Abstract
The key attributes of CD8+ T cell protective immunity in human immunodeficiency virus (HIV) infection remain unclear. We report that CD8+ T cell responses specific for Gag and, in particular, the immunodominant p24 epitope KK10 correlate with control of HIV-1 replication in human histocompatibility leukocyte antigen (HLA)–B27 patients. To understand further the nature of CD8+ T cell–mediated antiviral efficacy, we performed a comprehensive study of CD8+ T cells specific for the HLA-B27–restricted epitope KK10 in chronic HIV-1 infection based on the use of multiparametric flow cytometry together with molecular clonotypic analysis and viral sequencing. We show that B27-KK10–specific CD8+ T cells are characterized by polyfunctional capabilities, increased clonal turnover, and superior functional avidity. Such attributes are interlinked and constitute the basis for effective control of HIV-1 replication. These data on the features of effective CD8+ T cells in HIV infection may aid in the development of successful T cell vaccines.
Virus-specific CD8+ T cells are central players in the fight against HIV, yet their sole presence, even in large numbers, is not directly correlated with better control of HIV replication. Indeed, increasing evidence suggests that qualitative rather than quantitative aspects of CD8+ T cell immunity provide the key to antiviral efficacy (1, 2). Despite intense efforts, however, the precise attributes that confer certain CD8+ T cell populations with an advantage in controlling HIV remain unclear; understanding these issues is necessary for the rational design of effective T cell–based vaccines.
Multiple factors, including immunologic, genetic, viral, and environmental, can potentially contribute to the rate of HIV disease progression in the absence of antiretroviral treatment. Furthermore, the virus-specific CD8+ T cell response represents a constellation of constituent populations with respect to HLA restriction elements, targeted epitopes, and clonal diversity, even within a single infected individual. This profound heterogeneity hinders the accurate identification of CD8+ T cell characteristics associated with protection. However, the study of well-defined, more homogeneous CD8+ T cell populations known to have a protective role in HIV infection might facilitate the search for such protective attributes.
The HLA class I molecules B*2705 (B27) and B*5701 (B57) are consistently linked with slower rates of HIV-1 disease progression, but the basis for these associations remains unclear (3–5). Recent work suggests an active immunodomination of HLA-B27– and HLA-B57–restricted CD8+ T cell responses over other HLA allotypes (6); these two HLA molecules restrict >65% of the total HIV-1–specific CD8+ T cell response in individuals expressing these alleles (7). Remarkably, although HLA-B57–restricted CD8+ T cell populations can present several HIV-derived epitopes, the HLA-B27–restricted CD8+ T cells in HIV-1 patients usually target one particular immunodominant epitope in p24 Gag (residues 263–272), namely B27-KK10 (7, 8). Furthermore, the emergence of escape mutations within this immunodominant epitope coincides with increased viral replication and progression to AIDS (9, 10); in contrast, HLA-B57 individuals are able to control the virus even after HIV has escaped from the dominant B57-restricted CD8+ T cell response (11). Although it remains to be formally demonstrated, these data suggest that a single response to the immunodominant B27-KK10 epitope may account for the association of HLA-B*2705 with prolonged AIDS-free survival in HIV infection. Therefore, the study of B27-KK10–restricted CD8+ T cell characteristics represents a unique opportunity to explore the basis for the protective nature of virus-specific CD8+ T cells in HIV infection.
In this paper, we present a comprehensive study of B27-KK10–specific CD8+ T cells in comparison with HIV-specific cells restricted by other HLA class I molecules in treatment-naive HIV-infected slow or nonprogressors. For this purpose, we used multiparametric flow cytometry together with molecular clonotypic analysis to directly assess several attributes of HIV-specific CD8+ T cell populations ex vivo. Specifically, we examined the phenotype (differentiation status and replicative senescence), functional profile (effector cytokine production and degranulation), clonal diversity (TCRB gene usage), and antigen sensitivity (functional avidity) of epitope-specific CD8+ T cell populations. The data indicate a clear relationship between these parameters that distinguishes protective CD8+ T cell responses.
RESULTS
HLA-B27–restricted Gag-specific CD8+ T cells and superior control of HIV-1 replication
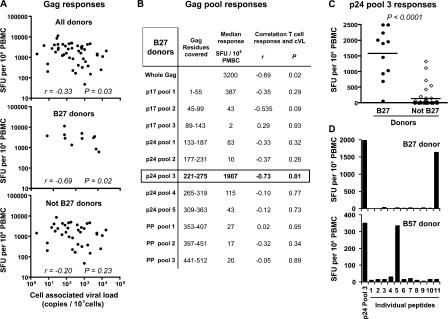
A recent population-based report has highlighted the stronger association of Gag-specific CD8+ T cell responses—compared with other HIV protein–specific responses—with the control of HIV-1 replication (12). To address further the potential importance of Gag-specific CD8+ T cells restricted through HLA-B27 in this association, whole Gag- or p24-specific CD8+ T cell magnitude was assessed, and its correlation with cell-associated HIV–DNA viral load (cVL) was analyzed in a cohort of untreated slow progressors (n = 47), which includes 11 HLA-B*2705 donors. cVL represents the number of infected cells harboring HIV-DNA, the direct targets of HIV-specific CD8+ T cells, and has been reported as a more sensitive marker of disease progression than plasma HIV–RNA load (pVL), at least once the viral setpoint is reached (13). As previously described (14), there was a modest but significant inverse correlation between Gag-specific CD8+ T cell numbers and cVL in these donors (Fig. 1 A). Interestingly, this inverse correlation became more significant if considering exclusively HLA-B27 donors and was lost when HLA-B27 donors were excluded from the analysis. The dissection of the total Gag-specific response according to the different peptide pools covering the Gag sequence revealed that the association between CD8+ T cell responses and cVL in HLA-B27 donors was held exclusively in p24 peptide pool 3, which also yielded the strongest responses compared with the other pools (Fig. 1 B). Among all donors, HLA-B27 patients presented particularly strong responses to this pool (Fig. 1 C); the few non–HLA-B27 donors presenting reactivity to pool 3 were found to be HLA-B57. Pool 3 covers aa 221–275 of Gag and, therefore, includes the HLA-B27–restricted immunodominant KK10 epitope (aa 263–272). To ensure that responses to pool 3 in HLA-B27 donors were indeed targeted to KK10, peptides from this pool were tested individually. Although pool 3–responding HLA-B57 donors targeted peptide 5 (which contains the previously identified HLA-B57 epitope TW10), HLA-B27 donors targeted peptide 11 (aa 261–275), which indeed contains KK10 (Fig. 1 D). These observations illustrate the weight held by certain CD8+ T cell populations (e.g., recognizing a single epitope) in associations between T cell responses and viral load. Together with the evidence from the literature (i.e., association between HLA-B*2705 and prolonged AIDS-free survival, as well as the immunodominance of B27-KK10–specific CD8+ T cells with the p24 Gag-specific populations) (3–10), these data provide strong support for the protective role played by KK10-specific CD8+ T cell responses in HLA-B27 donors and emphasize the importance of detailed analyses of this population to discover attributes associated with its antiviral efficacy.
Figure 1.
Association between Gag-specific CD8+ T cell responses and viral load in HLA-B27 individuals. (A) Total CD8+ T cells responses specific for the Gag protein were assessed in IFN-γ ELISPOT assays by stimulation with overlapping peptides of PBMCs from 47 antiretroviral therapy–naive HIV-infected donors, including 11 HLA-B27 donors. Magnitude (in SFU per 106 PBMCs) of responding cells specific for Gag was plotted as a function of cVL for all 47 donors, the 11 HLA-B27 donors, and the 36 non–HLA-B27 donors. Correlations were determined using the Spearman's rank test. (B) Subdivision of Gag-specific responses from HLA-B27 donors according to peptide pools. Median responses for the 11 HLA-B27 donors are shown for each pool, and correlations between T cell frequencies and cVL were calculated. (C) Frequency of T cells responding to p24 pool 3 in HLA-27 and non–HLA-B27 donors. Horizontal bars represent means. The p-value was calculated by the Mann-Whitney U test. (D) Representative examples of HLA-B27 and HLA-B57 donor responses to individual peptides (n = 11) included in p24 pool 3. Peptide 11 covers Gag residues 261–275. Gag-specific responses were assessed in duplicate, and CD8+ T cell–depleted PBMCs were used to verify that observed responses were CD8+ T cell mediated.
B27-KK10–specific CD8+ T cells display a polyfunctional profile
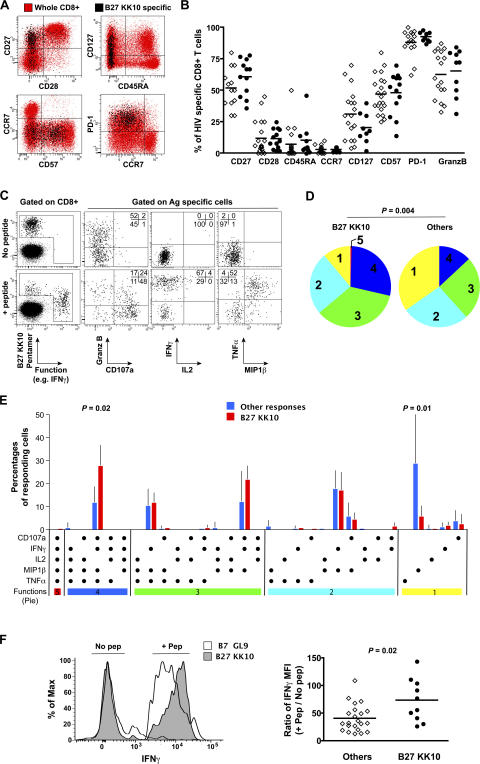
Because of the development of recombinant MHC class I–peptide complex technologies to identify HIV-specific CD8+ T cells physically, several studies have sought to find a relationship between the phenotype of such cells and their capacity to control viral replication. Skewed maturation, or the failure to reach advanced stages of differentiation (i.e., a CD45RA+/CD27− phenotype), has been proposed as one reason for the inability of CD8+ T cells to halt HIV disease progression (15, 16). Loss of IL-7 receptor (CD127) expression on virus-specific CD8+ T cells has also been proposed to indicate cellular impairment in relation to persisting viral replication (17). In addition, recent reports have associated the expression of the inhibitory receptor programmed death–1 (PD-1) on virus-specific CD8+ T cells with failure to control HIV (18, 19). In this study, the comprehensive assessment of a large panel of markers associated with T cell differentiation (CD27, CD28, CC chemokine receptor [CCR] 7, and CD45RA), function (CD127, PD-1, and granzyme B), and replicative senescence (CD57) using polychromatic flow cytometry revealed no significant differences between protective B27-KK10–specific CD8+ T cells and CD8+ T cells specific for other HIV epitopes from a series of 18 HIV-infected donors (Fig. 2, A and B). In both cases, the majority of the cells displayed a typical intermediate differentiation phenotype (CD27+/CD28−/CD45RA−/CCR7−) (20), with strong surface expression of PD-1, various levels of CD127 and CD57, and high intracellular levels of granzyme B.
Figure 2.
Phenotypic and functional assessment of B27-KK10–specific CD8+ T cells. (A) Representative staining for the expression of cell surface markers on B27-KK10–specific CD8+ T cells. HIV-specific cells identified using MHC class I–peptide complexes (black) are superimposed on the whole CD8+ T cell population in the same donor (red). Plots are gated on CD3+CD8+ cells. (B) Comparative marker expression between B27-KK10–specific CD8+ T cells (open diamonds) and other HIV-specific CD8+ T cells (closed circles). Horizontal bars show medians. (C) Representative example of simultaneous multifunctional assessment of B27-KK10–specific CD8+ T cells using nine-color flow cytometry. Cells were stimulated for 6 h in the presence of cognate peptide before intracellular staining. Percentages of cells in the different quadrants are shown. Plots are gated on CD3+CD8+ cells. (D) The pie charts depict the background-adjusted multifunctional behavior (one to five functions: CD107a, IFN-γ, TNF-α, IL-2, and MIP-1β) of B27-KK10–specific CD8+ T cell populations (n = 8) versus other HIV-specific CD8+ T cell populations (n = 7, including 2xA3-RY10, 2xB7-GL9, and 3xA2-SL9). For simplicity, responses are grouped by the number of functions (indicated by the numbers inside the pie charts and matched to the colored bars in E). The p-value was calculated by the permutation test. (E) Detailed functional responsiveness was analyzed for the B27-KK10–specific CD8+ T cell populations compared with the other HIV-specific CD8+ T cell populations. Each bar shows the mean percentages of cells displaying a particular combination of functions within the total of functional cells. p-values were calculated by the Mann-Whitney U test. (F) IFN-γ fluorescence of B27-KK10–specific CD8+ T cells and other HIV-specific CD8+ T cells upon stimulation with cognate peptides. Representative profiles are shown (left), and the IFN-γ MFI ratios (for antigen-specific CD8+ T cells upon stimulation/no activation) are presented for all populations studied (right). Horizontal bars represent means.
Virus-specific CD8+ T cells exert several effector functions, mediated through the production of soluble factors and cytolytic mechanisms, to tackle virus dissemination. We therefore assessed the capacity of B27-KK10–specific CD8+ T cells to produce effector cytokines (IFN-γ, TNF-α, macrophage inflammatory protein [MIP]–1β, and IL-2) and to release cytotoxic factors (up-regulation of CD107a together with degranulation of granzyme B) upon antigenic stimulation (Fig. 2 C). On average, we observed that 70% (±10% SE) of the different antigen-specific CD8+ T cell populations displayed at least one function. Recent data suggest that although the independent assessment of single functions may not be particularly informative, the simultaneous analysis of multiple parameters can provide a better picture of CD8+ T cell functional quality (21). Surprisingly, B27-KK10–specific CD8+ T cells presented a superior multifunctional profile in comparison with other HLA-restricted HIV-specific CD8+ T cells (Fig. 2 D). In particular, they showed significantly higher proportions of IFN-γ+/TNF-α+/MIP-1β+/CD107a+ cells (Fig. 2 E). Detection of IL-2 production was very limited, even by B27-KK10–specific CD8+ T cells. Remarkably, not only were B27-KK10–specific CD8+ T cells multifunctional, but they also produced more IFN-γ on a per-cell basis than CD8+ T cells specific for other HIV epitopes (as seen by measuring IFN-γ median fluorescence intensity (MFI; Fig. 2 F). This observation is consistent with recent studies showing an association between IFN-γ MFI and the degree of T cell polyfunctionality (22, 23). These findings suggest that protective CD8+ T cells are highly functional, which is in keeping with the maintenance of polyfunctional HIV-specific CD8+ T cells in HIV-infected nonprogressors (21).
Clonal dominance and turnover in B27-KK10–specific CD8+ T cell populations
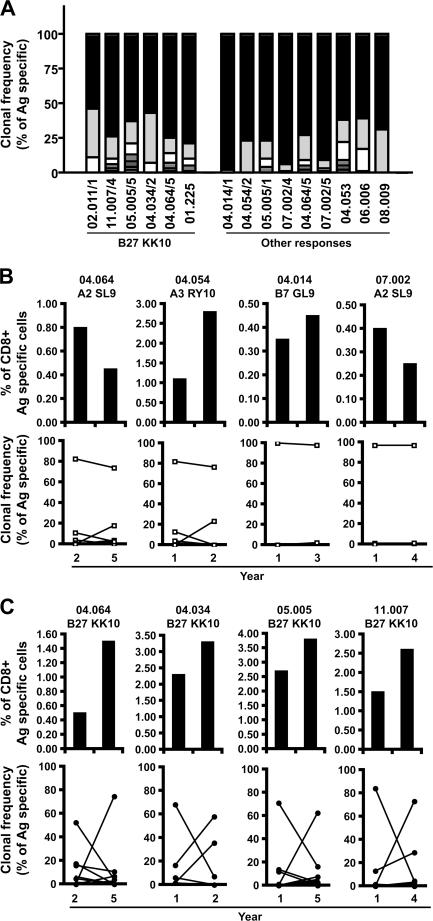
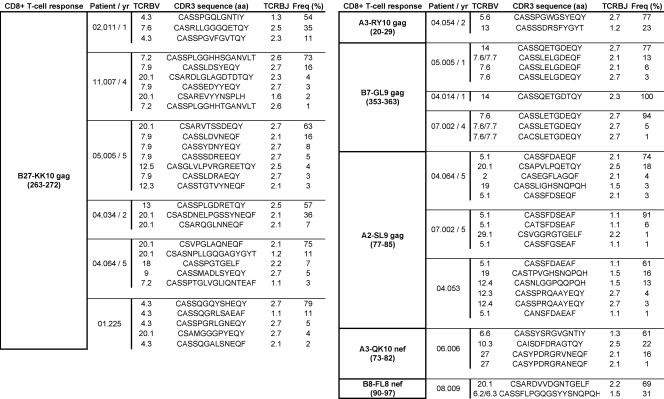
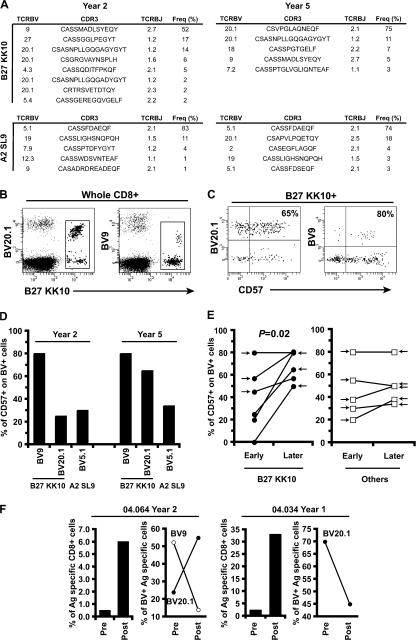
HIV-specific CD8+ T cell populations are generally oligoclonal (24), comprising several clonotypes specific for the same antigenic complex. The recognition properties and antiviral efficacy of such individual clonotypes, defined according to their distinct TCR usage, can vary considerably. To investigate a potential impact for clonotype recruitment in differentiating the bulk properties of HIV-specific CD8+ T cell populations, we performed a detailed molecular analysis of TCRB gene expression (25, 26) within B27-KK10–specific CD8+ T cell populations in comparison with CD8+ T cells specific for other HIV epitopes. Strikingly, we observed the consistent dominance of single clonotypes in all HIV-specific CD8+ T cell populations (Fig. 3 A and Table I). This observation held both for CD8+ T cell populations specific for B27-KK10 and for those specific for other antigens. Moreover, there was no evidence for preferential usage of a particular CDR3 motif among the various B27-KK10–specific CD8+ T cell clonotypes (all TCRB gene sequences are shown in Table S1, available at http://www.jem.org/cgi/content/full/jem.20070784/DC1). This observation suggests that the B27-KK10 complex can be recognized by diverse clonotypes with different binding modalities (27) and argues against the consistent selection of a particular TCR that confers exceptional properties.
Figure 3.
High turnover of B27-KK10–specific CD8+ T cell clonotypes. (A) Clonal composition of several CD8+ T cell populations specific for B27-KK10 or other epitopes. Clonotypic analysis was performed on FACS-sorted viable HIV-specific CD8+ T cells. The percent frequencies of single clonotypes within B27-KK10 or other HIV epitope–specific CD8+ T cell populations are shown; each bar represents one population from one donor and is subdivided into distinct clonotypes represented according to frequency. 6 B27-KK10 and 9 out of 13 other HIV epitope–specific CD8+ T cell populations are shown. Donor identification is provided under the bars, and a detailed clonotypic analysis (with sequences) is shown in Table I and Table S1. (B and C) Analysis of clonal dominance over two time points. The percent frequencies of HIV-specific CD8+ T cells, identified by cognate MHC class I–peptide multimeric complexes, within the total CD8+ population and clonal composition, represented as the frequency of individual clonotypes within sorted HIV-specific CD8+ T cell populations, are shown for four B27-KK10 (C) and four other HIV peptide–specific CD8+ T cell populations (B) separated by the indicated intervals. Donor identification and specific responses are indicated; a detailed clonotypic analysis with sequences is shown in Table S1.
Table I.
Clonal analysis of HIV-specific CD8+ T cell populations
BV usage, CD3 aa sequence, BJ usage, and percent frequency for 6 B27-KK10 and 9 out of 13 other HIV epitope–specific CD8+ T cell populations. The entire set of data is shown in Table S1.
We next wanted to assess the temporal pattern of clonal dominance. For this purpose, we analyzed historical cryopreserved samples from seven HIV-infected donors (Table II), from which four B27-KK10–specific CD8+ T cell populations versus four CD8+ T cell populations specific for other HIV epitopes could be studied at two different time points. A remarkable dichotomy was observed. Although the clonal dominance of CD8+ T cells specific for non–B27-KK10 epitopes did not alter over time (Fig. 3 B), the dominant clonotype in the B27-KK10–specific CD8+ T cell populations changed consistently in all four matched donors (Fig. 3 C); these differential dynamics occurred despite relatively limited variations in the magnitude of the respective HIV-specific T cell populations. These data suggest that B27-KK10–specific CD8+ T cell populations are characterized by a higher rate of clonal turnover compared with other HIV-specific CD8+ T cell populations.
Table II.
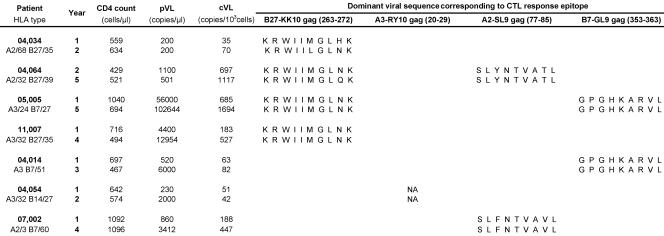
Patient characteristics and viral sequences
Clinical characteristics of HIV-1–infected patients and dominant (i.e., >80%) HIV-1 epitope sequences for which cognate HIV-specific CD8+ T cell responses were studied by clonotypic analysis at two time points. NA, nonavailable.
Relationship between clonal turnover and clonal senescence
Changes in clonal dominance may have two independent reasons: (a) the adaptation of B27-KK10–specific CD8+ T cell populations to viral variation (25), or (b) the replacement of clonotypes that approach replicative senescence. To explore the first possibility, global sequencing of the Gag gene was undertaken at both time points (Table II). In two HLA-B27 patients, viral variations could be seen in the KK10 epitope. In 04.034, an M268L mutation occurred that might account for a change in clonal dominance. In 04.064, an N271Q variation in KK10 was observed that, although confined to the C-terminal region of the epitope, might also affect TCR interaction (because the penultimate epitope peptide side chain appears to point out of the groove) (28). However, in two other HLA-B27 patients (05.005 and 11.007), we observed no changes in the KK10 epitope. The overall dataset indicated that viral variation could not be the universal cause for clonal turnover, which is consistent with earlier observations in primary HIV-1 infection (29).
To examine the alternative option that clonal turnover may relate to the replacement of clonotypes that approach replicative senescence, we assessed the expression of CD57 on dominant CD8+ T cell clonotypes directly ex vivo. CD57 expression is linked to the appearance of characteristics associated with replicative senescence, i.e., shortening of telomere length and reduction of proliferative capacity (30, 31). This association holds both when expressed on highly differentiated T cell subsets (i.e., the CD27−/CD28− compartment) and on less differentiated T cell subsets (i.e., the CD27+ compartment that includes the majority of HIV-specific CD8+ T cells; unpublished data). CD57 expression on T cells is therefore indicative of replicative history and provides information on earlier turnover. Dominant clonotypes within the HIV-specific CD8+ T cell populations studied in this paper exhibit distinct TCRBV gene usage (Fig. 4 A and Table S1). Thus, the use of anti-TCRBV antibodies provided the means to identify dominant clonotypes within HIV-specific CD8+ T cells by flow cytometry (Fig. 4 B) and assess the expression of CD57 in parallel (Fig. 4 C). The detection of both A2-SL9– and B27-KK10–specific populations in patient 04.064 provides an interesting example (Fig. 4 D). At the original time point, the dominant B27-KK10 clonotype (BV9) presented high levels of CD57 compared with both the subdominant B27-KK10 clonotype (BV20.1) and the dominant A2-SL9 clonotype (BV5.1). At the later time point, the BV9 clonotype became subdominant within the B27-KK10 population, replaced by the BV20.1 clonotype that already showed increased levels of CD57. Expression of CD57 on the dominant A2-SL9 clonotype remained low, and this clonotype persisted. In line with this observation, the overall analysis of TCRBV antibody–identified B27-KK10 dominant clonotypes among the patients studied at two time points (i.e., from four patients) revealed a significant increase in CD57 expression over time in relation to clonal turnover (Fig. 4 E). In contrast, no significant changes in either CD57 expression or clonal turnover were observed for the other HIV-specific CD8+ T cell populations (i.e., from five patients). Interestingly, the B27-KK10 dominant clonotypes (identified according to their TCRBV) from two donors lost their dominance upon peptide stimulation and in vitro expansion (Fig. 4 F). In the case of patient 04.064, we could even observe the preferential expansion of subdominant clonotypes (BV20.1) over the dominant one (BV9), which was in line with the ex vivo observations. Collectively, these data imply a decline of the replicative capacity for dominant B27 KK10 clonotypes and suggest that T cell turnover may be related to replicative senescence at a clonal level.
Figure 4.
Changes in CD57 expression on B27-KK10–specific CD8+ T cell clonotypes. (A) Clonal analysis (including TCRBV usage, CDR3 aa sequence, TCRBJ usage, and percent frequency) of the B27-KK10– and A2-SL9–specific CD8+ T cell populations present in patient 04.064 at two time points (representing a total of 292 TCRB sequences). Representative examples of B27-KK10 pentamer and anti-TCRBV costaining on CD8+ T cells (B), and of anti-TCRBV and CD57 costaining on B27-KK10–gated CD8+ T cells (patient 04.064, year 5; C) are shown. Numbers represent percentages of CD57+ cells within the BV+/pentamer+ populations. (D) CD57 expression on TCRBV+ B27-KK10– or A2-SL9–specific CD8+ T cell populations for patient 04.064. (E) CD57 expression on TCRBV+ B27-KK10– or other HIV-specific CD8+ T cell populations for the seven patients studied. Clonotypes that are dominant at one of the two time points (for B27-KK10) or the two time points (for the other) are presented; the arrows indicate the time of dominance. The p-value was calculated by the paired t test. (F) The percent frequency of B27-KK10–specific CD8+ T cells and the expression of TCRBV-characterizing dominant clonotypes before (Pre) or after (Post) 9 d of in vitro expansion in the presence of KK10 peptide and IL-2.
Importance of a single dominant clonotype on HIV replication control
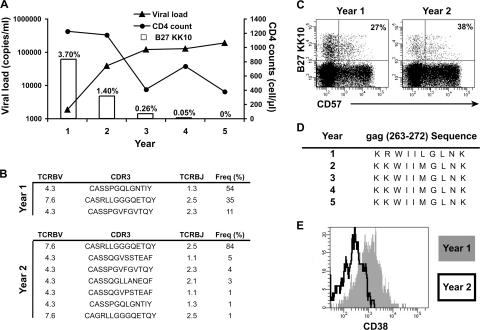
An irreversible loss of B27-KK10–specific CD8+ T cell–dominant clonotypes, because of clonal senescence, is likely to affect control of HIV replication. However, this remains a particularly difficult issue to address; because HIV-specific CD8+ T cell clonotypes approaching senescence can be replaced by new clonotypes from the pool of naive precursors, the assessment of markers like CD57 at a given time point offers no reliable information to predict the outcome for HIV control. We identified one HLA-B27 long-term survivor (02.011) who started progressing from one year (1) to another (2), as indicated by a rise in pVL together with a reduction in the magnitude of the B27-KK10 population and an ensuing drop in CD4+ T cell counts (Fig. 5 A). Loss of control of HIV replication was associated with a near-to-complete disappearance of the dominant B27-KK10 clonotype present before progression (Fig. 5 B). However, the B27-KK10–specific CD8+ T cell population at this time point expressed modest CD57 levels in comparison with declining clonotypes from other donors (Fig. 5 C). Instead, it appears that the demise of this clonotype was caused by loss of antigenic stimulus or neglect: sequencing of the cognate viral epitope revealed the rapid emergence of a variant bearing the R264K mutation (Fig. 5 D), which is known to cause defective binding to MHC class I and the loss of B27-KK10–specific CD8+ T cells associated with HIV disease progression (9, 10, 28, 32). The emergence of the R264K escape virus resulted in the failure to stimulate B27-KK10–specific CD8+ T cells (as seen with the reduced expression of the activation marker CD38 on the cell surface; Fig. 5 E) and the deletion of the dominant B27-KK10–specific CD8+ T cell clonotype, followed by uncontrolled viral replication. Importantly, loss of immune control occurred despite the presence at the same time (year 2) of at least three subdominant HIV-specific CD8+ T cell populations (all HLA-B7 restricted; specific for Gag 353–363, Env 843–851, and Nef 128–137) detected by IFN-γ ELISPOT (unpublished data). Although clonal turnover and senescence are not implicated here, this case is interesting in that it illustrates the importance of maintaining the activity of protective CD8+ T cell clonotypes in HIV infection: if the control exerted by such clonotypes is lost (because of viral escape as in the present example, or in the hypothetical case of clonal senescence in absence of renewal), uncontrolled viral replication can rapidly ensue in the face of persisting subdominant responses which are apparently unable to compensate.
Figure 5.
Disappearance of one B27-KK10–specific CD8+ T cell clonotype and loss of control of HIV replication. (A) 5-yr follow-up (pVL, CD4 counts, and percent frequency of B27-KK10–specific CD8+ T cells) in one patient (02.011) with sudden disease progression at year 2. (B) Clonal analysis (TCRBV usage, CDR3 aa sequence, TCRBJ usage, and percent frequency) of B27-KK10–specific CD8+ T cell populations is shown for years 1 and 2 (representing a total of 150 TCRB sequences). (C) CD57 expression on B27-KK10–specific CD8+ T cells. Percentages of pentamer+ cells that express CD57 are indicated. (D) KK10 viral sequence of the dominant (i.e., >80%) HIV-1 strain over time. (E) CD38 expression on B27-KK10–specific CD8+ T cells at years 1 and 2.
Higher functional avidity and superior control of HIV replication
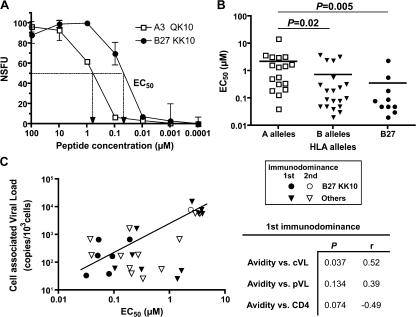
Increased T cell turnover is likely to be caused by a high level of antigenic exposure in relation to the quantity of antigen presented and/or the quality of the stimulus. The quantity of presented antigen may vary according to expression differences between the individual targeted HIV proteins (33). However, we observed differences in longitudinal clonal dominance patterns between CD8+ T cell populations specific for epitopes derived from the same protein (i.e., Gag); thus, an effect related to the quantity of presented antigen seems less likely. Alternatively, the quality of antigenic stimulation might differ between individual CD8+ T cell populations and could be related to the avidity of cognate antigen engagement on the cell surface. We therefore assessed the relative functional avidity of HIV-specific CD8+ T cell populations directly ex vivo, using serial peptide concentrations and defining EC50 as the exogenous peptide concentration yielding half-maximal counts in IFN-γ ELISPOT assays (Fig. 6 A). A significantly higher functional avidity of HLA-B–restricted responses over HLA-A–restricted responses was observed (Fig. 6 B); this dichotomy was driven, at least in part, by the preponderance of the B27-KK10 responses, which strikingly exhibited the highest avidity of all tested populations. This finding underlines the central influence of CD8+ T cell functional avidity on the control of HIV replication. Remarkably, when considering the immunodominant HIV-specific CD8+ T cell responses (identified through the screening of 49 CTL epitopes), we found an inverse correlation between their functional avidity and the patients' cVL (Fig. 6 C). The use of a limited number of CD8+ T cell epitopes makes it possible that some immunodominant responses were not detected; this could explain why some populations exhibited low functional avidities in the context of low cVL. Although it did not reach statistical significance, there was also a trend toward an inverse correlation between functional avidity and cVL when looking at highest subdominant responses. Interestingly, we could detect among these responses a subdominant B27-KK10 CD8+ T cell population, which showed a lower functional avidity (compared with immudominant B27-KK10 CD8+ T cell populations) and was actually associated with a higher cVL in this donor (Fig. 6 C, open circle). The observed correlation with avidity was stronger for cVL compared with pVL or CD4+ T cell count, consistent with a higher sensitivity of cVL as a marker of disease progression (13, 14). Overall, these observations suggest that the functional avidity of HIV-specific CD8+ T cells is directly related to their capacity to control HIV replication through the elimination of infected cells.
Figure 6.
CD8+ T cell functional avidity and control of viral replication. (A) Representative example of peptide titrations in IFN-γ ELISPOT assays (normalized SFC per 106 PBMCs) to assess functional avidity of KK10 or other HIV epitope–specific CD8+ T cells. Functional avidity is defined as the concentration required to achieve half-maximal recognition of the wild-type peptide (EC50). Error bar values are 95% confidence intervals. (B) Functional avidity of CD8+ T cell populations specific for HLA-A–restricted (including 2, 3, 11, 26, and 31) or HLA-B–restricted (including 7, 8, 27, 35, and 51) HIV epitopes (n = 20), as well as for B27-KK10. Horizontal bars represent means. p-values were calculated by the Mann-Whitney U test. (C) Correlation between functional avidity of immunodominant HIV-specific CD8+ T cell responses and HIV cVL. Immunodominant (n = 16, closed symbols) and the highest subdominant (n = 12, open symbols) responses are shown, as is a linear regression trend line. p-values and r values between cVL, pVL, CD4 count, and functional avidity are shown for the immunodominant responses. Spearman's rank test was used to determine correlations.
DISCUSSION
The identification of CD8+ T cell correlates of immune control in HIV infection has remained an elusive yet profoundly important goal that will immeasurably advance our quest for an effective vaccine. However, the heterogeneity that exists between individuals and between CD8+ T cell populations represents a major barrier to the precise identification of these attributes. To understand the mechanistic basis of the immune protection mediated by CD8+ T cells, we conducted a comprehensive study of CD8+ T cell populations specific for B27-KK10 that seem to confer favorable outcomes in infected individuals, as indicated by the literature and our data showing an inverse correlation between the magnitude of p24-specific CD8+ T cell responses and cVL. We found that B27-KK10–specific CD8+ T cells exhibited a more polyfunctional profile, demonstrated increased rates of clonal turnover associated with characteristics of senescence, and displayed superior functional avidity when compared with other HIV-specific CD8+ T cells. These features are interlinked and characterize protective CD8+ T cell–mediated immunity.
Polyfunctional capacity, or the ability to produce high levels of several soluble factors simultaneously, is emerging as a key factor of T cell effectiveness. Soluble factors secreted by T cells like cytokines, cytotoxins, or chemokines are central to the immune response, exerting direct effector capacities, or enabling further T cell recruitment or expansion. Two recent studies indicate that vaccine efficacy depends on the induction of polyfunctional T cells in both mice and humans (22, 23). Our data suggest that CD8+ T cells that achieve optimal control of HIV-1 replication also display polyfunctional capacities, in keeping with the original work from Betts et al. showing the maintenance of polyfunctional CD8+ T cells in long-term nonprogressors (21). Collectively, these data provide further support for the consideration of T cell polyfunctionality as a correlate of protective immunity.
Functional avidity, as defined in this paper, is a measure of the efficiency of antigen recognition by CD8+ T cells. In previous reports, high avidity CD8+ T cells have been shown to initiate the lysis of target cells more rapidly at any given antigen density and to be more effective at mediating viral clearance than low avidity cells (34–36). Thus, the efficiency with which CD8+ T cells are stimulated and exert their antiviral effects to eradicate HIV-infected cells is dependent on functional avidity. In addition, recent work in mouse models suggests a correlation between T cell functionality and avidity, so that the quality of the CD8+ T cell response in terms of cytokine profile is enhanced for highly avid T cells (37). Our results are in line with these findings. The superior polyfunctionality of B27-KK10–specific CD8+ T cell populations may therefore directly relate to their higher functional avidity, and these two features together may enable the more efficient eradication of HIV-infected cells. The high avidity of B27-KK10–specific CD8+ T cell populations may also explain the general immunodominance of this response over other populations and its importance in bearing the brunt of the immunological impact of HIV replication. This is in keeping with recent work suggesting a correlation between functional avidity and the magnitude of CD8+ T cell responses in HIV-1 infection (38), as well as with studies showing that immunodominant clonotypes have higher avidities compared with subdominant clonotypes in chronic CMV and EBV infections (39, 40). Thus, among the diversity of HIV-specific CD8+ T cells, it seems that those clonotypes that exhibit high levels of functional avidity will become immunodominant, presumably through competitive effects, and display superior functionality, thereby orchestrating more effective control of HIV replication.
However, high functional avidity also implies a stronger exposure of the cognate CD8+ T cells to activation signals derived from a given concentration of antigen. This effect will result in proportionately greater levels of CD8+ T cell stimulation and proliferation, thereby causing the increased rate of clonal turnover within B27-KK10–specific CD8+ T cell populations. However, CD8+ T cells, like any other immune cells, have a limited replicative lifespan in vivo (41). This implies that they can be driven toward replicative senescence over time with sustained activation and proliferation, as suggested both by our observations with B27-KK10–specific CD8+ T cell populations at the clonal level and in other persistent viral infections (39). Mathematical modeling has highlighted the potential role for cellular senescence in shaping the CD8+ T cell response to a persistent infection (42). Similarly, the incapacity of immunodominant HIV-specific CD8+ T cell populations to proliferate has been associated with progressive HIV disease (43). Our findings are also in line with work in mouse models showing that persisting lymphocytic choriomeningitis infection can lead to the disappearance of clonotypes with a near complete switch in the epitope-specific repertoire between acute and chronic infection (44), and that high dose antigen can be accompanied by the emergence of low avidity clonotypes (45). Collectively, this suggests that functional avidity can drive clonal turnover and senescence, and entails the need for continuous renewal of high avidity T cell clonotypes to maintain optimal control of HIV replication.
Although the case of patient 02.011 did not involve clonal senescence but loss of antigenic stimulation caused by viral escape, it offers an interesting illustration of the disastrous consequences that the irreversible disappearance of dominant CD8+ T cell clonotypes can have on the control of HIV replication. Constant output from the pool of naive T cell precursors is therefore a prerequisite to maintain an effective control of viral replication, to either replace important but senescent clonotypes or adapt the response to viral variations when possible. Recent data from the mouse models of persistent viral infection indicate that new naive cells are continuously being recruited during the chronic phase and that lack of thymic output results in the decline of virus-specific CD8+ T cells (46). However, as HIV-infected individuals reach advanced stages of the infection, their thymic output and the number of naive cells decline considerably (47, 48), which will affect their ability to renew senescent clones with high turnover, resulting in suboptimal control of HIV replication despite the presence of subdominant HIV-specific CD8+ T cell populations with lower rates of clonal turnover. Our work emphasizes the importance of studying clonal dynamics in the interpretation of cellular immune data. Although much work has concentrated on finding immunologic correlates of protection through the assessment of phenotypic markers on HIV-specific CD8+ T cells at a given time point, our study indicates that this is unlikely to have any predictive value regarding the outcome for viral control, because new clonotypes with different characteristics may be generated thereafter. The capacity to renew important clonotypes will be fundamental for the outcome of HIV-1 infection.
The identification of functional avidity as a central feature of the protective nature and life span of CD8+ T cells raises a question as to which factors determine the recruitment of high avidity clonotypes into any given antigen-specific CD8+ T cell population. Many variables, such as TCR avidity and density, CD8 coreceptor-mediated effects, membrane flexibility, and molecular topography can contribute to the overall functional avidity of a CD8+ T cell population. The study of B27-KK10–specific CD8+ T cell clonotypes described in this paper hints at another potential determinant. Specifically, we observed a high degree of clonotypic diversity at the level of primary sequence across the CDR loops when compared, for example, with A2-SL9– or B7-GL9–specific CD8+ T cell populations. One could speculate that a high degree of TCR diversity within antigen-experienced T cells provides the foundation to establish high avidity populations, as clonotypes with highly avid TCRs may arise more easily from such a large source of possibilities. Further work is necessary to clarify the potential relationship between functional avidity and TCR diversity, and to reveal new insights into the genetic, biophysical, and structural bases of these effects.
In conclusion, the comprehensive analysis of a unique CD8+ T cell population, known to mediate superior control of HIV-1 replication, enabled the identification of distinct attributes that characterize the protective immunity mediated by these cells. These findings represent a substantial step forward in our understanding of several allied immune phenomena and illuminate the biological basis of CD8+ T cell efficacy in HIV infection with associated implications for the evaluation of potential vaccine immunogens.
MATERIALS AND METHODS
Patients.
Samples were obtained from HIV-1–infected patients enrolled in the French Agence Nationale de la Recherche sur le Sida (ANRS) ALT (4) and IMMUNOCO (49) cohorts. Antiretroviral therapy–naive patients during the chronic phase of infection—with CD4 counts >400 cells per cubic millimeter, no clinical symptoms, and pVL ranging from 200 to 3.5 × 105 copies of HIV-1 RNA per milliliter—were selected for the study. The study was approved by the relevant local institutional (at the Hôpital Pitié-Salpêtrière) review board and ethics committee. PBMCs were separated from citrate anticoagulated blood and cryopreserved for subsequent studies. HLA genotyping was performed by amplification refractory mutation system–PCR using sequence-specific primers, as previously described (50). HLA-genotyped patients were screened for HIV-specific CD8+ T cell responses with IFN-γ ELISPOT assays using a panel of 49 known HLA class I–restricted viral epitope peptides. The 11 HLA-B27 patients studied were subtyped, and all were found to carry the HLA-B*2705 allele.
Quantification of cVL.
HIV-1 cVL was quantified in PBMCs using a real-time PCR assay combining the quantification of HIV-1 cVL and albumin gene DNA. This assay allows the detection of five copies of HIV-1 cVL in 106 PBMCs (13). This was performed with fluorescent TaqMan methodology on a sequence detection system (ABI Prism 7700; Applied Biosystems).
CD8+ T cell epitopes and IFN-γ ELISPOT assay.
Synthetic 15-mer peptides overlapping by 11 aa and spanning the entire HIV-1 Gag sequence were purchased from Neosystem and pooled into 11–15 peptide pools: 5 p24 pools, 3 p17 pools, and 3 PP (i.e., p2/p7/p1/p6) pools. In addition, 49 CD8+ T cell epitopes, 8–11 aa in length, derived from HXB2 Gag, Pol, Env, or Nef proteins were selected from the Los Alamos HIV molecular immunology database, according to the patients' HLA class I haplotypes, and synthesized by Syntem, or provided by ANRS. All peptides used in this study were >80% purity, as shown by HPLC profiles. For the ELISPOT assays, thawed PBMCs were plated at 105 cells per well in 96-well polyvinylidene plates (Millipore) precoated with capture anti–human IFN-γ mAb (Diaclone) at 4°C overnight. The final peptide concentration in screening assays was 2 μg/ml for overlapping peptides or 10 μM for optimal CTL epitopes. Plates were incubated overnight at 37°C/5% CO2 and developed according to the manufacturer's recommendations. Spots were counted using an automated ELISPOT reader (Carl Zeiss MicroImaging, Inc.), and data were expressed as spot-forming units (SFU) per 106 PBMCs. Results were considered positive if >50 SFU/106 PBMCs were stained after subtraction of the mean background obtained with cells alone. Functional avidity, which in this study refers to activation threshold in response to defined concentrations of exogenous peptide, was assessed by performing limiting peptide dilutions (from 100 to 0.001 μM) and determining the peptide concentration required to induce half-maximum IFN-γ production (number of spots) in ex vivo assays. Results were considered appropriate for analysis only when maximum IFN-γ production was >500 SFU/106 PBMCs (background subtracted) for both immunodominant or the highest subdominant responses.
Flow cytometry and reagents.
HLA-A*0301 QK10, HLA-A*0301 RY10, HLA-B*0702 GL9, and HLA-B*2705 KK10 pentamers were purchased from ProImmune; HLA-A*0201 SL9 tetramer was purchased from Beckman Coulter; and HLA-B*0801 FL9 tetramer was synthesized as previously described (51). PBMCs were stained with pretitrated concentrations of pentamer/tetramer (conjugated to PE or allophycocyanin [APC]) followed by a panel of antibodies, as previously described (52). Directly conjugated and unconjugated antibodies were as follows: CD3 (Cy5.5PerCP or Alexa Fluor 700), CD38 (APC), CD45RA (FITC), CCR7 (PE-Cy7), CD107a (Cy5-PE), IL-2 (APC), IFN-γ (Alexa Fluor 700), CD57 (FITC), and TNF-α (PE-Cy7; all from BD Biosciences); CD28 (PE–Texas red), CD45RA (PE–Texas red), TCRBV mAbs (5.1, 6.6, 9, 14, 13, and 20.1 conjugated to FITC or PE; all from Beckman Coulter); CD8 (Alexa Fluor 405), swine anti–goat (FITC), and granzyme B (PE–Texas red; all from Caltag); CD127 (purified) and MIP-1β (FITC; both from R&D Systems); PD-1 (PE; eBioscience); goat anti–mouse (PE; DakoCytomation); and CD27 (Alexa Fluor 700; BioLegend). Anti-CD57 (Qdot-545) and anti–PD-1 (PE) antibodies were provided by P. Chattopadhyay (Vaccine Research Center, Bethesda, MD) and G. Freeman (Dana-Farber Cancer Institute, Boston, MA), respectively. Cells were analyzed on a standard LSRII (equipped with blue, red, and violet lasers; BD Biosciences) to systematically perform eight to nine color stainings. The list mode data files were analyzed using FlowJo software (version 8.2; TreeStar, Inc.) and FACSDiva software (BD Biosciences). Multifunctional data were analyzed with PESTLE software (version 1.3.2) and SPICE software (version 3.1; obtained from M. Roederer, National Institutes of Health, Bethesda MD). Percent frequencies of multifunctional cells were calculated within the total population of detectable antigen-specific CD8+ T cells.
Cell stimulation for multifunctional assessment or in vitro expansion.
Purified PBMCs were thawed and rested overnight at 37°C in complete RPMI media (RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, l-glutamine, and antibiotics); viability was then examined by Trypan blue exclusion (typically ≥70% viable). For stimulation, at least 106 pentamer/tetramer prestained cells were incubated in the presence of 5 μM of specific peptide and 10 μl of anti-CD107a antibodies for 1 h at 37°C in a 5% CO2 incubator, followed by an additional 5 h in the presence of the secretion inhibitors monensin (2.5 μg/ml; Sigma-Aldrich) and Brefeldin A (5 μg/ml; Sigma-Aldrich). Negative controls were obtained in the absence of peptide and prestaining with pentamer/tetramer. Cytofix/Cytoperm (BD Biosciences) was used for permeabilization of the cells before staining for intracellular markers. For in vitro expansion, 105 PBMCs per well were cultivated in RPMI 1640 media supplemented with 8% heat-inactivated human serum in 96-well round-bottom plates in the presence of 1 μM KK10 peptide. 150 U/ml IL-2 was added after 2 d, and fresh medium was added on day 6. Cultures were harvested on day 9 to be stained with pentamers and surface antibodies.
Clonotype analysis.
Clonotype analysis was performed on live HIV-specific CD8+ T cells labeled with cognate MHC class I–peptide tetramers or pentamers, and sorted using a FACSAria (BD Biosciences) in a category III laboratory. Between 400 and 2,000 cells were collected for each population and directly sorted into 1.5-ml microtubes containing 100 μl RNAlater (Ambion). After cell lysis, mRNA was extracted (Oligotex kit; QIAGEN) and subjected to a nonnested, template-switch anchored RT-PCR using a 3′ TCRB constant region primer, as previously described (25, 26). Amplified products were ligated into pGEM-T Easy vector (Promega) and cloned by transformation of competent DH5α Escherichia coli. Selected colonies were amplified by PCR using standard M13 primers and sequenced from an insert-specific primer using fluorescent dye terminator chemistry (Applied Biosystems). A minimum of 50 clones was generated and analyzed per sample. Pseudogenes and “nonfunctional” sequences that could not be resolved after inspection of the individual chromatograms were discarded from the analysis. Nucleotide comparisons were used to establish clonal identity. Data analysis was performed using Sequencher software (version 4.6; Gene Codes). The international ImMunoGeneTics nomenclature system is used throughout this paper (25, 39). In total, 30 live populations were sorted to generate 1,914 TCRB sequences.
DNA sequencing of HIV-1 gag.
Whole cellular DNA was extracted from PBMC (3–5 millions of cells) using a QIAamp DNA blood kit (QIAGEN), according to the manufacturer's recommendations. First-round PCR was conducted to amplify the Gag region (nt 734–2,047) with the following primer pair: (forward) 5′-GACTGGTGAGTACGCCAAAA-3′ (GAG1) and (reverse) 5′-CTTCCTTTCCACATTTCCAACAGC-3′ (GAG2). Then, a second-round PCR (nt 812–1,920) was performed using the following primer pair: (forward) 5′-AAGCGGGGGAGAATTAGAT-3′ (GAG3) and (reverse) 5′-CATTATGGTAGCTGAAAT-3′ (GAG5). The PCR product was purified with Amicon's Microcon 100 (Millipore). The Gag region was sequenced (aa 20–363) using a cycle-sequencing reaction with the Big Dye terminator kit (Applied Biosystems) using the GAG3 and GAG5 primers. The sequences were aligned on the HXB2 reference sequence using Sequence Navigator software (Applied Biosystems).
Statistics.
Group medians and distributions were compared by the Mann-Whitney U test, the permutation test, or the paired t test. Associations between variables were determined by the nonparametric Spearman correlation test. P > 0.05 was considered not significant.
Online supplemental material.
Table S1 shows TCRB sequences from all FACS-sorted HIV-specific CD8+ T cell populations (n = 30) studied (representing a total of 1,914 sequences). Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070784/DC1.
Supplemental Material
Acknowledgments
We are very grateful to the staff and patients of the clinics that provided blood samples and to the ANRS cohorts ALT and IMMUNOCO study groups. We are indebted to Mario Roederer and Pratip Chattopadhyay for the use of PESTLE and SPICE software and for providing anti-CD57 antibodies conjugated to Qdot-545, as well as Gordon Freeman for providing anti–PD-1 antibodies. We thank Martha Nason and Stephano Corvasce for their help with the statistical analysis, as well as David Ambrozak for his help with FACS sorting.
This work was supported by the Institut National de la Santé et de la Recherche Médicale Avenir grant, the French ANRS, Sidaction, the National Institutes of Health, and the Medical Research Council (UK). D.A. Price is a Medical Research Council Senior Clinical Fellow. J.R. Almeida is supported by a fellowship from the Fundação para a Ciência e Tecnologia.
The authors have no conflicting financial interests.
Abbreviations used: APC, allophycocyanin; CCR, CC chemokine receptor; cVL, cell-associated HIV–DNA load; MFI, median fluorescence intensity; MIP, macrophage inflammatory protein; PD-1, programmed death–1; pVL, plasma HIV–RNA load; SFU, spot-forming unit.
References
- 1.Gea-Banacloche, J.C., S.A. Migueles, L. Martino, W.L. Shupert, A.C. McNeil, M.S. Sabbaghian, L. Ehler, C. Prussin, R. Stevens, L. Lambert, et al. 2000. Maintenance of large numbers of virus-specific CD8+ T cells in HIV-infected progressors and long-term nonprogressors. J. Immunol. 165:1082–1092. [DOI] [PubMed] [Google Scholar]
- 2.Betts, M.R., D.R. Ambrozak, D.C. Douek, S. Bonhoeffer, J.M. Brenchley, J.P. Casazza, R.A. Koup, and L.J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983–11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaslow, R.A., M. Carrington, R. Apple, L. Park, A. Munoz, A.J. Saah, J.J. Goedert, C. Winkler, S.J. O'Brien, C. Rinaldo, et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411. [DOI] [PubMed] [Google Scholar]
- 4.Magierowska, M., I. Theodorou, P. Debre, F. Sanson, B. Autran, Y. Riviere, D. Charron, and D. Costagliola. 1999. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood. 93:936–941. [PubMed] [Google Scholar]
- 5.O'Brien, S.J., G.W. Nelson, C.A. Winkler, and M.W. Smith. 2000. Polygenic and multifactorial disease gene association in man: Lessons from AIDS. Annu. Rev. Genet. 34:563–591. [DOI] [PubMed] [Google Scholar]
- 6.Scherer, A., J. Frater, A. Oxenius, J. Agudelo, D.A. Price, H.F. Gunthard, M. Barnardo, L. Perrin, B. Hirschel, R.E. Phillips, and A.R. McLean. 2004. Quantifiable cytotoxic T lymphocyte responses and HLA-related risk of progression to AIDS. Proc. Natl. Acad. Sci. USA. 101:12266–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altfeld, M., E.T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M.N. Johnston, N. Burgett, M.E. Swartz, A. Yang, G. Alter, et al. 2006. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson, J.D., G.S. Ogg, R.L. Allen, C. Davis, S. Shaunak, J. Downie, W. Dyer, C. Workman, S. Sullivan, A.J. McMichael, and S.L. Rowland-Jones. 2000. Direct visualization of HIV-1-specific cytotoxic T lymphocytes during primary infection. AIDS. 14:225–233. [DOI] [PubMed] [Google Scholar]
- 9.Goulder, P.J., R.E. Phillips, R.A. Colbert, S. McAdam, G. Ogg, M.A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217. [DOI] [PubMed] [Google Scholar]
- 10.Kelleher, A.D., C. Long, E.C. Holmes, R.L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, et al. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27–restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leslie, A.J., K.J. Pfafferott, P. Chetty, R. Draenert, M.M. Addo, M. Feeney, Y. Tang, E.C. Holmes, T. Allen, J.G. Prado, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289. [DOI] [PubMed] [Google Scholar]
- 12.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53. [DOI] [PubMed] [Google Scholar]
- 13.Rouzioux, C., J.B. Hubert, M. Burgard, C. Deveau, C. Goujard, M. Bary, D. Sereni, J.P. Viard, J.F. Delfraissy, and L. Meyer. 2005. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J. Infect. Dis. 192:46–55. [DOI] [PubMed] [Google Scholar]
- 14.Martinez, V., D. Costagliola, O. Bonduelle, N. Ngo, A. Schnuriger, I. Theodorou, J.P. Clauvel, D. Sicard, H. Agut, P. Debre, et al. 2005. Combination of HIV-1-specific CD4 Th1 cell responses and IgG2 antibodies is the best predictor for persistence of long-term nonprogression. J. Infect. Dis. 191:2053–2063. [DOI] [PubMed] [Google Scholar]
- 15.Champagne, P., G.S. Ogg, A.S. King, C. Knabenhans, K. Ellefsen, M. Nobile, V. Appay, G.P. Rizzardi, S. Fleury, M. Lipp, et al. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 410:106–111. [DOI] [PubMed] [Google Scholar]
- 16.van Baarle, D., S. Kostense, M.H. van Oers, D. Hamann, and F. Miedema. 2002. Failing immune control as a result of impaired CD8(+) T-cell maturation: CD27 might provide a clue. Trends Immunol. 23:586–591. [DOI] [PubMed] [Google Scholar]
- 17.Wherry, E.J., C.L. Day, R. Draenert, J.D. Miller, P. Kiepiela, T. Woodberry, C. Brander, M. Addo, P. Klenerman, R. Ahmed, and B.D. Walker. 2006. HIV-specific CD8 T cells express low levels of IL-7Ralpha: implications for HIV-specific T cell memory. Virology. 353:366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Day, C.L., D.E. Kaufmann, P. Kiepiela, J.A. Brown, E.S. Moodley, S. Reddy, E.W. Mackey, J.D. Miller, A.J. Leslie, C. DePierres, et al. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 443:350–354. [DOI] [PubMed] [Google Scholar]
- 19.Trautmann, L., L. Janbazian, N. Chomont, E.A. Said, S. Gimmig, B. Bessette, M.R. Boulassel, E. Delwart, H. Sepulveda, R.S. Balderas, et al. 2006. Upregulation of PD-1 expression on HIV-specific CD8(+) T cells leads to reversible immune dysfunction. Nat. Med. 12:1198–1202. [DOI] [PubMed] [Google Scholar]
- 20.Appay, V., P.R. Dunbar, M. Callan, P. Klenerman, G.M. Gillespie, L. Papagno, G.S. Ogg, A. King, F. Lechner, C.A. Spina, et al. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379–385. [DOI] [PubMed] [Google Scholar]
- 21.Betts, M.R., M.C. Nason, S.M. West, S.C. De Rosa, S.A. Migueles, J. Abraham, M.M. Lederman, J.M. Benito, P.A. Goepfert, M. Connors, et al. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 107:4781–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Precopio, M.L., M.R. Betts, J. Parrino, D.A. Price, E. Gostick, D.R. Ambrozak, T.E. Asher, D.C. Douek, A. Harari, G. Pantaleo, et al. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. J. Exp. Med. 204:1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darrah, P.A., D.T. Patel, P.M. De Luca, R.W. Lindsay, D.F. Davey, B.J. Flynn, S.T. Hoff, P. Andersen, S.G. Reed, S.L. Morris, et al. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13:843–850. [DOI] [PubMed] [Google Scholar]
- 24.Wilson, J.D., G.S. Ogg, R.L. Allen, P.J. Goulder, A. Kelleher, A.K. Sewell, C.A. O'Callaghan, S.L. Rowland-Jones, M.F. Callan, and A.J. McMichael. 1998. Oligoclonal expansions of CD8+ T cells in chronic HIV infection are antigen specific. J. Exp. Med. 188:785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douek, D.C., M.R. Betts, J.M. Brenchley, B.J. Hill, D.R. Ambrozak, K.L. Ngai, N.J. Karandikar, J.P. Casazza, and R.A. Koup. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 168:3099–3104. [DOI] [PubMed] [Google Scholar]
- 26.Price, D.A., S.M. West, M.R. Betts, L.E. Ruff, J.M. Brenchley, D.R. Ambrozak, Y. Edghill-Smith, M.J. Kuroda, D. Bogdan, K. Kunstman, et al. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 21:793–803. [DOI] [PubMed] [Google Scholar]
- 27.Turner, S.J., P.C. Doherty, J. McCluskey, and J. Rossjohn. 2006. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 6:883–894. [DOI] [PubMed] [Google Scholar]
- 28.Stewart-Jones, G.B., K. di Gleria, S. Kollnberger, A.J. McMichael, E.Y. Jones, and P. Bowness. 2005. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur. J. Immunol. 35:341–351. [DOI] [PubMed] [Google Scholar]
- 29.Pantaleo, G., H. Soudeyns, J.F. Demarest, M. Vaccarezza, C. Graziosi, S. Paolucci, M. Daucher, O.J. Cohen, F. Denis, W.E. Biddison, et al. 1997. Evidence for rapid disappearance of initially expanded HIV-specific CD8+ T cell clones during primary HIV infection. Proc. Natl. Acad. Sci. USA. 94:9848–9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenchley, J.M., N.J. Karandikar, M.R. Betts, D.R. Ambrozak, B.J. Hill, L.E. Crotty, J.P. Casazza, J. Kuruppu, S.A. Migueles, M. Connors, et al. 2003. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 101:2711–2720. [DOI] [PubMed] [Google Scholar]
- 31.Papagno, L., C.A. Spina, A. Marchant, M. Salio, N. Rufer, S. Little, T. Dong, G. Chesney, A. Waters, P. Easterbrook, et al. 2004. Immune activation and CD8+ T-cell differentiation towards senescence in HIV-1 infection. PLoS Biol. 2:E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goulder, P.J., C. Brander, Y. Tang, C. Tremblay, R.A. Colbert, M.M. Addo, E.S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, et al. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 412:334–338. [DOI] [PubMed] [Google Scholar]
- 33.Tsomides, T.J., A. Aldovini, R.P. Johnson, B.D. Walker, R.A. Young, and H.N. Eisen. 1994. Naturally processed viral peptides recognized by cytotoxic T lymphocytes on cells chronically infected by human immunodeficiency virus type 1. J. Exp. Med. 180:1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander-Miller, M.A., G.R. Leggatt, and J.A. Berzofsky. 1996. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc. Natl. Acad. Sci. USA. 93:4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedlik, C., G. Dadaglio, M.F. Saron, E. Deriaud, M. Rojas, S.I. Casal, and C. Leclerc. 2000. In vivo induction of a high-avidity, high-frequency cytotoxic T-lymphocyte response is associated with antiviral protective immunity. J. Virol. 74:5769–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derby, M., M. Alexander-Miller, R. Tse, and J. Berzofsky. 2001. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J. Immunol. 166:1690–1697. [DOI] [PubMed] [Google Scholar]
- 37.La Gruta, N.L., S.J. Turner, and P.C. Doherty. 2004. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J. Immunol. 172:5553–5560. [DOI] [PubMed] [Google Scholar]
- 38.Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H.S. Hewitt, L. Henry, et al. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094–4101. [DOI] [PubMed] [Google Scholar]
- 39.Price, D.A., J.M. Brenchley, L.E. Ruff, M.R. Betts, B.J. Hill, M. Roederer, R.A. Koup, S.A. Migueles, E. Gostick, L. Wooldridge, et al. 2005. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 202:1349–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trautmann, L., M. Rimbert, K. Echasserieau, X. Saulquin, B. Neveu, J. Dechanet, V. Cerundolo, and M. Bonneville. 2005. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 175:6123–6132. [DOI] [PubMed] [Google Scholar]
- 41.Pawelec, G., R.B. Effros, C. Caruso, E. Remarque, Y. Barnett, and R. Solana. 1999. T cells and aging (update February 1999). Front. Biosci. 4:D216–D269. [DOI] [PubMed] [Google Scholar]
- 42.Davenport, M.P., C. Fazou, A.J. McMichael, and M.F. Callan. 2002. Clonal selection, clonal senescence, and clonal succession: the evolution of the T cell response to infection with a persistent virus. J. Immunol. 168:3309–3317. [DOI] [PubMed] [Google Scholar]
- 43.Migueles, S.A., A.C. Laborico, W.L. Shupert, M.S. Sabbaghian, R. Rabin, C.W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, et al. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061–1068. [DOI] [PubMed] [Google Scholar]
- 44.Lin, M.Y., and R.M. Welsh. 1998. Stability and diversity of T cell receptor repertoire usage during lymphocytic choriomeningitis virus infection of mice. J. Exp. Med. 188:1993–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rees, W., J. Bender, T.K. Teague, R.M. Kedl, F. Crawford, P. Marrack, and J. Kappler. 1999. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 96:9781–9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vezys, V., D. Masopust, C.C. Kemball, D.L. Barber, L.A. O'Mara, C.P. Larsen, T.C. Pearson, R. Ahmed, and A.E. Lukacher. 2006. Continuous recruitment of naive T cells contributes to heterogeneity of antiviral CD8 T cells during persistent infection. J. Exp. Med. 203:2263–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Douek, D.C., R.D. McFarland, P.H. Keiser, E.A. Gage, J.M. Massey, B.F. Haynes, M.A. Polis, A.T. Haase, M.B. Feinberg, J.L. Sullivan, et al. 1998. Changes in thymic function with age and during the treatment of HIV infection. Nature. 396:690–695. [DOI] [PubMed] [Google Scholar]
- 48.Kalayjian, R.C., A. Landay, R.B. Pollard, D.D. Taub, B.H. Gross, I.R. Francis, A. Sevin, M. Pu, J. Spritzler, M. Chernoff, et al. 2003. Age-related immune dysfunction in health and in human immunodeficiency virus (HIV) disease: association of age and HIV infection with naive CD8+ cell depletion, reduced expression of CD28 on CD8+ cells, and reduced thymic volumes. J. Infect. Dis. 187:1924–1933. [DOI] [PubMed] [Google Scholar]
- 49.Chouquet, C., B. Autran, E. Gomard, J.M. Bouley, V. Calvez, C. Katlama, D. Costagliola, and Y. Riviere. 2002. Correlation between breadth of memory HIV-specific cytotoxic T cells, viral load and disease progression in HIV infection. AIDS. 16:2399–2407. [DOI] [PubMed] [Google Scholar]
- 50.Bunce, M., C.M. O'Neill, M.C. Barnardo, P. Krausa, M.J. Browning, P.J. Morris, and K.I. Welsh. 1995. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP). Tissue Antigens. 46:355–367. [DOI] [PubMed] [Google Scholar]
- 51.Altman, J.D., P.A.H. Moss, P.J.R. Goulder, D.H. Barouch, M.G. McHeyzer-Williams, J.I. Bell, A.J. McMichael, and M.M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science. 274:94–96. (published erratum appears in Science. 1998. 280:1821). [DOI] [PubMed] [Google Scholar]
- 52.Appay, V., and S.L. Rowland-Jones. 2002. The assessment of antigen-specific CD8+ T cells through the combination of MHC class I tetramer and intracellular staining. J. Immunol. Methods. 268:9–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.