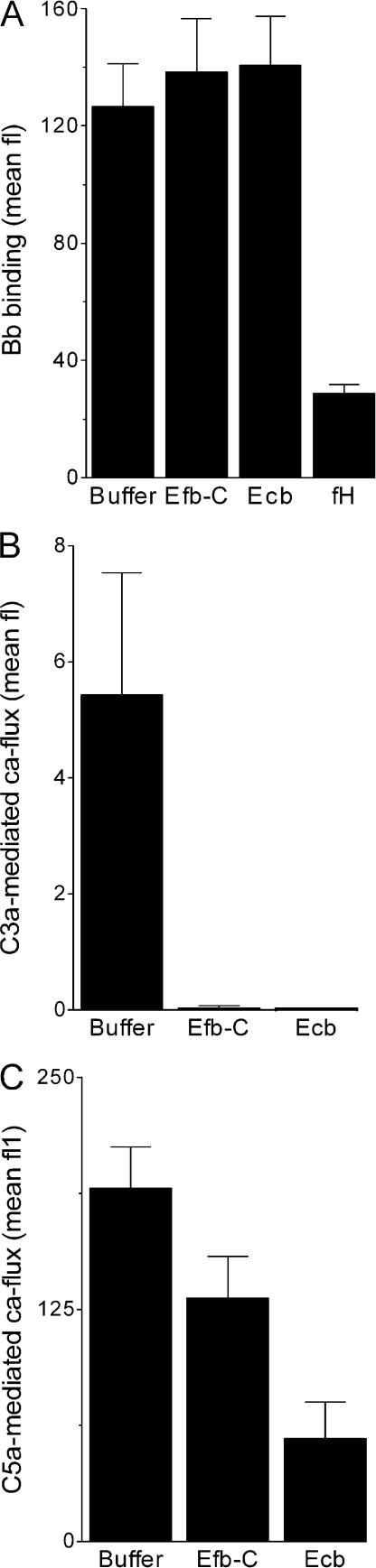
Because several human convertase inhibitors (e.g., factor H [fH] and decay-accelerating factor) promote dissociation of convertase complexes, we tested the decay-accelerating properties of Efb-C and Ecb. Surface-bound AP convertases, created by incubating C3b-coated bacteria with fB, fD, and properdin (38), were incubated with Efb-C, Ecb, and fH for 30 min at 37°C. Unlike fH, Efb-C and Ecb could not promote dissociation of Bb, indicating they do not act as decay-accelerating molecules (Fig. 7 A). To investigate whether Efb-C and Ecb affect substrate cleavage by convertases, zymosan-bound convertases were incubated with purified C3 or C5 in the presence or absence of Efb-C and Ecb. Formation of C3a and C5a was measured by neutrophil calcium mobilization. Efb-C and Ecb prevented both C3 cleavage by AP convertases (Fig. 7 B) and convertases-mediated C5 cleavage (Fig. 7 C). These data demonstrate that Efb-C and Ecb prevent convertase cleavage of C3 and C5.
Figure 7.
Efb-C and Ecb inhibit C3 and C5 cleavage by convertases. (A) Efb-C and Ecb do not promote decay of convertases. AP convertases were created on the bacterial surface using purified components. Subsequent incubation with inhibitors showed that Efb-C and Ecb do not dissociate convertases like fH. Surface-bound convertases were detected using anti-Bb antibodies and flow cytometry. (B and C) Efb-C and Ecb inhibit convertase activity. Zymosan-bound convertases were incubated with purified C3 or C5, and generated C3a or C5a was measured in a neutrophil calcium mobilization assay. (B) Efb-C and Ecb inhibit C3 cleavage by AP convertases. (C) Efb-C and Ecb inhibit C5 cleavage by convertases. Data shown represent the mean ± SE of three separate experiments. Mean fl, mean fluorescence.
Efb-C and Ecb inhibit complement in various species
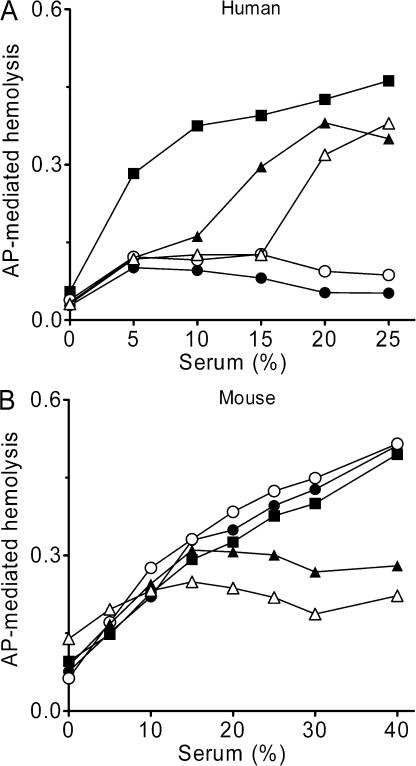
The species specificity of SCIN-B, SCIN-C, Efb-C, and Ecb was determined in an AP-mediated hemolytic assay in which MAC-dependent killing of rabbit erythrocytes is used as a readout for complement activity. As demonstrated for SCIN, SCIN-B and SCIN-C, strongly inhibited AP-mediated hemolysis in human serum (Fig. 8 A), whereas no inhibition was observed in mice (Fig. 8 B) and all other tested species (not depicted). In contrast, we found that Efb-C and Ecb not only blocked complement activation in humans but also in mouse (Fig. 8, A and B), rat, cow, sheep, goat, guinea pig, and dog serum. The fact that Ecb strongly blocks AP activation in mice allowed us for the first time to determine the complement inhibitory properties of an S. aureus–derived complement inhibitor in vivo.
Figure 8.
Efb-C and Ecb are not human specific. (A) SCIN-B, SCIN-C, Efb-C, and Ecb inhibit AP-mediated hemolysis of rabbit red blood cells in human serum. SCIN-B (○) and SCIN-C (•) completely block hemolysis at high serum concentrations. Efb-C (▴) and Ecb (Δ) also inhibit AP-mediated hemolysis but are less effective than SCIN-B or SCIN-C (all inhibitors at 10μg/ml). (B) Efb-C and Ecb inhibit AP-mediated hemolysis in mouse serum. Addition of 10 μg/ml Efb-C (▴) or Ecb (Δ) results in strong inhibition of MAC formation in mouse serum, whereas SCIN-B (○) and SCIN-C (•) had no effect. Graphs show one representative figure out of three separate experiments.
Ecb strongly blocks immune complex (IC)–mediated inflammation in mice
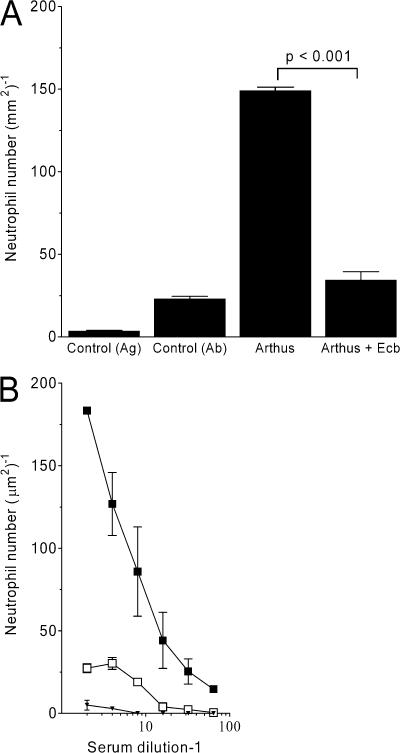
ICs are critical for the pathogenesis of several autoimmune diseases, including systemic lupus erythematosus, rheumatoid arthritis, and glomerulonephritis. In addition to the interaction with IgG Fc receptors, ICs also activate the CP and AP and attract various immune cells through the ligation of complement receptors. In the classical experimental model of soluble IC disease, the Arthus reaction, IC activation of local resident cells results in edema, hemorrhage, and neutrophil infiltration. We used the well-established reverse passive Arthus reaction peritonitis model to assess the in vivo activity of Ecb (39). In the absence of inhibitory protein, neutrophils accumulate in the peritoneal cavity within 6 h after IC challenge (Fig. 9 A). i.p. and i.v. injection of Ecb before IC challenge resulted in complete inhibition of neutrophil migration. Injection of Ecb alone did not induce IC peritonitis. The antiinflammatory effect of Ecb is similar to deletion of the C5aR or IgG Fc receptor (39).
Figure 9.
Ecb completely blocks IC-induced neutrophil influx in vivo. (A) In vivo complement inhibition by Ecb was tested in the reverse passive Arthus reaction peritonitis model. Neutrophils accumulate in the peritoneal cavity within 6 h after IC challenge. i.p. and i.v. injection of Ecb before IC challenge resulted in complete inhibition of neutrophil migration. Injection of Ecb alone did not induce IC peritonitis. P < 0.001 by analysis of variance (n = 10 mice per group). (B) Ecb completely blocks C5a-dependent migration of BM-derived mouse neutrophils in vitro. Preincubation of zymosan with C5-sufficient (▪) but not C5-deficient plasma (▾) results in neutrophil migration. In the presence of 50 μg/ml Ecb, C5a production in C5-sufficient plasma was blocked (□). Data shown represent the mean ± SE of three separate experiments.
To prove that Ecb also prevents C5a generation and C5a-mediated neutrophil migration in mice, zymosan was preincubated with EGTA plasma from C5-sufficient C57BL/6 mice and C5-deficient A/J mice to activate the AP. The plasma was then assessed for its ability to induce migration of BM-derived mouse neutrophils. Strong neutrophil migration occurred in response to C5-sufficient but not C5-deficient plasma (Fig. 9 B), demonstrating that neutrophil migration depends on AP-activated C5a. C5-sufficient plasma that had been activated in the presence of Ecb was strongly impaired in its capacity to induce neutrophil migration (Fig. 9 B). Collectively, these data show that Ecb inhibits the AP-induced activation of C5 necessary to induce neutrophil migration both in vitro and in vivo.
DISCUSSION
The human pathogen S. aureus successfully evades innate immune defenses by secretion of numerous small molecules that inhibit critical steps of the immune response. In this study, we identify SCIN-B, SCIN-C, Efb, and Ecb as four new staphylococcal complement modulators that block convertases, the central protease complexes of the complement cascade. Direct convertase modulators are specific for S. aureus and provide a very effective way of blocking complement. This paper shows that convertase inhibition is a very powerful mechanism to block inflammatory reactions initiated by complement both in vitro and in vivo.
The first IEC in S. aureus (IEC-1) encodes four important immune modulators, including SCIN and CHIPS (25). With the discovery of SCIN and CHIPS homologues, we are now confronted with the second IEC in S. aureus. Six molecules on IEC-2 (FLIPr [26], FLIPr-like [unpublished data], SCIN-B, SCIN-C, Efb, and Ecb) are now known to have immune evasive properties. The role of the other molecules on IEC-2 is the subject for further studies. The immune-modulating properties of SSL12–14 are very likely, because the SSLs, structural homologues of superantigens that lack superantigen activity, are believed to modulate various innate immune functions (31): SSL5 binds P-selectin glycoprotein ligand 1 to inhibit neutrophil rolling on epithelial cells (40), and SSL7 binds IgA and C5 and blocks IgA-FcR and C5 activation. (19). In contrast to the bacteriophage-located IEC-1, the borders and nature of IEC-2 are less clear. Because IEC-2 contains mobile elements and bacteriophage remnants, we believe horizontal gene transfer has played a role in its development. In sharp contrast to IEC-1, IEC-2 is not human specific, as indicated by its presence in the bovine S. aureus strain RF122 and the cross-reactivity of Efb and Ecb in several animal species.
In this paper, we highlight two different mechanisms used by S. aureus to regulate complement convertases. The SCIN molecules (SCIN, SCIN-B, and SCIN-C) specifically block C3 convertases (C4b2a and C3bBb) to prevent C3b/iC3b deposition, phagocytosis, and C5a generation. SCIN does not directly bind C3b but exclusively binds the activated C3 convertase, causing both stabilization and inactivation of this complex (22). Furthermore, S. aureus secretes C3d-binding molecules (Efb and Ecb) that specifically inactivate C3b-containing convertases, i.e., the AP C3 convertase (C3bBb) and the C5 convertases of CP/LP (C4b2aC3b) and AP (C3b2Bb). Because inhibition of the AP C3 convertase is not sufficient to block opsonization and phagocytosis in whole-serum assays, we believe that the major bacterial defense functions of Efb-C and Ecb are to block C5a production by C5 convertases and to down-regulate neutrophil responses. Because a previous report has indicated that Efb-C induces a conformational change in both C3b and C3, it was suggested that Efb-C blocks formation of a functional opsonin (41). However, our data indicate that Efb-C and Ecb do not directly block C3 cleavage and opsonin formation, because C3b deposition by the CP/LP was not affected at physiological inhibitor concentrations. Furthermore, the complement-dependent binding to bacteria demonstrates that Efb-C and Ecb act on the bacterial surface rather than in fluid phase. The finding that Efb-C and Ecb inhibit the CP/LP C5 convertase (C4b2aC3b) but not the C3 convertase (C4b2a) strongly suggests that they bind convertases via C3b. The conformational change in C3b (41) likely affects the activity of the protease complex. We observed that Efb-C and Ecb do not affect the stability of the complex but block cleavage of the substrate (C3 or C5). This, along with the fact that C3b molecules within C5 convertases provide the additional binding sites for C5 (15, 17, 18, 42–44), suggests that Efb-C and Ecb block substrate binding via C3b.
The identification of four novel convertase inhibitors in S. aureus illustrates the crucial role of convertase modulation for bacterial survival. It is remarkable that a single bacterium produces five different convertase inhibitors, stressing the importance of appropriate complement activation for S. aureus killing. In concert with several additional complement modulators (45), S. aureus contains an enormous arsenal of molecules that can block the complement cascade at different steps. These synergistic inhibitory pathways help the bacterium to prevent C5a-mediated neutrophil responses and/or phagocytosis. The fact that all staphylococcal complement modulators are produced in vivo suggests that they are beneficial for bacterial survival in the human host. The redundancy of this system is reminiscent of the mechanisms used by herpes viruses to escape antigen processing. Similar to S. aureus, herpes viruses evade host-mediated immunity by the blockade of different steps of the immune response, i.e., inhibition of different molecules critical for antigen processing (46, 47).
The identification of two nonhuman specific complement modulators allows us for the first time to study the role of complement modulation in bacterial pathogenesis in vivo. Ecb strongly prevented the development of neutrophil-mediated inflammation in a mouse model of IC-induced peritonitis. Our data suggest that the convertase-inhibitory molecules evolved in S. aureus are promising drug candidates for antiinflammatory therapy, though the occurrence of preexisting antibodies precludes their use in humans in their original form (45). However, as the active sites of SCIN and Efb-C comprise only minor parts of the molecule (25, 41), these molecules are promising lead compounds for the development of small nonimmunogenic complement inhibitors. The strong antiinflammatory effect of Ecb in vivo illustrates the therapeutic potential of such molecules in severe acute inflammatory disorders.
MATERIALS AND METHODS
Recombinant production of S. aureus proteins.
To enable protein expression with a cleavable N-terminal His tag, genes (without signal sequences) were cloned into the pRSETB vector (Invitrogen). Primers are listed in Table S1 (available at http://www.jem.org/cgi/content/full/jem.20070818/DC1). Cloning of SCIN homologues was performed as previously described (22). Efb, Efb-C, and Ecb were cloned by overlap extension PCR (24). In the first PCR reaction, a small region of pRSETB was amplified using XbaI forward and enterokinase (EK) cleavage site reverse (EK-reverse; Table S1) primers. The EK-reverse primers contain a 3′ overhang on the Efb genes. In a second PCR, efb and ecb were amplified from chromosomal S. aureus DNA using EK-forward primers (to provide a 5′ overhang on the EK site) and EcoRI-reverse primers (providing a 3′ EcoRI cleavage site). Both PCR products were mixed and amplified in a third PCR reaction using XbaI forward and EcoRI-reverse. PCR products and pRSETB were digested with XbaI and EcoRI before ligation. Transformation and expression were performed as previously described (21, 22). Proteins were expressed in E. coli and purified from bacterial lysates by nickel affinity chromatography (21). The His tag was removed by EK cleavage and separated by a second column passage. The His tag of Efb was not removed because EK cleavage resulted in unwanted proteolysis. PCR analyses of clinical S. aureus strains was performed as previously described (25) using the pRSETB cloning primers for orf-d, efb, and ecb and specifically designed primers for scb and scc (designated with “I” in Table S1).
Phagocytosis and complement activation on S. aureus.
Laboratory strain S. aureus Wood was used for phagocytosis and bacterial complement assays. Phagocytosis was performed using FITC-labeled S. aureus, human sera, and freshly isolated human neutrophils (22, 48). For C5a analyses, heat-killed S. aureus (to prevent formyl-Met-Leu-Phe production) was incubated with human sera in RPMI 1640 medium for 30 min at 37°C. Subsequently, collected supernatants were tested for neutrophil calcium mobilization, as previously described (23). C5a specificity of this assay was verified by preincubation of neutrophils with the C5a receptor antagonist CHIPS−30 (21). To analyze CD11b and CD62L expression, neutrophils were placed on ice for 10 min, after which expression of CD11b and CD62 was measured using PE-conjugated anti–human CD62L (BD Biosciences) and FITC-labeled anti–human CD11b (48) and flow cytometry. C3b deposition was performed by incubation of S. aureus with 10% human sera in HBS2+ (Hepes-buffered saline, 20 mM Hepes, 140 mM NaCl, 5 mM CaCl2, and 2.5 mM MgCl2), after which surface-bound C3b was detected with FITC-conjugated (Fab′)2 anti–human C3 (Protos Immunoresearch). Exclusive activation of the CP/LP was performed in fD-deficient serum (22). The AP was analyzed in serum supplemented with Mg-EGTA. Before calcium mobilization, Mg-EGTA–containing supernatants were supplemented with 12 mM CaCl2. To analyze binding, S. aureus was incubated with 10 μg/ml His–Efb-C or His-Ecb and human sera for 30 min at 37°C. His-tagged proteins were detected using mouse anti–His tag antibodies (QIAGEN), FITC-labeled goat anti–mouse IgG (DakoCytomation), and flow cytometry.
ELISAs.
Complement ELISAs were performed as previously described (36, 37), with minor modifications. Deposited C3b and C5b-9 were detected using anti-C3c WM1 (American Type Culture Collection) (48) and anti–C5b-9 (Abcam) antibodies, respectively, followed by peroxidase (PO)-conjugated goat anti–mouse IgG (Southern Biotechnology Associates, Inc.). Antibody distribution in humans was determined as previously described (20). To study binding of complement proteins by ELISA (20, 23), inhibitor-coated wells (10 μg/ml) were incubated with human C5, C4, fB, fH C3, C3b, C3d (all obtained from Calbiochem), or C3c (provided by B. Janssen, Utrecht University, Utrecht, Netherlands). Binding of complement proteins was detected using monoclonal antibodies against human C3c (WM1), C3d (Quidel Corp.), C5/C5b (HyCult Biotechnology), Bb (Quidel Corp.), fH (Quidel Corp.), or C4d (Quidel Corp.) and secondary PO-conjugated anti–mouse antibodies.
Convertase analyses.
Detection of bacterium-bound convertases after opsonization was performed as previously described for SCIN (22). To prepare C3b-covered bacteria, S. aureus was incubated with 20% human sera for 30 min in HBS2+, followed by a 30-min incubation in PBS at 37°C to dissociate surface-bound C2a/Bb (22). To create purified convertases on the bacterial surface, C3b-covered bacteria were incubated with 40 μg/ml fB, 1 μg/ml fD, and 4 μg/ml properdin (all obtained from Calbiochem) for 30 min at 37°C in HBS2+. To analyze stability, convertases were incubated with 20 μg/ml Efb-C, Ecb, or fH (Calbiochem) for 30 min at 37°C in HBS2+. Surface-bound Bb was detected by flow cytometry (22). Cleavage of C3 and C5 by surface-bound convertases was studied on zymosan. Convertases were created by incubating zymosan particles with serum in PBS supplemented with 2 mM NiCl2 (to increase convertase stability) for 30 min at 37°C. Washed zymosan particles were subsequently incubated with Efb-C, 10 μg/ml Ecb, or PBS for 5 min at 37°C before 4 μg/ml human C5 or C3 (Calbiochem) was added for 1 h at 37°C. For C3 activation, zymosan was opsonized in C2-deficient serum (Sigma-Aldrich) to exclusively form AP convertases. C5 cleavage was assessed using particles opsonized in normal serum. Formation of C5a or C3a was measured by analyzing supernatants in a calcium mobilization assay. AP hemolytic assays were performed as previously described (22).
Preparation of mouse EGTA plasma.
Blood was drawn from C57BL6 (n = 10) and A/J mice (n = 10) by cardiac puncture in syringes containing 0.2 M EGTA (Sigma-Aldrich) as an anticoagulant. Cells were pelleted by centrifugation in a swing-out rotor at 1,400 g for 10 min at 4°C.
Zymosan-induced complement activation.
2.5 ml EGTA plasma from either C57BL6 or A/J mice was heated to 37°C for 30 min in the presence or the absence of 50 μg/ml Ecb. The plasma was then treated with 1.335 mg PL-2-mercaptomethyl-3-guanidinoethylthio-propanoic acid (Calbiochem), 47 μl of 4 M 6-aminohexanoic acid (Fluka), 1 ml of washed zymosan A, and 187.5 μl of 0.2 M MgCl2 at 37°C for 1 h to activate the AP of complement and to block serum carboxypeptidase N. The activated plasma was centrifuged at 20,000 g for 1 h at 4°C to precipitate large plasma proteins. The supernatant, containing the anaphylatoxins, was stored at −80°C.
Neutrophil chemotaxis.
To obtain BM-derived neutrophils, femurs, tibias, and humeri of BALB/c mice were flushed with sterile PBS. The migration of BM-derived neutrophils was determined using a 48-well microchemotaxis chamber (Neuro Probe) with polycarbonate filters (pore size, 3 μm). Twofold serial dilutions (1:2 to 1:64) of Ecb-treated and untreated zymosan-activated plasma from C57BL6 and A/J mice were generated using 2% BSA-GBSS (Sigma-Aldrich) as diluent. 30 μl of the diluted plasma was added to the lower well to act as a chemoattractant to 50 μl of neutrophils (107 neutrophils per milliliter suspended in 2% BSA-GBSS buffer), which were added to the upper wells. Cells were allowed to migrate for 30 min at 37°C. Membranes were separated, fixed with methanol, and stained with Diff-Quick (BaxterDade). The numbers of neutrophils per square micrometer migrating beyond the lower surface of the membrane were determined microscopically (≥20 high power fields, 32× magnification).
Peritoneal Arthus reaction.
BALB/c mice were i.v. injected with 100 μl of OVA (20 mg/kg of body weight; Sigma-Aldrich), immediately followed by i.p. injection of 200 μl of rabbit anti-OVA IgG (800 μg per mouse; MP Biomedicals) in sterile PBS. In inhibition experiments, 100 μl Ecb was administered i.v. and i.p. (600 μg/ml) 30 min before initiation of the Arthus reaction. Further, Ecb was administered i.v. and i.p. using the same concentrations in the absence of the Arthus reaction (as a control). Mice of different treatment groups were killed 6 h after onset of the peritoneal Arthus reaction. The peritoneal cavity was lavaged with 6 ml of ice-cold PBS/0.1% BSA. Peritoneal cells were washed once with PBS, and the cell number was adjusted to 5 × 105 cells per milliliter. 50 μl of this cell suspension was used to prepare cytospin slides, which were stained with DiffQuick. Neutrophil numbers per square millimeter were calculated from ≥20 different microscopic fields. Animal care was provided in accordance with National Institutes of Health guidelines. Animal studies were approved by either the Bezirksregierung Hannover or the Cincinnati Children's Hospital Medical Center institutional animal care and use committee.
Accession numbers.
SCIN-B (NP_374275, NP_371683, YP_186032, YP_493754, YP_499659, YP_416506, YP_043217, NP_645858, and YP_040544), SCIN-C (YP_040544), ORF-D (YP_039686), Ecb (NP_374271, NP_371679, YP_186027, YP_493750, YP_499654, YP_416502, YP_043213, NP_645854, and YP_040541), and Efb (NP_374274, NP_371682, YP_186031, YP_493753, YP_499658, YP_416505, YP_043216, NP_645857, and YP_040543) are available from GenBank/EMBL/DDBJ under the accession numbers listed.
Online supplemental material.
Table S1 lists the primers used in this study. Fig. S1 A represents sequence alignments of SCIN, SCIN-B, SCIN-C, and ORF-D; Fig. S1 B represents sequence alignments of Efb, Efb-C, and Ecb. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070818/DC1.
Supplemental Material
Acknowledgments
This work was supported by grants from the Netherlands Genomics Initiative–Horizon (050-71-028), the Netherlands Organisation for Scientific Research (NWO)–TOP (9120.6020), NWO-VENI (916-76-037), and the National Institutes of Health (R01-AI59306 to J. Kohl).
The authors have no conflicting financial interests.
Abbreviations used: AP, CP, and LP, alternative, classical, and lectin pathway, respectively; CHIPS, chemotaxis inhibitory protein of S. aureus; Ecb, extracellular complement-binding protein; Efb, extracellular fibrinogen-binding protein; Efb-C, C3-binding domain of Efb; EK, enterokinase; fB, fD, and fH, factor B, D, and H, respectively; FLIPr, FPR-like 1 inhibitory protein; FPR, formylated peptide receptor; His, histidine; IC, immune complex; IEC, immune evasion cluster; MAC, membrane attack complex; ORF, open reading frame; PO, peroxidase; SCIN, staphylococcal complement inhibitor; SSL, staphylococcal superantigen like.
References
- 1.Neth, O., D.L. Jack, M. Johnson, N.J. Klein, and M.W. Turner. 2002. Enhancement of complement activation and opsonophagocytosis by complexes of mannose-binding lectin with mannose-binding lectin-associated serine protease after binding to Staphylococcus aureus. J. Immunol. 169:4430–4436. [DOI] [PubMed] [Google Scholar]
- 2.Wergeland, H., C. Endresen, O.B. Natas, P. Aasjord, and P. Oeding. 1984. Antibodies to Staphylococcus aureus peptidoglycan and lipoteichoic acid in sera from blood donors and patients with staphylococcal infections. Acta Pathol. Microbiol. Immunol. Scand. [B]. 92:265–269. [DOI] [PubMed] [Google Scholar]
- 3.Walport, M.J. 2001. Complement. Second of two parts. N. Engl. J. Med. 344:1140–1144. [DOI] [PubMed] [Google Scholar]
- 4.Walport, M.J. 2001. Complement. First of two parts. N. Engl. J. Med. 344:1058–1066. [DOI] [PubMed] [Google Scholar]
- 5.Muller-Eberhard, H.J. 1986. The membrane attack complex of complement. Annu. Rev. Immunol. 4:503–528. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, M.C., and V.M. Holers. 2005. Innate autoimmunity. Adv. Immunol. 86:137–157. [DOI] [PubMed] [Google Scholar]
- 7.Kohl, J., and M. Wills-Karp. 2007. Complement regulates inhalation tolerance at the dendritic cell/T cell interface. Mol. Immunol. 44:44–56. [DOI] [PubMed] [Google Scholar]
- 8.del Zoppo, G.J. 1999. In stroke, complement will get you nowhere. Nat. Med. 5:995–996. [DOI] [PubMed] [Google Scholar]
- 9.Ward, P.A. 2004. The dark side of C5a in sepsis. Nat. Rev. Immunol. 4:133–142. [DOI] [PubMed] [Google Scholar]
- 10.Kohl, J. 2006. The role of complement in danger sensing and transmission. Immunol. Res. 34:157–176. [DOI] [PubMed] [Google Scholar]
- 11.Gasque, P. 2004. Complement: a unique innate immune sensor for danger signals. Mol. Immunol. 41:1089–1098. [DOI] [PubMed] [Google Scholar]
- 12.Holmskov, U., S. Thiel, and J.C. Jensenius. 2003. Collections and ficolins: humoral lectins of the innate immune defense. Annu. Rev. Immunol. 21:547–578. [DOI] [PubMed] [Google Scholar]
- 13.Xu, Y., S.V. Narayana, and J.E. Volanakis. 2001. Structural biology of the alternative pathway convertase. Immunol. Rev. 180:123–135. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, T., M. Matsushita, and Y. Endo. 2004. The lectin-complement pathway–its role in innate immunity and evolution. Immunol. Rev. 198:185–202. [DOI] [PubMed] [Google Scholar]
- 15.Rawal, N., and M.K. Pangburn. 1998. C5 convertase of the alternative pathway of complement. Kinetic analysis of the free and surface-bound forms of the enzyme. J. Biol. Chem. 273:16828–16835. [DOI] [PubMed] [Google Scholar]
- 16.Hong, K., T. Kinoshita, P. Pramoonjago, Y.U. Kim, T. Seya, and K. Inoue. 1991. Reconstitution of C5 convertase of the alternative complement pathway with isolated C3b dimer and factors B and D. J. Immunol. 146:1868–1873. [PubMed] [Google Scholar]
- 17.Pangburn, M.K., and N. Rawal. 2002. Structure and function of complement C5 convertase enzymes. Biochem. Soc. Trans. 30:1006–1010. [DOI] [PubMed] [Google Scholar]
- 18.Rawal, N., and M. Pangburn. 2001. Formation of high-affinity C5 convertases of the alternative pathway of complement. J. Immunol. 166:2635–2642. [DOI] [PubMed] [Google Scholar]
- 19.Langley, R., B. Wines, N. Willoughby, I. Basu, T. Proft, and J.D. Fraser. 2005. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-Fc alpha RI binding and serum killing of bacteria. J. Immunol. 174:2926–2933. [DOI] [PubMed] [Google Scholar]
- 20.Rooijakkers, S.H., W.J. van Wamel, M. Ruyken, K.P. van Kessel, and J.A. van Strijp. 2005. Anti-opsonic properties of staphylokinase. Microbes Infect. 7:476–484. [DOI] [PubMed] [Google Scholar]
- 21.de Haas, C.J., K.E. Veldkamp, A. Peschel, F. Weerkamp, W.J. van Wamel, E.C. Heezius, M.J. Poppelier, K.P. van Kessel, and J.A. van Strijp. 2004. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 199:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rooijakkers, S.H., M. Ruyken, A. Roos, M.R. Daha, J.S. Presanis, R.B. Sim, W.J. van Wamel, K.P. van Kessel, and J.A. van Strijp. 2005. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6:920–927. [DOI] [PubMed] [Google Scholar]
- 23.Rooijakkers, S.H., M. Ruyken, J. van Roon, K.P. van Kessel, J.A. van Strijp, and W.J. van Wamel. 2006. Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell. Microbiol. 8:1282–1293. [DOI] [PubMed] [Google Scholar]
- 24.Rooijakkers, S.H., F.J. Milder, B.W. Bardoel, M. Ruyken, J.A. van Strijp, and P. Gros. 2007. Staphylococcal complement inhibitor: structure and active sites. J. Immunol. 179:2989–2998. [DOI] [PubMed] [Google Scholar]
- 25.van Wamel, W.J., S.H. Rooijakkers, M. Ruyken, K.P. van Kessel, and J.A. van Strijp. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188:1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prat, C., J. Bestebroer, C.J. de Haas, J.A. van Strijp, and K.P. van Kessel. 2006. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J. Immunol. 177:8017–8026. [DOI] [PubMed] [Google Scholar]
- 27.Boden, M.K., and J.I. Flock. 1992. Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman. Microb. Pathog. 12:289–298. [DOI] [PubMed] [Google Scholar]
- 28.Boden, M.K., and J.I. Flock. 1994. Cloning and characterization of a gene for a 19 kDa fibrinogen-binding protein from Staphylococcus aureus. Mol. Microbiol. 12:599–606. [DOI] [PubMed] [Google Scholar]
- 29.Lee, L.Y., X. Liang, M. Hook, and E.L. Brown. 2004. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb). J. Biol. Chem. 279:50710–50716. [DOI] [PubMed] [Google Scholar]
- 30.Lee, L.Y., M. Hook, D. Haviland, R.A. Wetsel, E.O. Yonter, P. Syribeys, J. Vernachio, and E.L. Brown. 2004. Inhibition of complement activation by a secreted Staphylococcus aureus protein. J. Infect. Dis. 190:571–579. [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald, J.R., S.D. Reid, E. Ruotsalainen, T.J. Tripp, M. Liu, R. Cole, P. Kuusela, P.M. Schlievert, A. Jarvinen, and J.M. Musser. 2003. Genome diversification in Staphylococcus aureus: Molecular evolution of a highly variable chromosomal region encoding the Staphylococcal exotoxin-like family of proteins. Infect. Immun. 71:2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frost, L.S., R. Leplae, A.O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732. [DOI] [PubMed] [Google Scholar]
- 33.Coenjaerts, F.E., A.M. Walenkamp, P.N. Mwinzi, J. Scharringa, H.A. Dekker, J.A. van Strijp, R. Cherniak, and A.I. Hoepelman. 2001. Potent inhibition of neutrophil migration by cryptococcal mannoprotein-4-induced desensitization. J. Immunol. 167:3988–3995. [DOI] [PubMed] [Google Scholar]
- 34.Lutz, H.U., and E. Jelezarova. 2006. Complement amplification revisited. Mol. Immunol. 43:2–12. [DOI] [PubMed] [Google Scholar]
- 35.Mollnes, T.E., O.L. Brekke, M. Fung, H. Fure, D. Christiansen, G. Bergseth, V. Videm, K.T. Lappegard, J. Kohl, and J.D. Lambris. 2002. Essential role of the C5a receptor in E. coli-induced oxidative burst and phagocytosis revealed by a novel lepirudin-based human whole blood model of inflammation. Blood. 100:1869–1877. [PubMed] [Google Scholar]
- 36.Roos, A., L.H. Bouwman, J. Munoz, T. Zuiverloon, M.C. Faber-Krol, F.C. Fallaux-van den Houten, N. Klar-Mohamad, C.E. Hack, M.G. Tilanus, and M.R. Daha. 2003. Functional characterization of the lectin pathway of complement in human serum. Mol. Immunol. 39:655–668. [DOI] [PubMed] [Google Scholar]
- 37.Seelen, M.A., A. Roos, J. Wieslander, T.E. Mollnes, A.G. Sjoholm, R. Wurzner, M. Loos, F. Tedesco, R.B. Sim, P. Garred, et al. 2005. Functional analysis of the classical, alternative, and MBL pathways of the complement system: standardization and validation of a simple ELISA. J. Immunol. Methods. 296:187–198. [DOI] [PubMed] [Google Scholar]
- 38.Hourcade, D.E. 2006. The role of properdin in the assembly of the alternative pathway C3 convertases of complement. J. Biol. Chem. 281:2128–2132. [DOI] [PubMed] [Google Scholar]
- 39.Godau, J., T. Heller, H. Hawlisch, M. Trappe, E. Howells, J. Best, J. Zwirner, J.S. Verbeek, P.M. Hogarth, C. Gerard, et al. 2004. C5a initiates the inflammatory cascade in immune complex peritonitis. J. Immunol. 173:3437–3445. [DOI] [PubMed] [Google Scholar]
- 40.Bestebroer, J., M.J. Poppelier, L.H. Ulfman, P.J. Lenting, C.V. Denis, K.P. van Kessel, J.A. van Strijp, and C.J. de Haas. 2007. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood. 109:2936–2943. [DOI] [PubMed] [Google Scholar]
- 41.Hammel, M., G. Sfyroera, D. Ricklin, P. Magotti, J.D. Lambris, and B.V. Geisbrecht. 2007. A structural basis for complement inhibition by Staphylococcus aureus. Nat. Immunol. 8:430–437. [DOI] [PubMed] [Google Scholar]
- 42.Rawal, N., and M.K. Pangburn. 2000. Functional role of the noncatalytic subunit of complement C5 convertase. J. Immunol. 164:1379–1385. [DOI] [PubMed] [Google Scholar]
- 43.Rawal, N., and M.K. Pangburn. 2003. Formation of high affinity C5 convertase of the classical pathway of complement. J. Biol. Chem. 278:38476–38483. [DOI] [PubMed] [Google Scholar]
- 44.Rawal, N., and M.K. Pangburn. 2007. Role of the C3b-binding site on C4b-binding protein in regulating classical pathway C5 convertase. Mol. Immunol. 44:1105–1114. [DOI] [PubMed] [Google Scholar]
- 45.Rooijakkers, S.H., and J.A. van Strijp. 2006. Bacterial complement evasion. Mol. Immunol. 44:23–32. [DOI] [PubMed] [Google Scholar]
- 46.Novak, N., and W.M. Peng. 2005. Dancing with the enemy: the interplay of herpes simplex virus with dendritic cells. Clin. Exp. Immunol. 142:405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ploegh, H.L. 1998. Viral strategies of immune evasion. Science. 280:248–253. [DOI] [PubMed] [Google Scholar]
- 48.Veldkamp, K.E., K.P. van Kessel, J. Verhoef, and J.A. van Strijp. 1997. Staphylococcal culture supernates stimulate human phagocytes. Inflammation. 21:541–551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.