Abstract
Dendritic cells (DCs) play a critical role in the initiation, maintenance, and resolution of an immune response. DC survival is tightly controlled by extracellular stimuli such as cytokines and Toll-like receptor (TLR) signaling, but the intracellular events that translate such extracellular stimuli into life or death for the DC remain poorly understood. The endoplasmic reticulum (ER) stress, or unfolded protein response (UPR), is a signaling pathway that is activated when unfolded proteins accumulate in the ER. The most conserved arm of the UPR involves IRE1α, an ER transmembrane kinase and endoribonuclease that activates the transcription factor XBP-1 to maintain ER homeostasis and prevent activation of cell death pathways caused by sustained ER stress. We report that XBP-1 is essential for DC development and survival. Lymphoid chimeras lacking XBP-1 possessed decreased numbers of both conventional and plasmacytoid DCs with reduced survival both at baseline and in response to TLR signaling. Overexpression of XBP-1 in hematopoietic progenitors rescued and enhanced DC development. Remarkably, in contrast to other cell types we have examined, the XBP-1 pathway was constitutively activated in immature DCs.
Signals emanating from the ER induce a transcriptional program that enables cells to survive ER stress. This highly coordinated response is essential for the folding, processing, export, and degradation of all proteins emanating from the ER during stressed and normal conditions. Examples of physiological conditions that require the unfolded protein response (UPR) include plasma cell differentiation (1) and pancreatic β cell function (2). Adaptation of tumor cells to hypoxic conditions and glucose deprivation also induces the ER stress response (3). Additionally, there is increasing evidence of protein misfolding in neurodegenerative diseases such as Huntington's, Alzheimer's, and prion-related diseases (4).The UPR exists in all eukaryotes and consists of multiple signaling pathways, the most conserved of which is mediated by IRE1. Upon sensing unfolded proteins, IRE1 oligomerizes, is activated by autophosphorylation, and uses its endoribonuclease activity to excise an intron from the transcription factor Hac1p in yeast or its mammalian homologue, XBP-1, a cyclic-AMP response element binding protein/activating transcription factor family member first isolated in our laboratory (5). This unconventional mRNA splicing event results in the conversion of the inactive 267–amino-acid unspliced XBP-1 (XBP-1u) protein to an active 371–amino-acid spliced XBP-1 (XBP-1s) protein (6–8). We have previously shown that XBP-1 is essential for the differentiation of highly secretory cells, including embryonic hepatocytes, exocrine pancreatic acinar cells, and plasma cells (1, 9–11).
Much has been learned about the factors that control DC differentiation. The fms-related tyrosine kinase 3 ligand (Flt3L) (12, 13) and GM-CSF (14, 15) are well known positive regulators of DC development. A more recent study described the involvement of Toll-like receptor (TLR) stimulation in hematopoietic cell proliferation and subsequent DC differentiation (16). Additionally, several intracellular signaling molecules and transcription factors—including Gfi1, Id2, Ikaros, IFN regulatory factor 2 (IRF-2), IRF-4, IRF-8, relB, Runx3, Spi-B, and STAT3—that affect the development of individual DC subsets in vivo have been reported (17–24). The control of DC survival plays an important role in regulating T cell activation and function. Components of the immune system involved in DC survival include TLR stimulation and engagement of CD40 on the DC by CD154 expressed on activated T cells. Studies with inflammatory cytokines and tumor necrosis–related activation-induced cytokine/receptor activator for NF-κB demonstrated an augmentation of T cell priming through an enhanced DC survival response (25, 26). The intracellular signaling pathway mediated by the NF-κB family has been shown to be responsible for the enhancement of DC survival by these stimuli (27, 28). Because the ER stress response functions to regulate the balance between homeostasis and apoptosis, we asked whether the UPR and, in particular, the IRE1/XBP-1 branch of the UPR might contribute to the differentiation and survival of the DC (29).
RESULTS AND DISCUSSION
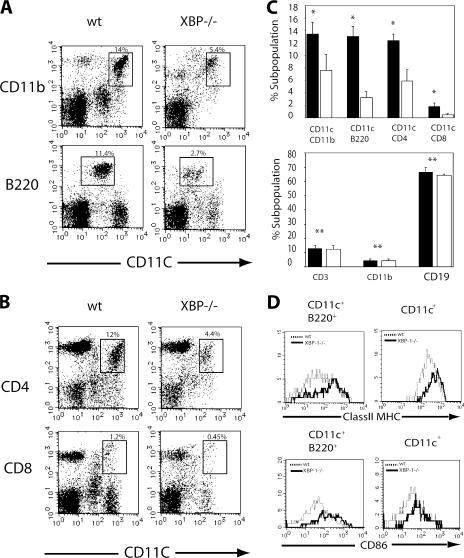
Flow cytometric analysis of DC-enriched low density fractions from spleens of XBP-1/RAG-2−/− chimeric mice and control 129/RAG-2−/− chimeric or 129/SvImJ mice was performed using the surface markers CD11c and CD11b. The total number of spleen cells was the same in XBP-1/RAG-2−/− and control mice. However, the percentage of CD11c+ CD11b+ DCs was markedly reduced in XBP-1/RAG-2−/− mice compared with control animals (Fig. 1 A). The CD11c+ population was further subdivided by the expression of CD4 and CD8 into two subsets (CD11chiCD4−CD8+ and CD11chiCD4+CD8). Both subsets were decreased in XBP-1/RAG-2−/− versus control chimeric mice (Fig. 1, B and C). Interestingly, the most profound reduction was observed in a third subset of DCs, the plasmacytoid DCs (pDCs). This DC subset is characterized by the surface phenotype CD11cintB220+DX5− and displays extensive ER expansion and high level secretion of the cytokine IFN-α (Fig. 1 A) (30). As we previously reported, XBP-1/RAG-2−/− versus control chimeric mice had similar numbers of CD19+, CD3+, and CD11b+ CD11c− cells when compared with WT (Fig. 1 C, bottom) (1). In the absence of stimulation, CD11c+ DCs express low levels of co-stimulatory molecules such as CD86 and moderate levels of MHC class II. However, splenic DCs from XBP-1/RAG-2−/− mice constitutively expressed higher levels of these activation markers (Fig. 1 D). This hyperactivated state was most profound in the pDC (CD11cintB220+) population. Thus, XBP-1 RAG-2−/− mice had reduced numbers of both conventional and pDCs, and these DCs were hyperactivated.
Figure 1.
XBP-1–deficient lymphoid chimeras have reduced numbers of conventional and pDCs. (A) Low density mouse spleen cells from WT or XBP-1−/− animals were isolated and depleted of CD19+ and DX5+ cells by immunomagnetic bead negative selection. DCs were analyzed by flow cytometry for CD11b or B220 staining on CD11c+ cells (CD19−DX5−). (B) CD4 and CD8α staining of CD11c+ (CD19−DX5−)-enriched low density mouse spleen cells from WT or XBP-1−/− animals. (C) Percentage of splenic DC (top) and lymphocyte (bottom subsets from WT (shaded bar) or XBP-1−/− (open bar) animals. The data represent the mean + SEM (n = 3). Values in A represent the percentage of each DC subset from low density mouse spleen cells isolated and depleted of CD19+ and DX5+ cells by immunomagnetic bead negative selection. Values for lymphocyte subsets in B represent percentages from unenriched low density spleen preparations. *, P < 0.02 when comparing WT with XBP-1 samples; **, NS. (D) Activated phenotype of XBP-1−/− splenic DCs. Enriched low density populations from WT and XBP-1−/− animals were gated for CD11c+ and CD11c+B220+ subsets and analyzed for CD86 and MHC class II expression. Numbers indicate the percentage of cells in each CD11c+ subset. The experiment shown is representative of three independent experiments performed.
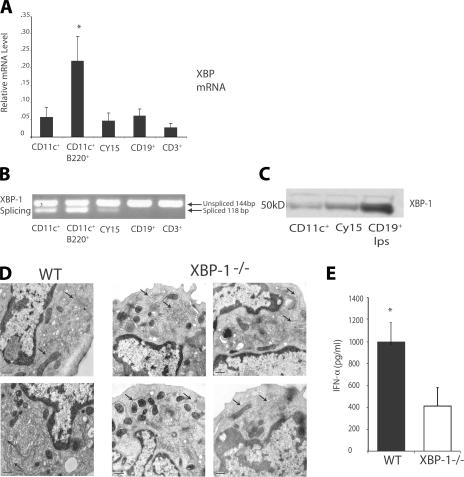
To understand how XBP-1 controls DC number, we examined XBP-1 mRNA expression and splicing by real-time PCR to assess whether total levels of XBP-1 mRNA and/or the extent of XBP-1 splicing correlated with DC differentiation. As shown in Fig. 2 A, total XBP-1 mRNA transcripts including both the unspliced and spliced versions of XBP-1 were expressed at high levels in the pDC subset relative to immature conventional DCs and unactivated T and B lymphocytes. This expression was not further increased upon stimulation with TLR ligands such as LPS and CpG oligonucleotides (ODNs; unpublished data). However, to our surprise, analysis of the relative amounts of XBP-1s versus XBP-1u transcripts by RT-PCR revealed that freshly isolated DC subsets spontaneously exhibited high levels of XBP-1 splicing relative to unactivated T and B lymphocytes (Fig. 2 B). We also examined the transformed mouse DC line CY15 (31) and found that it also exhibited high levels of XBP-1 splicing. Protein levels of the spliced version of XBP in DCs and the CY15 DC line were determined by Western blot analysis and were elevated, consistent with the mRNA expression and splicing experiments (Fig. 2 C). These results stand in marked contrast to the low levels of XBP-1 mRNA and absent XBP-1 splicing observed in mature B cells before their differentiation to plasma cells (11). However, like the XBP-1–deficient terminally differentiated B cells and pancreatic acinar cells (10, 32), pDCs without XBP-1 lacked the highly elaborate rough ER with multiple layers of closely spaced cisternae seen in WT pDCs; instead, the ER in the mutant pDCs was poorly developed and had few, disorganized cisternae (Fig. 2 D). Further, such XBP-1–deficient pDCs had markedly reduced production of the major pDC cytokine, IFN-α (Fig. 2 E).
Figure 2.
Constitutive expression and splicing of XBP-1 in DCs. (A) Enriched low density spleens cell were sorted for CD3+, CD19+, CD11c+, and CD11c+B220+ (DX5−) cell subsets. Purified subsets and the transformed DC line CY15 were analyzed by RT-PCR using XBP-1 and β-actin–specific probes. *, P < 0.05 when comparing with all other groups. The data represent mean values ± SD from three independent experiments. (B) Total RNA from purified subsets (shown in A) and CY15 cells was isolated for RT-PCR analysis. Primers spanning the splice junction in mice Xbp-1 were used to amplify products of unspliced and spliced mRNA. PCR products were separated by electrophoresis on a 3% agarose gel and visualized by ethidium bromide staining. (C) Immunoblot analysis of XBP-1s protein in CD11c+ and CY15 cells and splenic B cells stimulated with LPS. Results are representative of three separate experiments. (D) Electron micrographs of FACS-sorted WT and XBP-1−/− CD11c+B220+ pDCs (arrows indicate ER cisternae). (E) IFN-α production of FACS-sorted WT and XBP-1−/− CD11c+B220+ pDCs in response to 1 μM CpG for 24 h. The data represent the mean + SEM (n = 3). *, P < 0.002 when comparing WT with XBP-1−/− samples. Bars, 50 nm.
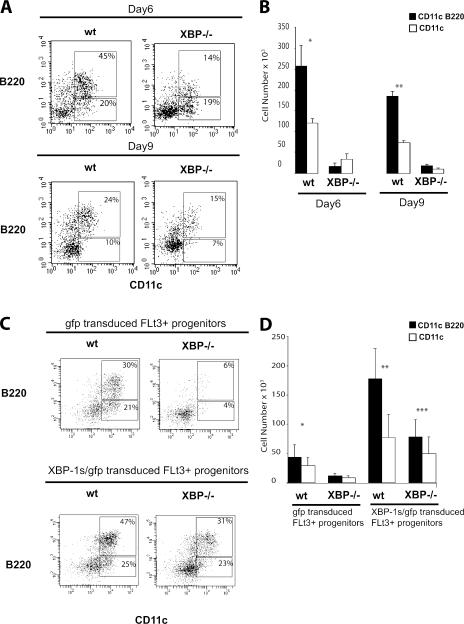
The chimeric mice used in this study were produced by the injection of XBP-1−/− embryonic stem cells into C57BL/6–RAG-2−/− blastocysts. Hence, it was likely that the defect found in DCs and pDCs was cell autonomous for XBP-1−/− hematopoietic precursor cells. Therefore, we considered that high basal level expression of XBP-1s might influence DC development or survival. We tested this hypothesis by using a Flt3L-based in vitro culture system to generate DCs. Lin−IL-7Rα−Thy1.1−c-Kit+Ly9+ BM cells were sorted from 129/RAG2−/− and XBP-1/RAG-2−/− chimeric mice. BM cultures were analyzed for cell number and CD11c+ cell production at days 6 and 9. As expected, phenotypic analysis showed that WT BM cultures were able to produce CD11c+ and CD11c+B220+ cells. Consistent with the in vivo data described above, XBP-1−/− BM cultures produced fewer CD11c+ or CD11c+B220+ cells (Fig. 3, A and B). Indeed, XBP-1−/− BM cultures were mainly composed of cells that stained exclusively for CD11c+ but not cells expressing both CD11c+ and B220 by day 6. By day 9 of culture, XBP-1−/− BM cultures did yield CD11c+ and B220+ cells; however, the numbers were greatly reduced (Fig. 3, A and B).
Figure 3.
Decreased development and survival of XBP-1−/− DCs. (A) c-Kit+lin−–enriched BM cells were sorted and cultured for 6 d in msFlt3L- and SCF-supplemented media. Flow cytometric analysis of WT and XBP-1−/− BM cells stained for CD11c and B220. Numbers in gates represent percentages of gated cells. (B) Graph depicts numbers of WT and XBP-1−/− DCs derived from 105 BM cells cultured with msFlt3L and SCF at day 6. The data represent mean values ± SD from three independent experiments. * and **, P < 0.01 when comparing CD11c and CD11c/B220 WT with XBP-1−/− samples. (C) Flow cytometric analysis of GFP + cells derived from control-GFP– and XBP-1s–GFP–transduced Flt3+ progenitors cultured for 6 d in Flt3L- and SCF-supplemented media. Values represent percentages of total plotted cells. Results are representative of three independent experiments. (D) Graph depicts numbers of pDCs (shaded bars) and DCs (open bars) derived from 0.75 × 105 GFP-transduced Flt3+ progenitors and XBP-1s–GFP–transduced Flt3+ progenitors cultured with Flt3L and SCF for 6 d. Bars represent mean values ± SD from three independent experiments. *, P < 0.04 when comparing WT gfp with XBP-1−/− gfp samples; **, P < 0.04 when comparing WT gfp with WT XBP-1s–gfp samples; ***, P < 0.02 when comparing XBP-1−/− XBP-1s–gfp with WT gfp and XBP-1−/− gfp samples.
A previous study demonstrated that in vitro and in vivo pDC and DC differentiation potential is restricted to Flt3-expressing hematopoietic progenitors (33). To determine if XBP-1 may influence the ability of pDCs and DCs to develop from these committed progenitors, we used a bicistronic retroviral transduction system to transduce the spliced form of mouse XBP-1 into progenitor cells. This approach also enabled us to test whether ectopically expressed XBP-1s in XBP-1−/− progenitors can rescue the developmental defect. Lin−IL-7R−Thy1.1−c-Kit+Ly9+ BM cells were sorted into Flt3− and Flt3+ cell fractions. Of note, there was no difference in the percentage of Flt3+ progenitors between WT and XBP-1−/− BM cells (unpublished data). Therefore, the absence of XBP-1 did not affect differentiation to the Flt3+ progenitor stage. To study the effects of enforced XBP-1 expression on pDC and DC development, Flt3− and Flt3+ progenitors were transduced with control-GFP or XBp-1s–GFP retroviruses and cultured in mouse Flt3L– and stem cell factor (SCF)–supplemented media. Cultures were analyzed for cell number and the presence of pDCs and DCs at day 6. As expected, control GFP-transduced or XBP-1s–GFP–transduced Flt3− progenitors gave rise to no pDCs or DCs (unpublished data). As seen in untransduced progenitors, GFP-transduced WT progenitors produced significantly greater numbers of both pDCs and DCs when compared with GFP-transduced XBP-1−/− BM progenitors (Fig. 3, C and D). Interestingly, XBP-1s–GFP–transduced Flt3+ WT progenitors produced a significantly higher percentage of and total numbers of both pDCs and DCs compared with GFP-transduced Flt3+ progenitors (Fig. 3, C and D). Surprisingly, XBP-1s–GFP–transduced Flt3+ XBP-1−/− progenitors produced significantly higher total numbers of both pDCs and DCs compared with GFP-transduced Flt3+ progenitors from WT and XBP-1−/− BM. Collectively, these results indicate that ectopic expression of XBP-1s into WT and XBP-1−/− Flt3+ hematopoietic progenitors can enhance and rescue the development of pDCs and DCs.
Our results showed that there was an especially marked impairment in the production of the pDC subset. We wondered if XBP-1 might regulate both the development and lifespan of DCs. Therefore, we next examined whether XBP-1 controlled DC survival. Because it has been reported that TLR components of the innate immune system promote DC survival, we examined ligands for two of the best characterized members of the TLR family: the TLR4 ligand LPS, derived from the Gram-negative bacterium Escherichia coli, and the TLR9 ligand CpG DNA16 (34, 35). To examine DC survival, we cultured cells in the absence and presence of TLR stimulation. As shown in Fig. 4 A, without stimulation (nCPG) ∼45% of WT DCs underwent apoptosis at 24 h, as indicated by staining for annexin V and 7-amino-actinomycin D (7-AAD) staining. This was in marked contrast to XBP−/− DCs, which exhibited increased apoptosis with ∼65% cells staining positive for annexin V and 7-AAD at 24 h. In addition, viable cells were counted at 24 h to assess cell survival. Consistent with the increased apoptosis, 46% of WT DCs were still viable at 24 h, whereas only 24% of XBP−/− DCs were still viable at that time. As reported earlier in this paper, treatment with TLR4 and TLR9 ligands increased WT DC survival at 24 h with ∼60–70% survival rate (34, 35). However, XBP−/− DCs were resistant to survival signals given by TLR engagement (Fig. 4 B).
Figure 4.
DC lines with reduced XBP-1 activity are more sensitive to ER stress-induced apoptosis and exhibit decreased survival in vivo. (A) Increased apoptosis of XBP-1−/− DCs. WT and XBP-1−/− DCs. Apoptotic cell death was analyzed by flow cytometric staining with annexin V and 7-AAD. Numbers in gates represent percentages of gated cells. Data are representative of three to four independent experiments. (B) Failure to rescue apoptosis of XBP-1−/− DCs in response to TLR signaling. 3 × 105 WT and XBP-1−/− pDCs were incubated with 1 μM CpG DNA or nCpG DNA, or 1 μg/ml LPS, for 24 h. Viable WT and XBP-1−/− cells were counted after Trypan blue dye exclusion at 24 h. Data are representative of three to four independent experiments. * and **, P < 0.01; and ***, P < 0.025 when comparing WT with XBP-1−/− samples. *, P < 0.01 when comparing WT versus ** and ***; *, NS when comparing XBP-1 versus ** and ***. (C) CY15 cells that express control GFP or dnXBP-1–GFP were treated with the indicated amount of tunicamycin for 24 h. Apoptotic cell death was analyzed by flow cytometric staining with annexin V and 7-AAD. (D) 106 control GFP or dnXBP-1–GFP CY15 cells were injected s.c. into RAG1−/− BALB/c mice. Mice were killed 30 d later, and the percentage of CY15-GFP cells in the respective organs was determined by flow cytometry. The data represent mean values ± SD from three independent experiments. *, P < 0.01; and **, P < 0.05 when comparing with XBP-1−/− samples.
To further test the function of the XBP-1 arm of the UPR in DCs, we generated transformed DC lines functionally deficient in XBP-1 by transducing Cy15 cells with a potent dominant-negative XBP-1 (dnXBP-1) retrovirus or gfp-control retrovirus. Because dnXBP-1 does not possess the C-terminal destabilization motif present in XBP-1u, it is stably expressed and efficiently inhibits XBP-1s–induced transactivation. Suppression of XBP-1 activity by dnXBP-1 markedly inhibits the expression of known XBP-1–dependent UPR target genes such as ERdj4 and p58IPK (36, 37). No effect on cell proliferation was observed at baseline. However, upon treatment with the ER stressor tunicamycin, dnXBP-1 Cy15 cells displayed considerably increased apoptosis when compared with control GFP-transduced Cy15 cells, demonstrating that the IRE1–XBP-1 pathway contributes to the survival of transformed DCs under ER stress conditions (Fig. 4 C). We should note that Cy15 cells transduced with a potent XBP-1 siRNA were difficult to stably maintain in culture, consistent with an essential function for XBP-1 in DC survival.
To test whether blockade of the XBP-1 signaling pathway might alter tumor growth in vivo, CY15-gfp and CY15–dnXBP-1 cells were injected into BALB/c-RAG2−/− mice. By 4 wk, all animals injected with 106 CY15-gfp cells had developed large numbers of macroscopically visible metastases in the spleen, liver, lung, and kidney. In contrast, the growth and metastasis of CY15 cells expressing the dnXBP-1 protein were greatly decreased. Although macroscopically visible metastases were present, the number of nodules was markedly reduced, and by flow cytometry, the number of malignant CY15–dnXBP-1 cells was ∼10–20-fold less than the control CY15-gfp–transduced cells in the various tissues examined (Fig. 4 D). Consistent with the in vitro studies in Fig. 4 C, injection of XBP-1 siRNA knockdown Cy15 cells led to the outgrowth of metastases containing XBP-1 siRNA–nonexpressing Cy15 cells.
In the present study, we provide the first evidence for a critical function of the ER stress response in the survival of DCs. Chimeric mice lacking the UPR transcription factor XBP-1 had markedly reduced numbers of the four DC subsets, with the most severe reduction seen in the pDC compartment. Our data suggested that the disappearance of XBP-1–deficient DCs reflected an increased sensitivity to apoptotic cell death during differentiation, two processes that are intimately linked to the lifespan of the DC and play a key role in controlling immune responses (25). In addition, retroviral transduction of the spliced form of mouse XBP-1 into progenitor cells into WT and XBP-1−/− Flt3+ hematopoietic progenitors not only enhanced pDC and DC development in WT progenitors but also rescued the development of pDCs and DCs in XBP-1−/− progenitors. These results provide strong evidence for a key function of XBP-1 in pDC and DC differentiation. The unexpected finding that IRE1 was constitutively active in freshly isolated DCs led to the prediction that its inhibition might result in increased apoptosis of transformed DCs. Indeed, the growth and metastasis of a malignant DC line was greatly compromised when XBP-1 activity was reduced. This is consistent with previous results from our laboratory that have shown the proapoptotic proteins BAX and BAK as essential components of the UPR that interact with the IRE1 cytosolic domain under stress conditions to modulate IRE1 signaling. However, levels of transcripts encoding BCL-2 family members and BH3-only proteins were normal in XBP-1−/− splenic DCs (unpublished data).
The extraordinarily high levels of XBP-1 transcripts in pDCs and, more importantly, the constitutive UPR activation, as indicated by the presence of abundant XBP-1s protein in DCs, is without precedent in other tissues. This was particularly noticeable in the pDC subset and manifested by the more profound reduction of pDCs compared with conventional DCs. pDCs are a distinct DC population with spherical plasmacytoid morphology that, interestingly, resembles plasma cells, the only known lymphoid cells that require XBP-1. During B cell differentiation to the plasma cell, a remodeling process occurs in which the ER undergoes massive expansion to accommodate the large quantities of newly synthesized Ig for secretion. Similarly, after activation by bacterial DNA or by viral infection, pDCs develop multiple cytoplasmic extensions and an expanded ER, and produce large amounts of IFN-α. Therefore, it makes sense that a vigorous UPR is required in this cell; however, it functions at an earlier stage, as indicated by constitutive UPR activation, suggesting that the pDC is being readied for prompt IFN-α secretion in the setting of viral infection. Indeed, ER expansion and IFN-α levels are diminished in XBP-1–deficient pDCs (Fig. 2, D and E). Although conventional DCs are not known to possess plasmacytoid morphology or secrete excessive cytokines, basal level XBP-1 splicing demonstrates considerable activation of this vital arm of the UPR. Our data suggest that a basal XBP-1 activation state allows for optimal DC function. In the absence of XBP-1, DC homeostasis is thus dysregulated, resulting in impaired survival during unstimulated and stimulated conditions. As mentioned earlier, at this point in our studies we have no evidence to support cross talk between the ER stress and TLR signaling pathways in DCs via protein load, as TLR stimulation of pDCs and DCs fails to either increase XBP-1 mRNA or splicing. Hence, we suggest that the XBP-1 survival pathway acts at an earlier stage and is distinct from the TLR-induced NF-κB–dependent pathway. Consistent with this function at an earlier stage is the inability of TLR engagement to rescue the impaired survival of XBP-1–deficient DCs. In these studies, we only provide evidence for the effect of XBP-1 deficiency on cell death induced by the ER stress response; future studies will aim to examine other forms of DC survival/death. Identifying the factors that evoke constitutive UPR activation before DC maturation, excessive protein secretion, and ER expansion will be of great interest. This pathway offers a novel target for the development of small molecules that can extend or shorten the lifespan of a DC to achieve tolerance or immunity and that may be therapeutic in the setting of malignant histiocytosis.
MATERIAL AND METHODS
Mice.
129SV6, C57BL/6, and BALB/c RAG1−/− mice were purchased from Taconic and used at 6–8 wks of age. The generation and screening of XBP-1/RAG-2−/− chimeric mice were done as previously described (1). All mice were housed in a pathogen-free facility at the Harvard School of Public Health, and all animal studies were performed according to institutional and National Institutes of Health guidelines for animal use and care.
Reagents and cell lines.
The sequences of phosphorothioate ODNs (QIAGEN) are as follows: TCCATGACGTTCCTGATGCT for stimulatory CpG ODN and TTCATGAGCTTCCTGATGCT for control (nCpG) ODN. Endotoxin-free LPS (InvivoGen). The CY15 DC line was provided by T. Blankenstein (Institute of Immunology, Berlin, Germany).
Purification and isolation of DCs.
DCs were isolated as previously described (38). In brief, CD11c+ DCs were isolated from collagenase-treated spleen preparations and enriched by centrifugation in a cell separation medium (Accudenz; Accurate Chemicals). B lymphocytes and NK cells were depleted by using coated magnetic beads (anti-CD19 and anti-DX5, respectively) before FACS sorting (Miltenyi Biotec). DC subsets were sorted and analyzed using a FACSAria (Becton Dickinson) and DIVA software (Becton Dickinson).
Flow cytometry.
Cells were stained with the following mouse reactive antibodies: anti-CD16/32 (2.4G2); anti–MHC class II (IA/IE; 2G9); anti-CD8α (53-6.7); anti-CD11c (HL3); anti-CD4 (GK1.5); anti-B220 (RA3-6B2); anti-CD11b (M1/70); anti-CD86 (GL1); and pan-NK (DX5). To distinguish host from donor DCs in chimaeras, we used the surface marker LY9.1 (30C7), carried by the 129 but not the B6 strain (39). For apoptosis assays, cells were stained with PE-conjugated annexin V and 7-AAD (BD Biosciences) according to the manufacturer's instructions. Analyses of stained cells were performed on a FACSCalibur (BD Bioscience) with CellQuest software (BD Bioscience). All antibodies were purchased from BD Bioscience.
In vitro DC differentiation and survival assay.
Hematopoietic progenitors were isolated as previously described (33), with minor changes. BM cells were isolated and enriched for c-Kit+ cells by using APC-conjugated c-Kit antibodies (BD Biosciences) and APC microbeads (Miltenyi Biotec). Cells were then stained with mAbs for lineage markers (CD3ε, 145-2C11; CD4, GK1.5; CD8, 53-6.7; B220, RA3-6B2; CD19, MB19-1; CD11b, M1/70; Gr-1, RB6-8C5; and TER119, TER119), IL-7Rα− (A7R34), and Thy1.1− (19XE5; BD Biosciences). LY9+c-Kit+Lin− BM progenitors were sorted and analyzed using a FACSAria with DIVA software. For XBP-1/RAG-2−/− chimeric mice, the embryonic stem cells were derived from the 129 strain. Therefore, we used the surface marker Ly9.1, which is an allele that is carried by the 129 but not the B6 strain (Rag2−/− background) to purify donor from host progenitors. Progenitors were cultured in RPMI 1640 supplemented with 10% FCS, 10−4 M 2-βME, sodium pyruvate, and antibiotics (100 ng/ml msFlt3L and 10 ng/ml msSCF; R&D Systems). Media was replaced every 3 d with new media and cytokines. For survival assays, 3 × 105 FACS-sorted CD11c+ cells per well were plated at 1 μM CpG DNA or nCpG DNA, or 1 μg/ml LPS, for 24 h. Apoptosis assays using annexin V and 7-AAD were performed as described in Flow cytometry and expressed as the percentage of apoptotic cells. DC survival was quantified by using Trypan blue dye exclusion at 24 h. Statistical significance was evaluated by using the Student's t test.
Retroviral transduction of hematopoietic progenitors.
Hematopoietic progenitors were transduced as previously described (33), with minor changes. The full-length mouse XBP-1s RV-gfp was used to make retrovirus supernatants, as previously described (11). Supernatants were used to transduce GP+E-86 cells. After 48 h, the GFP-expressing GP+E-86 cells were FACS sorted and expanded. For transduction of FLT3R+c-Kit+Lin− BM progenitor cells, GP+E-86 cells were irradiated (20 Gy) and plated in 24-well plates at 0.75 × 105 cells per well for 24 h. Sorted progenitor cells were transduced by co-culture with GP+E-86 for 18 h in IMDM (Invitrogen) containing 2% FCS, 4 μg/ml polybrene (Sigma-Aldrich), 100 ng/ml mouse Flt3L− protein (R&D Systems), 10 ng/ml mSCF (R&D Systems), and 10 ng/ml mIL-11 (R&D Systems). Transduced cells were removed by gently dispersing and placed in Flt3L differentiation cultures.
RNA, ELISA, and protein analysis.
Total RNA was isolated from tissues using TRIzol reagent (Invitrogen). cDNA was synthesized from RNA samples using the iScript cDNA synthesis kit containing oligo (dT) and random hexamer primers (Bio-Rad Laboratories). Quantitative real-time PCR reactions using SYBR green fluorescent reagent were run in an ABI PRISM 7700 system (Applied Biosystems). The relative amounts of mRNAs were calculated from the comparative threshold cycle (Cτ) values by using β-actin as control. Primer sequences designed by Primer Express software (Applied Biosystems) have been previously described (10). Whole-cell lysates of DCs were prepared in RIPA buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS). Western blot analysis was performed as previously described (11), with anti–XBP-1 pAb. ELISA for IFN-α was done by using 5 μg/ml of capture mAb, 200 U/ml of secondary pAb (PBL Biomedical Laboratories), and a 1:1,000 dilution of alkaline phosphatase–conjugated anti–rabbit IgG (Jackson ImmunoResearch Laboratories). Statistical significance was evaluated using the Student's t test.
Electron microscopy.
Cells were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate, pH 7.4, for 1 h at room temperature. After washing in cacodylate buffer, cells were postfixed for 1 h in 1% osmium tetroxide in the same buffer at room temperature, washed again, and stained “en bloc” in 2% uranyl acetate in 0.05 M sodium maleate, pH 5.2, for 1 h at room temperature. After dehydration in a graded series of ethanol, cells were embedded in epoxy resin (Embed 812; Electron Microscopy Sciences). Ultrathin sections were cut on a ultramicrotome (Reichert), collected on formvar- and carbon-coated grids, stained with 2% uranyl acetate and lead citrate, and viewed on an electron microscope (Tecnai 12 BioTWIN; FEI Company).
In vitro and in vivo growth of CY15 cells.
Retroviral transduction to produce GFP and dnXBP-1–GFP CY15 cells was performed as previously described (36). Assessment of metastasis of CY15-GFP and dnXBP-1 cells was performed as previously described (31), with minor modifications. In brief, 106 CY15 cells were injected s.c. into RAG1−/− BALB/c mice, and mice were killed 30 d later. The liver, lung and kidney, and spleen were harvested and digested with collagenase treatment (as described for DCs) for 4 h at 37°C with occasional shaking. The percentage of CY15-GFP cells in each organ was determined by flow cytometry on a FACSCalibur with CellQuest software. Statistical significance was evaluated using the Student's t test.
Acknowledgments
We thank Ira Mellmann for his contribution to the transmission electron microscopy on the pDCs, Ann-Hwee Lee and Marc Wein for thoughtful comments on the manuscript, Claudio Hetz for valuable technical advice, Landy Kangaloo and Jacobo Ramirez for technical assistance and animal care, and Ryan Ghan for expert manuscript preparation.
This work was supported by National Institutes of Health grants AI32412 and AI56296 (to L.H. Glimcher) and a fellowship award from the Irvington Foundation (to N.N. Iwakoshi).
The authors have no conflicting financial interests.
References
- 1.Reimold, A.M., N.N. Iwakoshi, J. Manis, P. Vallabhajosyula, E. Szomolanyi-Tsuda, E.M. Gravallese, D. Friend, M.J. Grusby, F. Alt, and L.H. Glimcher. 2001. Plasma cell differentiation requires transcription factor XBP-1. Nature. 412:300–307. [DOI] [PubMed] [Google Scholar]
- 2.Lipson, K.L., S.G. Fonseca, S. Ishigaki, L.X. Nguyen, E. Foss, R. Bortell, A.A. Rossini, and F. Urano. 2006. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 4:245–254. [DOI] [PubMed] [Google Scholar]
- 3.Koumenis, C. 2006. ER stress, hypoxia tolerance and tumor progression. Curr. Mol. Med. 6:55–69. [DOI] [PubMed] [Google Scholar]
- 4.Paschen, W., and T. Mengesdorf. 2005. Endoplasmic reticulum stress response and neurodegeneration. Cell Calcium. 38:409–415. [DOI] [PubMed] [Google Scholar]
- 5.Liou, H.C., M.R. Boothby, P.W. Finn, R. Davidon, N. Nabavi, N.J. Zeleznik-Le, J.P. Ting, and L.H. Glimcher. 1990. A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science. 247:1581–1584. [DOI] [PubMed] [Google Scholar]
- 6.Shen, X., R.E. Ellis, K. Lee, C.Y. Liu, K. Yang, A. Solomon, H. Yoshida, R. Morimoto, D.M. Kurnit, K. Mori, and R.J. Kaufman. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 107:893–903. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891. [DOI] [PubMed] [Google Scholar]
- 8.Calfon, M., H. Zeng, F. Urano, J.H. Till, S.R. Hubbard, H.P. Harding, S.G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. [DOI] [PubMed] [Google Scholar]
- 9.Reimold, A.M., A. Etkin, I. Clauss, A. Perkins, D.S. Friend, J. Zhang, H.F. Horton, A. Scott, S.H. Orkin, M.C. Byrne, et al. 2000. An essential role in liver development for transcription factor XBP-1. Genes Dev. 14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, A.H., G.C. Chu, N.N. Iwakoshi, and L.H. Glimcher. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24:4368–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwakoshi, N.N., A.H. Lee, P. Vallabhajosyula, K.L. Otipoby, K. Rajewsky, and L.H. Glimcher. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321–329. [DOI] [PubMed] [Google Scholar]
- 12.McKenna, H.J., K.L. Stocking, R.E. Miller, K. Brasel, T. De Smedt, E. Maraskovsky, C.R. Maliszewski, D.H. Lynch, J. Smith, B. Pulendran, et al. 2000. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 95:3489–3497. [PubMed] [Google Scholar]
- 13.Maraskovsky, E., K. Brasel, M. Teepe, E.R. Roux, S.D. Lyman, K. Shortman, and H.J. McKenna. 1996. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J. Exp. Med. 184:1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caux, C., B. Vanbervliet, C. Massacrier, C. Dezutter-Dambuyant, B. de Saint-Vis, C. Jacquet, K. Yoneda, S. Imamura, D. Schmitt, and J. Banchereau. 1996. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to GM-CSF + TNF-α. J. Exp. Med. 184:695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sallusto, F., and A. Lanzavecchia. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J. Exp. Med. 179:1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagai, Y., K.P. Garrett, S. Ohta, U. Bahrun, T. Kouro, S. Akira, K. Takatsu, and P.W. Kincade. 2006. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 24:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu, L., A. D'Amico, K.D. Winkel, M. Suter, D. Lo, and K. Shortman. 1998. RelB is essential for the development of myeloid-related CD8alpha− dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 9:839–847. [DOI] [PubMed] [Google Scholar]
- 18.Rathinam, C., R. Geffers, R. Yucel, J. Buer, K. Welte, T. Moroy, and C. Klein. 2005. The transcriptional repressor Gfi1 controls STAT3-dependent dendritic cell development and function. Immunity. 22:717–728. [DOI] [PubMed] [Google Scholar]
- 19.Hacker, C., R.D. Kirsch, X.S. Ju, T. Hieronymus, T.C. Gust, C. Kuhl, T. Jorgas, S.M. Kurz, S. Rose-John, Y. Yokota, and M. Zenke. 2003. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 4:380–386. [DOI] [PubMed] [Google Scholar]
- 20.Tamura, T., P. Tailor, K. Yamaoka, H.J. Kong, H. Tsujimura, J.J. O'Shea, H. Singh, and K. Ozato. 2005. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 174:2573–2581. [DOI] [PubMed] [Google Scholar]
- 21.Schiavoni, G., F. Mattei, P. Sestili, P. Borghi, M. Venditti, H.C. Morse III, F. Belardelli, and L. Gabriele. 2002. ICSBP is essential for the development of mouse type I interferon–producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med. 196:1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson, K.L., H. Perkin, C.D. Surh, S. Venturini, R.A. Maki, and B.E. Torbett. 2000. Transcription factor PU.1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J. Immunol. 164:1855–1861. [DOI] [PubMed] [Google Scholar]
- 23.Laouar, Y., T. Welte, X.Y. Fu, and R.A. Flavell. 2003. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 19:903–912. [DOI] [PubMed] [Google Scholar]
- 24.Zenke, M., and T. Hieronymus. 2006. Towards an understanding of the transcription factor network of dendritic cell development. Trends Immunol. 27:140–145. [DOI] [PubMed] [Google Scholar]
- 25.Josien, R., H.L. Li, E. Ingulli, S. Sarma, B.R. Wong, M. Vologodskaia, R.M. Steinman, and Y. Choi. 2000. TRANCE, a tumor necrosis factor family member, enhances the longevity and adjuvant properties of dendritic cells in vivo. J. Exp. Med. 191:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong, B.R., R. Josien, S.Y. Lee, B. Sauter, H.-L. Li, R.M. Steinman, and Y. Choi. 1997. TRANCE (tumor necrosis factor [TNF]–related activation-induced cytokine), a new TNF family member predominantly expressed in T cells, is a dendritic cell–specific survival factor. J. Exp. Med. 186:2075–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouaaz, F., J. Arron, Y. Zheng, Y. Choi, and A.A. Beg. 2002. Dendritic cell development and survival require distinct NF-κB subunits. Immunity. 16:257–270. [DOI] [PubMed] [Google Scholar]
- 28.Ardeshna, K.M., A.R. Pizzey, S. Devereux, and A. Khwaja. 2000. The PI3 kinase, p38 SAP kinase, and NF-kappaB signal transduction pathways are involved in the survival and maturation of lipopolysaccharide-stimulated human monocyte-derived dendritic cells. Blood. 96:1039–1046. [PubMed] [Google Scholar]
- 29.Hetz, C., P. Bernasconi, J. Fisher, A.H. Lee, M.C. Bassik, B. Antonsson, G.S. Brandt, N.N. Iwakoshi, A. Schinzel, L.H. Glimcher, and S.J. Korsmeyer. 2006. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 312:572–576. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y.J. 2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23:275–306. [DOI] [PubMed] [Google Scholar]
- 31.Kammertoens, T., R. Willebrand, B. Erdmann, L. Li, Y. Li, B. Engels, W. Uckert, and T. Blankenstein. 2005. CY15, a malignant histiocytic tumor that is phenotypically similar to immature dendritic cells. Cancer Res. 65:2560–2564. [DOI] [PubMed] [Google Scholar]
- 32.Shaffer, A.L., M. Shapiro-Shelef, N.N. Iwakoshi, A.H. Lee, S.B. Qian, H. Zhao, X. Yu, L. Yang, B.K. Tan, A. Rosenwald, et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. [DOI] [PubMed] [Google Scholar]
- 33.Onai, N., A. Obata-Onai, R. Tussiwand, A. Lanzavecchia, and M.G. Manz. 2006. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon–producing and dendritic cell development. J. Exp. Med. 203:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, Y., S.W. Lee, and Y.C. Sung. 2002. Cutting Edge: CpG DNA inhibits dendritic cell apoptosis by up-regulating cellular inhibitor of apoptosis proteins through the phosphatidylinositide-3′-OH kinase pathway. J. Immunol. 168:5–8. [DOI] [PubMed] [Google Scholar]
- 35.Rescigno, M., M. Martino, C.L. Sutherland, M.R. Gold, and P. Ricciardi-Castagnoli. 1998. Dendritic cell survival and maturation are regulated by different signaling pathways. J. Exp. Med. 188:2175–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, A.H., N.N. Iwakoshi, K.C. Anderson, and L.H. Glimcher. 2003. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA. 100:9946–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, A.H., N.N. Iwakoshi, and L.H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lugo-Villarino, G., S.C. Ito, D.M. Klinman, and L.H. Glimcher. 2005. The adjuvant activity of CpG DNA requires T-bet expression in dendritic cells. Proc. Natl. Acad. Sci. USA. 102:13248–13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ting, C.N., M.C. Olson, K.P. Barton, and J.M. Leiden. 1996. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 384:474–478. [DOI] [PubMed] [Google Scholar]