Abstract
Lymphoid organs contain a B220+CD11c+NK1.1+ cell population that was recently characterized as a novel dendritic cell (DC) subset that functionally overlaps with natural killer (NK) cells and plasmacytoid DCs (PDCs). Using Siglec-H and NK1.1 markers, we unambiguously dissected B220+CD11c+ cells and found that PDCs are the only professional interferon (IFN)-α–producing cells within this heterogeneous population. In contrast, B220+CD11c+NK1.1+ cells are a discrete NK cell subset capable of producing higher levels of IFN-γ than conventional NK cells. Unlike DCs, only a minute fraction of B220+CD11c+NK1.1+ cells in the spleen expressed major histocompatibility complex class II ex vivo or after stimulation with CpG. Consistent with being a NK cell subset, B220+CD11c+NK1.1+ cells depended primarily on interleukin 15 and common cytokine receptor γ chain signaling for their development. In terms of function, expression of distinctive cell surface receptors, and location in lymphoid organs, NK1.1+B220+CD11c+ appear to be the murine equivalent of human CD56bright NK cells.
Plasmacytoid DCs (PDCs) are a rare cell population in lymphoid organs that specialize in secreting type I IFNs, i.e. IFN-α/β, in response to DNA and RNA viruses. In mice, PDCs were initially identified within the CD11c+ DC population as CD11clowB220+Gr1+CD11b− cells (1). Because classical DCs do not express B220, and Gr1 expression on PDCs can be variable, the complex PDC phenotype has often been simplified and PDCs have been identified in many studies as B220+CD11c+CD11b− cells or B220+CD11c+ cells. However, there is increasing evidence that B220+CD11c+ cells are heterogeneous and include more than PDCs. Indeed, it was recently found that B220+CD11c+ cells include a hybrid cell type, which was termed IFN-producing killer DCs (IKDCs) (2, 3).
IKDCs were shown to display molecular and functional characteristics that overlap with those of NK cells, PDCs, and DCs. IKDCs express NK1.1, lack Gr1, and express MHC class II. As NK cells, IKDCs produce IFN-γ and kill typical NK target cells using NK cell–activating receptors. Upon stimulation with CpG oligonucleotides, IKDCs curtail their NK-like activity and acquire DC-like antigen-presenting activity through up-regulation of MHC class II and co-stimulatory molecules. The ability of IKDCs to produce IFN-α/β is controversial. In one report, IKDCs were found to produce substantial amounts of IFN-α (3). In two other studies, secretion of IFN-α was found to be restricted to PDCs (2, 4). Thus, it is unclear whether there is functional overlap between PDCs and IKDCs. The developmental pathway of IKDCs is also unclear (5, 6). IKDCs were virtually absent in IL-2 receptor β chain–deficient mice (IL-2Rβ−/−) (3). At odds with this report, IKDCs were found in mice lacking the common cytokine receptor γ chain (γc) (2), which is required for IL-2Rβ signaling. Moreover, IKDCs were found to derive from a unique progenitor (7). Thus, it is unclear whether IKDCs belong to an NK cell or a DC lineage.
Because of the constitutive heterogeneity of B220+CD11c+ cells, the partial overlap of IKDC functions with those of PDCs, DCs, and NK cells, surface markers that univocally define distinct cell types have become essential to dissect their relationship, development, and functions. Three PDC-specific cell surface molecules have been recently identified. Ly49Q and bone marrow stromal antigen 2 are both specific for PDCs under steady-state conditions but are up-regulated by type I IFNs and IFN-γ during viral infections or after administration of Toll-like receptor (TLR) ligands (8–10). However, Siglec-H appears to be specific for PDCs under steady-state and stimulatory conditions (11–13). In this study, we found that the heterogeneous population of B220+CD11c+ cells can be resolved into three distinct cell subsets based on the expression of Siglec-H and NK1.1. Siglec-H+NK1.1− cells are the only true PDCs that produce IFN-α in response to TLR9 stimulation and require FLT3L for development. A second Siglec-H−NK1.1− subset mostly consists of CD19+ Ig+ B cells. Finally, Siglec-H−NK1.1+ cells correspond to IKDCs. In contrast to previous reports, we found that these cells do not produce IFN-α and do require the γc and IL-15 for development. Thus, these cells belong to the NK cell lineage. We detected MHC class II only on a very small percentage of B220+CD11c+NK1.1+ NK cells in the spleen. Although in lymph nodes the percentage of MHC class II+ NK cells was markedly higher than in the spleen, MHC class II expression did not correlate with CD11c and/or B220 expression. Importantly, we found that B220+CD11c+NK1.1+ NK cell phenotype and function resemble those of human CD56bright NK cells, which are present in lymphoid organs and effectively produce IFN-γ. We conclude that B220+CD11c+Siglec-H−NK1.1+ cells have no marked functional or developmental overlap with PDCs or DCs, belong to the NK cell lineage, and include a mature NK cell subset specialized in cytokine secretion.
RESULTS AND DISCUSSION
Heterogeneity of B220+CD11c+ cells in lymphoid organs
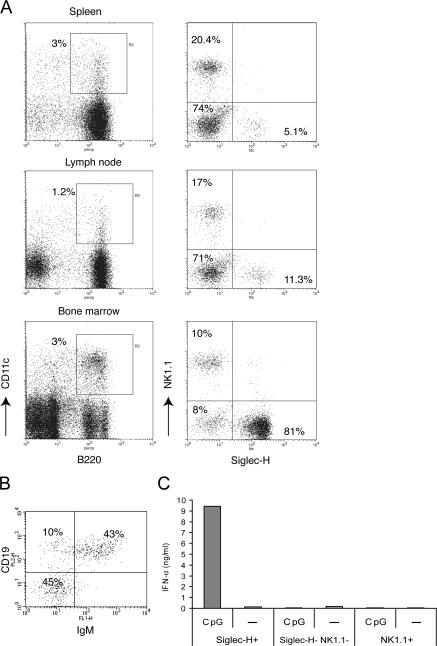
To investigate the nature of B220+CD11c+ cells in lymphoid organs, we analyzed Siglec-H and NK1.1 expression within these cells. Siglec-H is a PDC marker that, in contrast to bone marrow stromal antigen 2 (120G8, 927, and PDCA1) and Ly49Q, maintains specificity after viral infection or stimulation with TLR ligands (8, 10–12). NK1.1 is expressed on conventional NK cells and a recently described hybrid cell type, termed IKDC, which appears to share functional features with NK cells, PDCs, and DCs (2, 3). B220+CD11c+ cells in the spleen, lymph nodes, and bone marrow included three distinct subsets: (a) a Siglec-H+NK1.1− subset that corresponds to PDC; (b) a Siglec-H−NK1.1+ subset that fulfills the definition of IKDC (B220+CD11c+NK1.1+ cells); and (c) a Siglec-H−NK1.1− subset (Fig. 1 A). This latter subset was highly represented in the spleen and lymph node, where it mostly included mature CD19+IgM+ B cells (not depicted). In bone marrow, this subset was less abundant and exhibited further heterogeneity, as ∼40–50% of the cells expressed CD19 and IgM (Fig. 1 B). To determine which cell subset is capable of producing IFN-α in response to TLR9 stimulation, we sorted the three cell subsets and measured IFN-α released in culture supernatant after stimulation with CpG oligodeoxyribonucleotide (ODN). Siglec-H+NK1.1− cells were the only cells capable of producing copious amounts of IFN-α (up to 10 ng/ml from 105 cells). In contrast, Siglec-H−NK1.1+ and Siglec-H−NK1.1− cells did not produce detectable levels of IFN-α. Collectively, these results provide evidence of phenotypic and functional heterogeneity within B220+CD11c+ cells in lymphoid organs and define the Siglec-H+NK1.1− cell subset as the professional IFN-α producer cell type.
Figure 1.
Heterogeneity of B220+CD11c+ cells in lymphoid organs. (A) Four-color analysis of spleen, lymph node, and bone marrow cells. B220+CD11c+ double-positive cells (gate R2) were divided into three subsets based on expression of Siglec-H and NK1.1. One subset expresses Siglec-H, a marker of PDCs. A second subset expresses NK1.1. A third subset is Siglec-H−NK1.1− and includes mature B cells in the spleen and lymph nodes (not depicted). Percentages of each subset are indicated. Each spleen contained 5–8 × 107 cells. From two femurs of an 8–16-wk-old mouse we recovered 2–4 × 107 bone marrow cells. A Gate (R1) was applied on live cells based on FSC and SSC. FL-1/FL-2 double-positive autofluorescent cellular debris was removed from the analysis by an exclusion gate (R3). (B) The Siglec-H−NK1.1− subset displays further heterogeneity in bone marrow. About 40–55% of the cells in different experiments are CD19+IgM+ B cells. However, ∼40–50% of the remaining cells do not express lineage markers. Plots shown in A and B are representative of at least three to four separate experiments with concordant results. (C) Siglec-H+NK1.1−, NK1.1+Siglec-H−, and Siglec-H−NK1.1− were FACS sorted from the spleens of C57BL/6 mice, and identical numbers (105) of cells were stimulated with CpG ODN 2216. After overnight culture, IFN-α secretion was determined in culture supernatants. Results are representative of three independent experiments.
Siglec-H−NK1.1+ cells are endowed with high IFN-γ secretion capacity
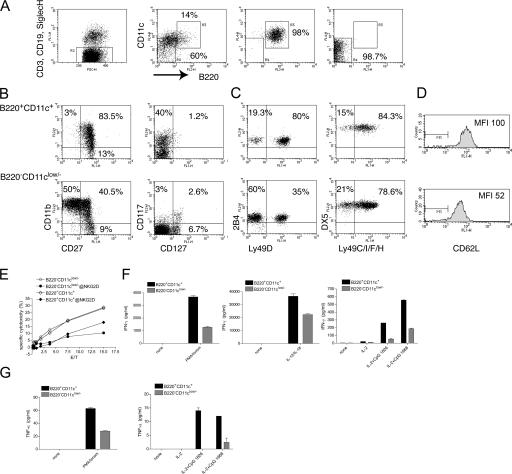
To compare B220+CD11c+ NK1.1+ cells with conventional NK cells, we enriched DX5+ cells from spleens by magnetic positive selection and sorted B220+CD11c+ and B220−CD11clow/− cells, excluding contaminating CD3+, CD19+, and Siglec-H+ cells (Fig. 2 A). By using CD11c and B220 antibodies conjugated to allophycocyanin and PerCP-Cy5.5 for sorting, we were able to further characterize the two cell populations using FITC- and RPE-conjugated antibodies. B220+CD11c+ cells were highly enriched in cells coexpressing the TNF receptor family member CD27, which is the receptor for CD70, and the integrin αM (CD11b) (Fig. 2 B). They also included high percentages of cells expressing the growth factor receptor c-kit (CD117), but not the IL-7Rα chain (CD127), which was recently reported as a marker for NK cells of thymic origin (14). In contrast, B220−CD11clow/− cells contained a high percentage of CD11b+CD27− NK cells, few CD117+ cells (5–8% in different experiments), and some CD127+ cells. B220+CD11c+ cells also exhibited a unique repertoire of activating and inhibitory receptors. As compared with B220−CD11clow/−, they contained two- to threefold more cells expressing the activating receptor Ly49D, which recognizes both allogeneic and xenogenic MHC class I (15, 16), and expressed slightly higher levels of 2B4 (Fig. 2 C). B220+CD11c+ cells also expressed higher level of the lymphocyte homing molecule L-selectin (CD62L) (Fig. 2 D). Several other Ly49 family receptors were similarly expressed by the two subsets, with the exception of a slight but reproducible increase in the percentage of Ly49C/I+ cells within the B220−CD11clow/− population (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20070991/DC1). The receptors NKG2A/C/E and the integrin CD49 (DX5) were also equally expressed (Fig. S1 and Fig. 2).
Figure 2.
B220+CD11c+NK1.1+ cells are enriched in CD27+CD11b+ cells endowed with high IFN-γ secretion capacity. (A) Splenic DX5+ cells were preenriched by DX5 MACS separation. Cells were then stained with a combination of FITC-conjugated anti-CD3, anti-CD19, and anti–Siglec-H antibodies to remove contaminating T cells, B cells, and PDCs (R2). In addition, they were stained with allophycocyanin-conjugated CD11c and PerCP-Cy5.5–conjugated B220. B220+CD11c+ (R3) and B220−CD11clow/− (R4) cells were FACS sorted at high purity. One sorting experiment representative of seven is illustrated. (B) Sorted B220+CD11c+ cells (top panels) are highly enriched in CD11b+CD27+ (75–90% in different experiments) and c-kit+ (CD117, 35–45% in different experiments) but not in IL-7Rα+ (CD127) NK cells. One experiment representative of three is shown. (C) B220+CD11c+ cells (top panels) are enriched in Ly49D+ NK cells, whereas other Ly49 family members have a similar distribution between the two subsets. 2B4 is slightly higher in B220+CD11c+ cells. One experiment representative of three is shown. (D) B220+CD11c+ NK cells express more L-selectin than B220−CD11clow/− NK cells. (E) B220+CD11c+ NK cells kill YAC cells as efficiently as B220−CD11clow/− NK cells. Killing is partially blocked by an anti-NKG2D antibody in both subsets. One of two experiments with identical results is shown. (F) B220+CD11c+ cells produce higher amounts of IFN-γ than B220−CD11clow/− NK cells in response to PMA/ionomycin, IL-12 plus IL-18, and CpG ODN1826 and ODN1668. 2 × 104 cells for each condition and subset were assayed in 100 μl of cell culture medium. The supernatants were analyzed after overnight culture. (G) TNF-α was measured after stimulation with PMA/ionomycin and CpG, as described above. No TNF-α secretion was detected upon stimulation with IL-18 plus IL-12. In E and F, one of two experiments with identical results is shown.
Comparison of the cytolytic capacity of B220+CD11c+ and B220−CD11clow/− NK cells revealed that these NK cell subsets killed the classical target YAC-1 ex vivo equally well (Fig. 2 E). Reduction of YAC-1 killing by an anti-NKG2D antibody demonstrated that cytotoxicity was partly mediated by the activating receptor NKG2D in both subsets. We also compared the ability of the two cell subsets to secrete cytokines in response to different activating stimuli. Both subsets produced IFN-γ in response to PMA/ionomycin and IL-12/IL-18 (Fig. 2 F), although B220+CD11c+ cells produced two- to fourfold more IFN-γ than B220−CD11clow/− cells. In addition, both cell subsets produced IFN-γ and TNF-α in response to CpG stimulation, although B220+CD11c+ cells were again much more efficient than B220−CD11clow/− cells (Fig. 2, F and E, and Fig. S1). Collectively, these results demonstrate that B220+CD11c+ NK cells are enriched in CD27+CD11b+ NK cells that secrete cytokines more effectively than conventional B220−CD11clow/− NK cells, although they are equivalent in lytic capacity.
A minor subset of B220+CD11c+NK1.1+ cells express MHC class II
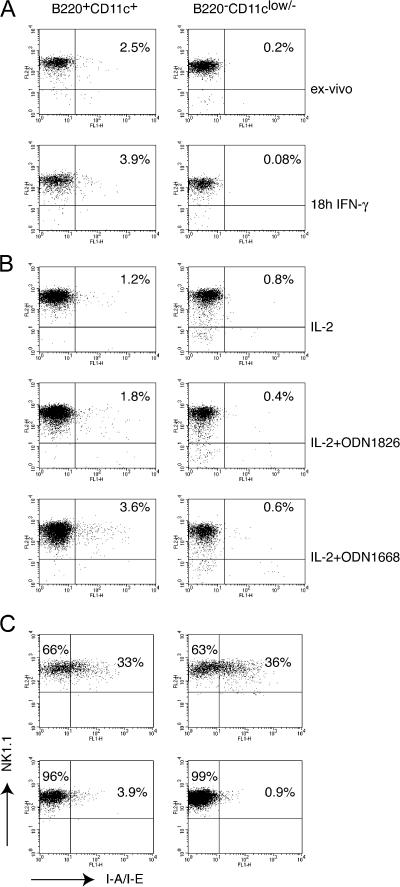
It has been proposed that B220+CD11c+NK1.1+ cells are related to DCs because they express MHC class II (2, 3). To determine whether MHC class II is a unique feature of B220+CD11c+NK1.1+ cells, we assessed MHC class II expression on B220+CD11c+ and B220−CD11clow/− NK cells sorted from the spleen. An antibody specific for I-A and I-E detected MHC class II on few B220+CD11c+ NK cells (between 1 and 5%) (Fig. 3 A and Fig. S2, which is available at http://www.jem.org/cgi/content/full/jem.20070991/DC1). Treatment of sorted cells with IFN-γ, a known inducer of MHC class II, did not result in a marked increase of MHC expression (Fig. 3 A). However, stimulation of sorted cells with CpG 1668 induced approximately a twofold increase in the percentage of MHC class II+ cells (Fig. 3 B). We conclude that very few spleen NK1.1+B220+CD11c+ cells express MHC class II, and this expression appears to be controlled by an IFN-γ–independent TLR-dependent mechanism. In peripheral lymph nodes many more NK1.1+ NK cells expressed cell surface MHC class II than in the spleen. However, MHC class II expression did not correlate with CD11c and B220 expression (Fig. 3 C).
Figure 3.
MHC class II expression in NK cells sorted from the spleen and lymph nodes. (A) Sorted B220+CD11c+ and B220−CD11clow/− splenic cell subsets were stained with anti-NK1.1 and anti–I-A/I-E, either ex vivo or after overnight culture with 5 μg/ml IFN-γ. Between 1 and 5% of B220+CD11c+ cells expressed MHC class II in at least six different experiments performed. IFN-γ did not induce a significant up-regulation of MHC class II. One of two experiments with identical outcome is shown. Notably, B220+CD11c+ cells reproducibly expressed slightly higher levels of NK1.1. (B) CpG stimulation slightly increased the percentage of MHC class II+ cells within the B220+CD11c+ cell subset. CpG ODN1668 was more efficient than CpG ODN1826; however, the increment in MHC class II+ cells never exceeded a threefold increase. One of two experiments with similar results is illustrated. (C) Inguinal and laterocervical lymph nodes and spleens of RAG2−/− mice were dissected, and DX5+ cells were enriched by magnetic selection. Cells were stained with B220, CD11c, NK1.1, and MHC class II. Gates were applied on the B220+CD11c+NK1.1+ and B220−CD11clow/−NK1.1+ subsets as represented in Fig. 2 A. Although lymph nodes exhibit higher percentages of MHC class II+ NK1.1 cells than spleen, MHC class II expression does not correlate with B220 and/or CD11c. One of three experiments with identical results is shown.
B220+CD11c+NK1.1+ cells belong to the NK cell lineage
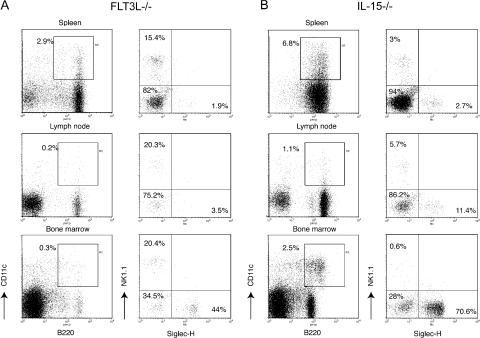
To determine whether B220+CD11c+NK1.1+ cells follow DC or NK cell developmental pathways, we assessed which cytokines are required in vivo for B220+CD11c+NK1.1+ development. FLT3L is a cytokine essential for the development of classical DC subsets and PDC (17, 18), whereas NK cells are less FLT3L dependent (19). Analysis of heterogeneity of B220+CD11c+ cells in FLT3L-deficient mice showed that Siglec-H+NK1.1− PDCs were selectively reduced in the spleen and lymph nodes of these mice (Fig. 4 A and Fig. S3, which is available at http://www.jem.org/cgi/content/full/jem.20070991/DC1). On the contrary, the presence of NK1.1+Siglec-H− was minimally affected, suggesting that B220+CD11c+NK1.1+ cells do not follow a PDC/DC developmental pathway. Although in the bone marrow the relative percentage of Siglec-H+NK1.1− was only partly reduced, and that of NK1.1+Siglec-H− cells was conserved, all B220+CD11c+ cells were severely reduced (∼10–15-fold in different mice). Therefore, the total numbers of bone marrow Siglec-H+NK1.1− and NK1.1+Siglec-H− cells are also reduced (Fig. S3).
Figure 4.
B220+CD11c+NK1.1+ cell development is largely FLT3-L independent but requires IL-15. Siglec-H+NK1.1−, Siglec-H−NK1.1+, and Siglec-H−NK1.1− subsets in the spleen, lymph nodes, and bone marrow of FLT3L−/− mice or IL-15–deficient mice were gated within the B220+CD11c+ population as described in Fig. 1. (A) In the absence of FLT3L, Siglec-H+ PDCs are significantly reduced in the spleen and lymph nodes. Although in bone marrow the percentage of Siglec-H+ cells was only partly reduced, and that of NK1.1+ cells was not affected, the entire B220+CD11c+ population was severely reduced (∼10–15-fold). Percentages of each subset are indicated. An FLT3L−/− spleen contained 3.5–7 × 107 cells. From two femurs of an 8–16 wk-old FLT3L−/− mouse we recovered 1.5–3 × 107 bone marrow cells. (B) Lack of IL-15 resulted in a severe decrease of the B220+CD11c+NK1.1+ subset in all the lymphoid organs analyzed. Percentages of each subset are indicated. An IL-15−/− spleen contained 4.5–7.5 × 107 cells. From two femurs of an 8–16-wk-old IL-15−/− mice we recovered 2.5–4 × 107 bone marrow cells. Data presented in this figure were acquired in parallel with data from the B6 WT mice presented in Fig. 1 and are representative of at least four separate experiments with two mice for each genotype.
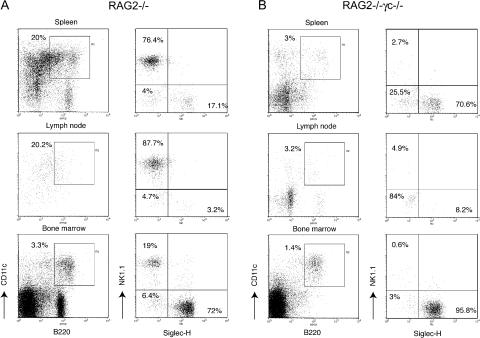
It has been shown that IL-2Rβ−/− mice lack B220+CD11c+NK1.1+ cells (3). IL-2Rβ is the receptor for IL-15, a cytokine required for NK cell development (20). Thus, we examined B220+CD11c+ heterogeneity in IL-15−/− mice and found that B220+CD11c+NK1.1+ cells were severely reduced, whereas PDCs and Siglec-H−NK1.1− cells were not particularly affected (Fig. 4 B and Fig. S3). This result corroborated the hypothesis that the development of B220+CD11c+NK1.1+ cells depends on IL-15/IL-15R signaling. This conclusion was at odds with a previous report showing that B220+CD11c+NK1.1+ cells do not develop in mice lacking γc, which is required for IL-15R signaling. To address this discrepancy, we analyzed RAG2−/− and RAG2−/−γc−/− mice. PDCs were present in both RAG2−/− and RAG2−/−γc−/− mice (Fig. 5 and Fig. S3). Siglec-H−NK1.1− cells were almost absent in the spleens and lymph nodes of RAG2−/− and RAG2−/−γc−/− mice, corroborating the notion that this subset contains mainly B cells. In contrast, B220+CD11c+NK1.1+ cells were dramatically reduced in the lymphoid organs of RAG2−/−γc−/− but not RAG2−/− mice (Fig. 5 and Fig. S3).
Figure 5.
Development of B220+CD11c+NK1.1+ cells requires γc signaling. Siglec-H+NK1.1−, Siglec-H−NK1.1+, and Siglec-H−NK1.1− subsets were evaluated in the spleen, lymph nodes, and bone marrow of RAG2−/− and RAG2−/−γc−/− double-deficient mice. (A) Percentages of Siglec-H−NK1.1− cells were strongly reduced in RAG2−/− mice, consistent with their B cell identity. Siglec-H+NK1.1− and Siglec-H−NK1.1+ were both present. Percentages of each subset are indicated. Each RAG2−/− spleen contained 1.5–3 × 107 cells. From two femurs of a RAG2−/− mouse we recovered 1.5–4 × 107 bone marrow cells. (B) B220+CD11c+NK1.1+ cells in the spleen, lymph nodes, and bone marrow of RAG2−/−γc−/− mice were dramatically reduced. Each RAG2−/−γc−/− spleen contained 1–2.5 × 107 cells. From two femurs of a RAG2−/−γc−/− mouse we recovered 1–2.5 × 107 bone marrow cells. Data presented in this figure were acquired in parallel with data from the B6 WT mice presented in Fig. 1 and are representative of at least three separate experiments with two mice for each genotype.
Notably, a few residual NK1.1+ cells were present in the spleens of RAG2−/−γc−/− mice. To define the nature of these cells, we enriched DX5+ cells from RAG2−/−γc−/− spleens by positive selection. Few live cells (0.5–1 × 104/spleen) copurified with cellular debris (Fig. S4 A, available at http://www.jem.org/cgi/content/full/jem.20070991/DC1). Only 5–10% of these cells were CD3−CD19−NK1.1+ and coexpressed CD11c and B220, and to some extent also MHC class II ex vivo (Fig. S4 B). Collectively, our results indicate that most B220+CD11c+NK1.1+ cells require IL-15/IL-15R signaling for their development, supporting the notion that these cells belong to NK cell lineage. However, very few B220+CD11c+NK1.1+ cells do develop in the absence of γc. The number of cells recovered from RAG2−/−γc−/− spleens was ∼1% of the B220+CD11c+NK1.1+ cells purified from WT mice. Whether these cells are a novel cell type or a rare precursor cell and what the function of these cells is in vivo remains to be determined.
Concluding remarks
Dissection of the heterogeneous B220+CD11c+ cell population has allowed us to address important questions concerning the nature of B220+CD11c+NK1.1+ cells as well as possible functional overlaps with PDCs and DCs. We found that PDCs but not B220+CD11c+NK1.1+ cells produce IFN-α upon stimulation with CpG. We also demonstrated that FLT3L is required for normal PDC numbers in the bone marrow and peripheral lymphoid organs, but it is not essential for B220+CD11c+NK1.1+ cell development. Therefore, PDCs and B220+CD11c+NK1.1+ cells are distinct with respect to secretion of type I IFN and developmental requirements.
Are B220+CD11c+ NK cells any different from other NK cells? We found that most B220+CD11c+ are CD27+CD11b+. A previous study showed that CD27 is a marker for immature NK cells as well as a subset of mature NK cells that home to secondary lymphoid organs and have highly cytolytic and IFN-γ secretion capacity. In contrast, mature CD11b+CD27− NK cells home to peripheral tissues and have limited effector functions (21). Our results demonstrate that CD27+CD11b+ NK cells are highly enriched within the B220+CD11c+ NK population. Moreover, their phenotypes and functions are reminiscent of those of human CD56bright NK cells, which are preferentially found in lymphoid organs (22). Both human CD56bright and murine CD27+CD11b+ NK cells express high levels of the lymph node homing receptor CD62L, the growth factor receptor c-kit, have unique MHC class I receptor repertoires, and secrete IFN-γ more effectively than other NK cells. Because the counterligand of CD27, CD70, is highly expressed by DCs in the T cell area of lymphoid organs (23), B220+CD11c+ NK cells may represent a subset of NK cells that preferentially interact with DCs.
B220+CD11c+NK1.1+ cells were proposed as a novel DC type partially on the basis of their phenotype, particularly the expression of CD11c, B220, and MHC class II. We found that MHC class II is not expressed on most B220+CD11c+ NK cells in the spleen, and it is not induced upon exposure to IFN-γ. However, we did find few MHC class II+ cells within the B220+CD11c+ NK subset, and the percentage of these MHC class II+ cells increased to two- to threefold upon stimulation with CpG. Although peripheral lymph nodes harbored many more MHC class II+ NK1.1+ cells, MHC class II expression did not correlate with that of CD11c and B220. Thus, MHC class II expression may reflect an activation stage of NK cells induced by the lymph node microenvironment. Interestingly, activated human NK cells have been shown to express MHC class II (24) and T cell co-stimulatory molecules such as OX40L (25, 26) and to function as nonprofessional antigen-presenting cells (24). The impact of activated MHC class II+ NK cells on immune responses in vivo remains to be determined. Because NK cells express the low affinity Fc receptor for Ig, CD16, it is tempting to speculate that MHC class II+ NK cells may capture antigen–antibody complexes through CD16 and present processed antigens onto MHC class II. We observed that MHC class II can be induced by CpG but not IFN-γ. Thus, MHC class II expression on some NK cells appears to be uniquely induced by TLR9. Interestingly, human NK cells also express TLR9 and are activated by CpG (27). Thus, NK cell–mediated antigen presentation may be particularly relevant in autoimmune diseases such systemic lupus erithematosus, where cellular DNA in complex with antinuclear antibodies can be captured through CD16 and directed to an endosomal compartment to trigger TLR9 signaling.
B220+CD11c+NK1.1+ cells were designated IKDC also because of their presence in mice that lack γc, which is essential for NK cell but not DC development. However, we have demonstrated that most B220+CD11c+NK1.1+ cells require IL-15 and γc signaling for development, as do conventional NK cells. Consistent with this result, a recent study demonstrated that IKDC development, like NK cell development, requires the Id-2 transcriptional inhibitor (7). Thus, although B220+CD11c+NK1.1+ cells may originate from a Lin−Sca-1+c-KitHiThy1.1−L-selectin+ lymphoid progenitor that may be distinct from the common NK cell progenitor (7), most B220+CD11c+NK1.1+ cells appear to belong to the NK cell lineage and have no obvious developmental overlap with DCs.
MATERIALS AND METHODS
Reagents and mice.
C57BL/6 mice were purchased from The Jackson Laboratory. Mice lacking FLT3L, IL-15, or RAG-2 were purchased from Taconic Farms. Mice doubly deficient in RAG-2 and common cytokine receptor γc mice were provided by W.M. Yokoyama and R.D. Schreiber (Washington University, St. Louis, MO). All animal studies were approved by the Washington University animal studies committee. Antibodies against CD11b, CD11c, CD19, NK1.1, B220, CD3, Ly49-A, Ly49-D, Ly49-C/I, CD27, 2B4, CD62L, DX5, NKG2A/C/E, and I-A/I-E were purchased from BD Biosciences. Antibodies against Ly49C/I/F/H and CD127 were purchased from eBioscience. The anti–Siglec-H antibody was described previously (13). Sorting was performed on a MoFlo cell sorter (DakoCytomation) or a FACSVantage (BD Biosciences). Flow cytometry analysis was performed on a FACSCalibur or FACScan (BD Biosciences). Data were analyzed using the CELLQuest Pro software (BD Biosciences).
Identification, isolation, and culture of cell subsets.
Mouse spleen and lymph nodes were mechanically disrupted and digested with collagenase D (Sigma-Aldrich). Splenocytes and bone marrow were treated with red blood cell lysing buffer (Sigma-Aldrich). Cell suspensions were filtered and treated with FcR blocking antibody before staining. To assay for type I IFN production, sorted cell populations were cultured for 18 h at 106 cells/ml and stimulated with CpG ODN2216 at 10 μg/ml. Cell culture supernatants were assayed for IFN-α production by ELISA (PBL Interferonsource). To FACS sort NK cell subsets, DX5+ cells were preenriched by magnetic positive selection with DX5 microbeads (Miltenyi Biotec). Cytotoxicity was measured by standard chromium release assay. To assay for IFN-γ and TNF-α production, 2 × 104 cells were stimulated overnight with IL-2, IL-12 (1 ng/ml; PeproTech), IL-18 (10 ng/ml; PeproTech), PMA (10−7 M; Sigma-Aldrich), ionomycin (0.5 μg/ml; Sigma-Aldrich), and CpG ODN1826 and CpG ODN1668 (12 μg/ml; QIAGEN) in 100 μl of complete medium. Supernatants were collected and cytokines measured by BD Cytometric Bead Arrays (mouse inflammation kit and mouse Th1/Th2 kit).
Online supplemental material.
Fig. S1 shows expression of NK cell receptors and CpG-induced IFN-γ production in B220+CD11c+ and B220−CD11clow/− NK cells. Fig. S2 shows expression of MHC class II and CD122 on B220+CD11c+ and B220−CD11clow/− NK cells. Fig. S3 shows absolute numbers of B220+CD11c+ cell subsets in the spleen and bone marrow of WT, FLT3-L−/−, IL-15−/−, RAG2−/−, and RAG2−/−γc−/− mice. Fig. S4 shows characterization of DX5+ cells from the spleen of RAG2−/−γc−/− mice. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070991/DC1.
Supplemental Material
Acknowledgments
We would like to thank Suzanne Schloemann in the Flow Cytometry Core of the Department of Pathology and Immunology; William Eades, Jacqueline Hughes, and Chris Holley in the Siteman Cancer Center High Speed Sorter Core Facility for flow cytometry, excellent cell sorting, and advice; Susan Gilfillan for reading the manuscript and advice; Wayne M. Yokoyama and Robert D. Schreiber for generous supply of reagents; and Jason Mills, Victoria Ramsey, and Anja Fuchs for advice and technical assistance in performing gene expression profiling.
A.L. Blasius was supported by the Cancer Research Institute training grant T32 CA009547. This work was supported by National Institutes of Health grants AI056139 and CA109673-02.
The authors have no conflicting financial interests.
W. Barchet's present address is Institute for Clinical Biochemistry and Pharmacology, University Hospital, University of Bonn, 53105 Bonn, Germany.
References
- 1.Asselin-Paturel, C., A. Boonstra, M. Dalod, I. Durand, N. Yessaad, C. Dezutter-Dambuyant, A. Vicari, A. O'Garra, C. Biron, F. Briere, and G. Trinchieri. 2001. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat. Immunol. 2:1144–1150. [DOI] [PubMed] [Google Scholar]
- 2.Taieb, J., N. Chaput, C. Menard, L. Apetoh, E. Ullrich, M. Bonmort, M. Pequignot, N. Casares, M. Terme, C. Flament, et al. 2006. A novel dendritic cell subset involved in tumor immunosurveillance. Nat. Med. 12:214–219. [DOI] [PubMed] [Google Scholar]
- 3.Chan, C.W., E. Crafton, H.N. Fan, J. Flook, K. Yoshimura, M. Skarica, D. Brockstedt, T.W. Dubensky, M.F. Stins, L.L. Lanier, et al. 2006. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nat. Med. 12:207–213. [DOI] [PubMed] [Google Scholar]
- 4.Vremec, D., M. O'Keeffe, H. Hochrein, M. Fuchsberger, I. Caminschi, M. Lahoud, and K. Shortman. 2007. Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood. 109:1165–1173. [DOI] [PubMed] [Google Scholar]
- 5.Spits, H., and L.L. Lanier. 2007. Natural killer or dendritic: what's in a name? Immunity. 26:11–16. [DOI] [PubMed] [Google Scholar]
- 6.Shortman, K., and J.A. Villadangos. 2006. Is it a DC, is it an NK? No, it's an IKDC. Nat. Med. 12:167–168. [DOI] [PubMed] [Google Scholar]
- 7.Welner, R.S., R. Pelayo, K.P. Garrett, X. Chen, S.S. Perry, X.H. Sun, B.L. Kee, and P.W. Kincade. 2007. Interferon-producing killer dendritic cells (IKDC) arise via a unique differentiation pathway from primitive c-kitHiCD62L+ lymphoid progenitors. Blood. 109:4825–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasius, A.L., E. Giurisato, M. Cella, R.D. Schreiber, A.S. Shaw, and M. Colonna. 2006. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol. 177:3260–3265. [DOI] [PubMed] [Google Scholar]
- 9.Asselin-Paturel, C., G. Brizard, J.J. Pin, F. Briere, and G. Trinchieri. 2003. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J. Immunol. 171:6466–6477. [DOI] [PubMed] [Google Scholar]
- 10.Toyama-Sorimachi, N., Y. Omatsu, A. Onoda, Y. Tsujimura, T. Iyoda, A. Kikuchi-Maki, H. Sorimachi, T. Dohi, S. Taki, K. Inaba, and H. Karasuyama. 2005. Inhibitory NK receptor Ly49Q is expressed on subsets of dendritic cells in a cellular maturation- and cytokine stimulation-dependent manner. J. Immunol. 174:4621–4629. [DOI] [PubMed] [Google Scholar]
- 11.Blasius, A.L., M. Cella, J. Maldonado, T. Takai, and M. Colonna. 2006. Siglec-H is an IPC-specific receptor that modulates type I IFN secretion through DAP12. Blood. 107:2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, J., A. Raper, N. Sugita, R. Hingorani, M. Salio, M.J. Palmowski, V. Cerundolo, and P.R. Crocker. 2006. Characterization of Siglec-H as a novel endocytic receptor expressed on murine plasmacytoid dendritic cell precursors. Blood. 107:3600–3608. [DOI] [PubMed] [Google Scholar]
- 13.Blasius, A., W. Vermi, A. Krug, F. Facchetti, M. Cella, and M. Colonna. 2004. A cell-surface molecule selectively expressed on murine natural interferon-producing cells that blocks secretion of interferon-alpha. Blood. 103:4201–4206. [DOI] [PubMed] [Google Scholar]
- 14.Vosshenrich, C.A., M.E. Garcia-Ojeda, S.I. Samson-Villeger, V. Pasqualetto, L. Enault, O. Richard-Le Goff, E. Corcuff, D. Guy-Grand, B. Rocha, A. Cumano, et al. 2006. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat. Immunol. 7:1217–1224. [DOI] [PubMed] [Google Scholar]
- 15.George, T.C., L.H. Mason, J.R. Ortaldo, V. Kumar, and M. Bennett. 1999. Positive recognition of MHC class I molecules by the Ly49D receptor of murine NK cells. J. Immunol. 162:2035–2043. [PubMed] [Google Scholar]
- 16.Furukawa, H., K. Iizuka, J. Poursine-Laurent, N. Shastri, and W.M. Yokoyama. 2002. A ligand for the murine NK activation receptor Ly-49D: activation of tolerized NK cells from beta 2-microglobulin-deficient mice. J. Immunol. 169:126–136. [DOI] [PubMed] [Google Scholar]
- 17.Gilliet, M., A. Boonstra, C. Paturel, S. Antonenko, X.L. Xu, G. Trinchieri, A. O'Garra, and Y.J. Liu. 2002. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 195:953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brawand, P., D.R. Fitzpatrick, B.W. Greenfield, K. Brasel, C.R. Maliszewski, and T. De Smedt. 2002. Murine plasmacytoid pre-dendritic cells generated from Flt3 ligand-supplemented bone marrow cultures are immature APCs. J. Immunol. 169:6711–6719. [DOI] [PubMed] [Google Scholar]
- 19.McKenna, H.J., K.L. Stocking, R.E. Miller, K. Brasel, T. De Smedt, E. Maraskovsky, C.R. Maliszewski, D.H. Lynch, J. Smith, B. Pulendran, et al. 2000. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 95:3489–3497. [PubMed] [Google Scholar]
- 20.Waldmann, T.A., and Y. Tagaya. 1999. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annu. Rev. Immunol. 17:19–49. [DOI] [PubMed] [Google Scholar]
- 21.Hayakawa, Y., and M.J. Smyth. 2006. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J. Immunol. 176:1517–1524. [DOI] [PubMed] [Google Scholar]
- 22.Cooper, M.A., T.A. Fehniger, and M.A. Caligiuri. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22:633–640. [DOI] [PubMed] [Google Scholar]
- 23.Soares, H., H. Waechter, N. Glaichenhaus, E. Mougneau, H. Yagita, O. Mizenina, D. Dudziak, M.C. Nussenzweig, and R.M. Steinman. 2007. A subset of dendritic cells induces CD4+ T cells to produce IFN-γ by an IL-12–independent but CD70-dependent mechanism in vivo. J. Exp. Med. 204:1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roncarolo, M.G., M. Bigler, J.B. Haanen, H. Yssel, R. Bacchetta, J.E. de Vries, and H. Spits. 1991. Natural killer cell clones can efficiently process and present protein antigens. J. Immunol. 147:781–787. [PubMed] [Google Scholar]
- 25.Hanna, J., T. Gonen-Gross, J. Fitchett, T. Rowe, M. Daniels, T.I. Arnon, R. Gazit, A. Joseph, K.W. Schjetne, A. Steinle, et al. 2004. Novel APC-like properties of human NK cells directly regulate T cell activation. J. Clin. Invest. 114:1612–1623. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Zingoni, A., T. Sornasse, B.G. Cocks, Y. Tanaka, A. Santoni, and L.L. Lanier. 2004. Cross-talk between activated human NK cells and CD4+ T cells via OX40-OX40 ligand interactions. J. Immunol. 173:3716–3724. [DOI] [PubMed] [Google Scholar]
- 27.Sivori, S., M. Falco, M. Della Chiesa, S. Carlomagno, M. Vitale, L. Moretta, and A. Moretta. 2004. CpG and double-stranded RNA trigger human NK cells by Toll-like receptors: induction of cytokine release and cytotoxicity against tumors and dendritic cells. Proc. Natl. Acad. Sci. USA. 101:10116–10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.