Abstract
Although physiological and pharmacological evidence suggests the presence of multiple tetrodotoxin-resistant (TTX-R) Na channels in neurons of peripheral nervous system ganglia, only one, SNS/PN3, has been identified in these cells to date. We have identified and sequenced a novel Na channel α-subunit (NaN), predicted to be TTX-R and voltage-gated, that is expressed preferentially in sensory neurons within dorsal root ganglia (DRG) and trigeminal ganglia. The predicted amino acid sequence of NaN can be aligned with the predicted structure of known Na channel α-subunits; all relevant landmark sequences, including positively charged S4 and pore-lining SS1–SS2 segments, and the inactivation tripeptide IFM, are present at predicted positions. However, NaN exhibits only 42–53% similarity to other mammalian Na channels, including SNS/PN3, indicating that it is a novel channel, and suggesting that it may represent a third subfamily of Na channels. NaN transcript levels are reduced significantly 7 days post axotomy in DRG neurons, consistent with previous findings of a reduction in TTX-R Na currents. The preferential expression of NaN in DRG and trigeminal ganglia and the reduction of NaN mRNA levels in DRG after axonal injury suggest that NaN, together with SNS/PN3, may produce TTX-R currents in peripheral sensory neurons and may influence the generation of electrical activity in these cells.
Keywords: ion channels/dorsal root ganglia neurons/trigeminal ganglia/pain
Voltage-gated Na channels in rat brain are composed of three subunits: the pore-forming α-subunit (260 kDa), which is sufficient to generate Na current flow across the membrane, and two auxiliary subunits, β1 (36 kDa) and β2 (33 kDa), which can modulate the properties of the α-subunit (1, 2). Nine distinct α-subunits have been identified in the rat (ref. 3 and references therein, refs. 4–6), and homologues have been cloned from various mammalian species, including humans (3). Specific α-subunits are expressed in a tissue- and developmentally specific manner (7). Aberrant expression patterns or mutations of voltage-gated sodium channel α-subunits underlie a number of human and animal disorders (1, 8–11).
Multiple voltage-gated Na currents, some tetrodotoxin-sensitive (TTX-S) and others TTX-resistant (TTX-R) (12–15), have been observed in dorsal root ganglia (DRG) neurons, which express multiple Na channel α-subunit mRNAs (16). The complex Na current profile in DRG neurons influences their excitability (13, 17–19) and may contribute to ectopic or spontaneous firing in these cells after injury to their axons (8, 20, 21).
Excitability and Na current density are altered in neurons after axonal injury (22–25). After axotomy, rat DRG neurons display dramatic changes in their TTX-R and TTX-S Na currents and in their Na channel mRNA profile; these changes include an attenuation of TTX-R and enhancement of TTX-S Na currents (21, 26), down-regulation of SNS/PN3 transcripts and up-regulation of αIII transcripts (20), and moderate elevation in the levels of αI and αII mRNAs (27). Inflammatory modulators also up-regulate TTX-R current (28–30) and SNS/PN3 transcripts (30) in C type DRG neurons. These results suggest that changes in the Na current profile may contribute to neuropathic and inflammation-evoked pain.
DRG neurons, particularly C type neurons, are unique in expressing high levels of TTX-R Na current (12–15, 21) but until recently, the identity of the channels that produce these currents was unknown. A recently identified Na channel α-subunit (SNS/PN3), which produces a slowly inactivating TTX-R current when expressed in Xenopus oocytes, is expressed preferentially in small peripheral sensory neurons, which include nociceptive (C type) neurons (4, 6). However, physiological and pharmacological evidence (15, 31, 32) suggests the presence of multiple TTX-R Na channels in DRG and nodose ganglia neurons. We report here the identification of a distinct Na channel α-subunit with limited homology to existing subunits, which we term NaN. NaN is predicted to produce a TTX-R current and is expressed preferentially in small-diameter peripheral nervous system neurons. Consistent with its predicted role as a TTX-R Na channel in sensory neurons, NaN is down-regulated after axotomy.
MATERIALS AND METHODS
RNA Preparation and Reverse Transcription.
Total cellular RNA was isolated from L4-L5 DRG dissected from adult Sprague–Dawley rats by the single-step guanidinium isothiocyanate/acid phenol procedure (33). Poly(A)+ RNA was purified from about 300 μg (28 animals) of total DRG RNA (Promega). Half of the purified RNA was used for preparation of Marathon cDNA (CLONTECH) and the other half was used for Northern blot analysis (see below).
First-strand cDNA was synthesized from total RNA as described previously (34). Marathon first- and second-strand cDNA were synthesized by using poly(dT) primer (CLONTECH). The final cDNA product ligated to the adapter primers was diluted 1:250 in Tricine/EDTA buffer and used as template in 5′ and 3′ rapid amplification of cDNA ends (RACE).
PCR.
For the initial identification of NaN-specific fragment, we used generic primers (F1–4; R1–3) designed against highly conserved sequences in domain 1 (D1) of α-subunits (35). NaN sequences 3′-terminal to this fragment were amplified by using NaN-specific and two degenerate generic reverse primers, R4 and R5. The sequence of R4 (5′-ACYTCCATRCANWCCCACAT-3′; Y = T or C, R = A or G; W = A or T, N = A, C, G, or T) and R5 (5′-AGRAARTCNAGCCARCACCA) primers was based on the amino acid sequence MWV/DCMEV, located just N-terminal to domain II S6, and AWCWLDFL, which forms the N-terminal portion of domain III S3 segment, respectively. Amplification was carried out in two stages by using a PTC-200 programmable thermal cycler (34).
Primary RACE amplification was performed in 50 μl final volume using 4 μl diluted DRG marathon cDNA template, 0.2 μM marathon AP-1 and NaN-specific primers, 3.5 units of Expand Long Template enzyme mixture in buffer 3, and 3.0 mM MgCl2 (Boehringer Mannheim). Extension period was adjusted at 1 min/800 bp based on the expected product. 5′ RACE amplification was performed by using Marathon AP-1 (5′-CCATCCTAATACGACTCACTATAGGGC-3′)/NaN-specific R6 (5′-TCTGCTGCCGAGCCAGGTA-3′) primers; nested PCR amplification under similar conditions used marathon AP-2 (5′-ACTCACTATAGGGCTCGAGCGGC-3′)/NaN-specific R7 (5′-CTGAGATAACTGAAATCGCC-3′) primers. 3′ RACE was performed similarly by using NaN-specific F5 (5′-AACATAGTGCTGGAGTTCAGG-3′)/Marathon AP-1 primers; nested PCR was performed by using NaN-specific F6 (5′-GTGGCCTTTGGATTCCGGAGG-3′)/Marathon AP-2 primers. Amplification was via (i) initial denaturation at 92°C for 2 min; (ii) 35 cycles of denaturation at 92°C for 20 sec, annealing at 60°C for 1 min, and elongation at 68°C; and (iii) elongation at 68°C for 5 min. Nested amplification under similar conditions used 2 μl of a 1/500 diluted primary product. Secondary RACE amplification products from five independent reactions were typically pooled, band-isolated, and used as templates for sequencing by primer walking, with analysis using lasergene (DNAstar, Madison, WI) and blast (National Library of Medicine) software.
Tissue Distribution.
Tissue-specific expression of NaN was investigated by reverse transcription–PCR (RT-PCR) and Northern blot analysis. NaN-specific forward (5′-CCCTGCTGCGCTCGGTGAAGAA-3′) and reverse (5′-GACAAAGTAGATCCCAGAGG-3′) primers, which amplify nucleotides 765-1156 (392 bp), were used for RT-PCR under cycling conditions described previously (34).
Approximately 1 μg of poly(A)+ rat DRG RNA, digoxigenin (DIG)-labeled RNA molecular weight marker 11 (Boehringer Mannheim), and 0.24–9.5 kb RNA ladder (GIBCO) with ethidium bromide were electrophoresed in a 0.8% agarose/2.2 M formaldehyde gel and RNA was transferred overnight to positively charged nylon membrane (Boehringer Mannheim) by capillary action (33).
A rat multiple-tissue Northern blot (CLONTECH) of poly(A)+ RNA from heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testis was hybridized along with the DRG blot by using the DIG-labeled, NaN-specific antisense riboprobe, described below. Blots were prehybridized for 3 hr at 60°C in Dig Easy Hyb solution (Boehringer Mannheim), hybridized overnight with probe (100 ng/ml) at 60°C, and washed under stringent conditions, and the hybridization signal was detected by chemiluminescence using disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-ylphenyl phosphate (CSPD) as substrate (Boehringer Mannheim). The blots then were washed immediately in prehybridization solution at room temperature for 30 min, prehybridized for 2 hr at 60°C in fresh solution, and reprobed for β-actin mRNA by using a DIG-labeled riboprobe (100 ng/ml, Boehringer Mannheim) under similar conditions.
Axotomy.
Adult Sprague–Dawley female rats were anaesthetized with ketamine (40 mg/kg) and xylazine (2.5 mg/kg) i.p. Sciatic nerves were exposed on the right side, ligated with 4-0 sutures proximal to the pyriform ligament, transected, and placed in a silicon cuff to prevent regeneration (20). Seven days post axotomy (dpa), the rats were anesthetized, and control (contralateral) and axotomized (ipsilateral) L4/5 DRG were processed for quantitative RT-PCR and in situ hybridization.
Quantitative RT-PCR.
Total cellular RNA was isolated from pooled L4-L5 DRGs of 5 rats, from the respective sides, and reverse transcription and quantitation were performed as described previously (36). PCR conditions for the simultaneous linear amplification of NaN and GAPDH, which was used as an endogenous internal control to compensate for sample-to-sample variation, were determined empirically. To prevent inhibition of the amplification of NaN by excess glyceraldehyde-3-phosphate dehydrogenase (GAPDH) templates, we delayed addition of GAPDH primers (37) for five cycles. GAPDH and NaN primers were used at final concentrations of 0.75 and 3.75 μM, respectively. Control PCR with water or RNA template produced no amplification products (data not shown). Amplification conditions were (i) denaturation at 94°C for 3 min, annealing at 60°C for 2 min, and elongation at 72°C for 2 min; (ii) four cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 1 min, and elongation at 72°C for 1 min, followed by pause at 20°C to add GAPDH primers; (iii) 23 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 1 min, and elongation at 72°C for 1 min; and (iv) elongation at 72°C for 10 min.
In Situ Hybridization.
DIG-labeled sense and antisense riboprobes recognizing NaN nucleotide sequences 1371–1751 (GenBank numbering) were prepared by in vitro transcription. Transcript yield and integrity were determined by comparison with a control DIG-labeled RNA (Boehringer Mannheim) on 2% agarose/2.2 M formaldehyde gel. Tissue section (14 μm) preparation, in situ hybridization conditions, and quantitation of cell diameter (calculated from area in cells displaying nuclear diameters >50% cell diameter) were as described previously (16, 20, 36). Four separate experiments were analyzed for the expression of NaN mRNA in control and axotomized DRG neurons, and statistical significance was determined by using Student’s t test.
RESULTS
Restriction Enzyme Analysis Predicts the Presence of an Additional Na Channel.
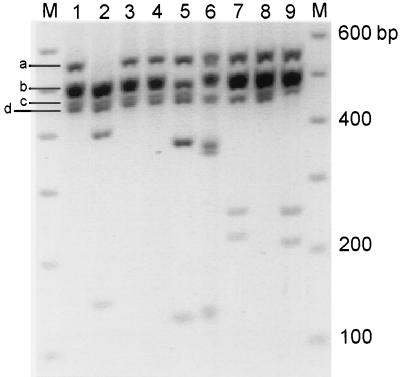
Restriction enzyme profile analysis (35) of amplification products from domain 1 (D1) of voltage-gated Na channel subunits from DRG (Fig. 1) reveals multiple amplification products (lane 1); bands “a”, “b,” and “c” are consistent with the presence of αI (558 bp), αII, and αIII (561 bp; band a), Na6 and PN1 (507 and 501 bp, respectively; band b), and SNS/PN3 (479 bp; band c). The presence of band d was unexpected and suggested the amplification of a novel product. Lanes 2–7 show the result of cutting this DNA with EcoRV, EcoNI, AvaI, SphI, BamHI, and AflII, respectively. As expected, αI product appears to constitute most of band a (lane 2). αII (lane 3) and αIII (lane 4) are not evident by this analysis; however, their very low level of expression was demonstrated previously upon individual amplification (16). Na6 and PN1 products (lanes 5 and 6) are in agreement with the predicted results (35). The doublet migrating faster than the 400-bp marker in lane 6 is from the presence of a restriction site for BamHI in both PN1 and SNS/PN3. SNS/PN3 comprises band c, which is cleaved by AflII (lane 7). NaG (SCL-11) is not detected in this assay (lane 8), but was demonstrated when amplified individually (38).
Figure 1.
Restriction enzyme profile analysis of Na channel domain 1 RT-PCR products from DRG. “M” lanes contain the 100-bp ladder marker (Pharmacia). Lane 1 contains the amplification product from DRG cDNA. Lanes 2–9 show the result of cutting this DNA with EcoRV, EcoNI, AvaI, SphI, BamHI, AflII, XbaI, and EcoRI, which are specific to subunits αI, -II, -III, Na6, PN1, SNS/PN3, NaG, and NaN, respectively. The image was digitized by using gelbase 7500 system (Ultraviolet Products) and printed in black and white dye sublimation mode.
Identification of NaN.
The novel species “d” was amplified reproducibly by F1 and R3 primers (data not shown). The sequence of this PCR product defined a template that corresponds to nucleotides 608-1075 of NaN (GenBank numbering). Digestion by EcoRI was determined to be characteristic of NaN (Fig. 1, lane 9). Using NaN-specific and generic primers, the sequence was extended to DIIIS3. The remaining sequence was obtained from overlapping 5′ and 3′ RACE products.
The final sequence of 5,875 bp predicts an ORF of 1,765 aa (Fig. 2), 40 bp and 536 bp of 5′ and 3′ untranslated sequences, respectively. A polyadenylation signal is present at position 5855, followed, 10 nt downstream, by a poly(A) tail. Northern analysis of DRG poly(A)+ RNA shows a single band corresponding to transcripts about 6.5-kb long (Fig. 3B), suggesting a much longer 5′ untranslated sequence. The first ATG codon matches the vertebrate consensus sequence at −3, +4 positions (39) and is predicted to initiate translation of NaN. Na channel subfamily 1 subunits meet this criterion; the initiator ATG of subfamily 2 members shows a suboptimal sequence at the +4 position. Unlike subfamily 1 members, and in common with subfamily 2 members, NaN does not have an out-of-frame ATG codon at positions −8 to −6.
Figure 2.
Predicted amino acid sequence of NaN. DI–DIV represent the four domains of Na channels, with the putative transmembrane segments underlined. The serine residue of DI-SS2 predicted to underlie the TTX-R phenotype (S355), the PKC phosphorylation site in L3 (T1321), and the tripeptide IFM in L3 involved in fast inactivation (40) are in bold and larger type, and are underlined.
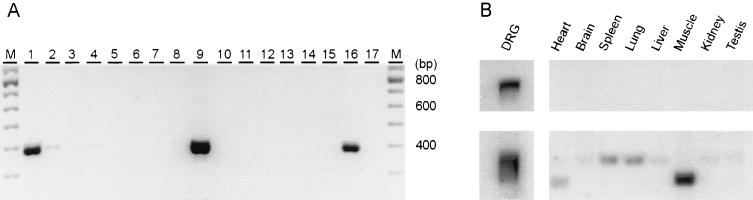
Figure 3.
Tissue distribution of NaN in adult rat by RT-PCR (A) and Northern blot analysis (B). (A) “M” lanes contain the 100-bp ladder marker (Pharmacia). Amplification product from DRG (lanes 1 and 16), trigeminal ganglia (lane 9), cerebral hemispheres (lane 2), and retina (lane 4) are consistent with a predicted size of 392 bp. No detectable signal is seen in cerebellum (lane 3), optic nerve (lane 5), spinal cord (lane 6), sciatic nerve (lane 7), superior cervical ganglia (lane 8), skeletal muscle (lane 10), cardiac muscle (lane 11), adrenal gland (lane 12), uterus (lane 13), liver (lane 14), kidney (lane 15), or water (lane 17). (B Upper) An antisense riboprobe hybridized specifically to a single transcript (about 6.5 kb) in poly(A)+ DRG RNA. No similar hybridization signal was seen in multiple-tissue Northern blot (CLONTECH) lanes containing poly(A)+ RNA from heart, brain, spleen, lung, liver, muscle, kidney, and testis. (B Lower) β-actin (1.6 kb and 2.0 kb) hybridization signal. Heart and muscle lanes show an actin signal at 1.6 kb, consistent with hybridization to the α- or γ-actin forms in these tissues (Boehringer Mannheim).
The predicted amino acid sequence of NaN can be aligned with the predicted primary structures of all known Na channel α-subunits. All of the relevant landmark sequences of voltage-gated Na channels, including the positively charged S4 and the putative pore-lining SS1-SS2 segments, are present at the predicted positions in each of the four domains, indicating that NaN is a member of the Na channel family. However, similarity to known rat Na channels is only 42–50% (Table 1). Intracellular loop L3 shows the highest (51–91%) similarity. The inactivation tripeptide IFM (40) is conserved within L3 of NaN as is the consensus PKC phosphorylation site (T1301; S in all subfamily 1 channels). Intracellular loop L2 shows the lowest (13–20%) similarity compared with other channels, with only 18% similarity to SNS/PN3 (Table 1). Multiple predicted phosphorylation sites also are present in the L1 and L2 loops of NaN. The S4 segments, the voltage sensors of Na channels (41), of NaN contain positively charged residues in the predicted pattern. The number and spacing of such charged residues in the S4 segments of domains I and IV are identical to those in subfamily 1; however, the S4 segments of NaN domains II and III display an intermediate number compared with subfamilies 1 and 2.
Table 1.
Amino acid similarity between NaN and rat Na channel α-subunits
| Subunit | % amino acid similarity
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | N | DI | L1 | D2 | L2 | D3 | L3 | D4 | C | |
| αI | 47 | 59 | 51 | 30 | 55 | 15 | 61 | 85 | 60 | 51 |
| αII | 47 | 59 | 51 | 26 | 55 | 18 | 63 | 85 | 60 | 54 |
| αIII | 48 | 58 | 52 | 24 | 57 | 16 | 63 | 85 | 60 | 53 |
| Na6 | 47 | 54 | 50 | 20 | 57 | 14 | 61 | 87 | 59 | 52 |
| PN1 | 47 | 55 | 56 | 29 | 56 | 17 | 62 | 85 | 59 | 53 |
| μI | 50 | 53 | 52 | 24 | 56 | 13 | 62 | 81 | 60 | 53 |
| rH1 | 49 | 59 | 53 | 36 | 57 | 20 | 66 | 89 | 59 | 57 |
| SNS | 47 | 62 | 54 | 32 | 54 | 18 | 62 | 91 | 61 | 52 |
| SCL-11 | 42 | 50 | 35 | 13 | 38 | 14 | 45 | 51 | 42 | 41 |
Percentage amino acid similarity was determined after aligning NaN and previously identified rat Na channels by using the clustal method (Megalign of lasergene software, DNAstar). DI–D4 represent the four domains of transmembrane segments and their linkers; L1–L3 represent the intracellular loops linking the four domains; N and C represent N and C termini, respectively. Pairwise alignment of NaN and these channels, using gap software (GCG), produced higher identity, 39–55% (SNS/PN3, 54%; rH1, 55%), and similarity, 49–64% (SNS/PN3, 62%; rH1, 64%). By comparison, SNS/PN3 is 65% identical to rH1 (4). All subunit sequences are based on the Genbank database [accession nos. X03638 (αI), X03639 (αII), Y00766 (αIII), L39018 (Na6), U79568 (PN1), M26643 (μI), M27902 (rH1), X92184 (SNS), and Y09164 (SCL-11)].
NaN Is Expressed Preferentially in C Type DRG and Trigeminal Ganglia Neurons.
Screening by RT-PCR (Fig. 3A) and Northern blot analysis (Fig. 3B) shows that NaN is expressed preferentially in DRG and trigeminal ganglia neurons. Lanes 1, 2, 4, 9, and 16 (Fig. 3A) show a single amplification product close to the 400-bp marker, in agreement with the predicted size of 392 bp. Abundant amplification products were obtained from DRG (lanes 1 and 16) and trigeminal ganglia (lane 9). Very faint bands are detectable in cerebral hemisphere and retina (lanes 2 and 4, respectively). NaN was not detected in cerebellum, optic nerve, spinal cord, sciatic nerve, superior cervical ganglia, skeletal muscle, cardiac muscle, adrenal gland, uterus, liver, or kidney (lanes 3, 5–8, and 10–15, respectively). The attenuated NaN signal in cerebral hemisphere and retina, as well as the absence of this signal in the remaining tissues, is not a result of degraded RNA or of PCR inhibitors in the cDNA templates because GAPDH amplification products were obtained in parallel PCRs (data not shown). Northern blot analysis (Fig. 3B) shows a NaN hybridization signal only in DRG, and its absence from multiple other neuronal and nonneuronal samples. The absence of a signal in brain is not surprising given the weak amplification by PCR (Fig. 3A, lane 2).
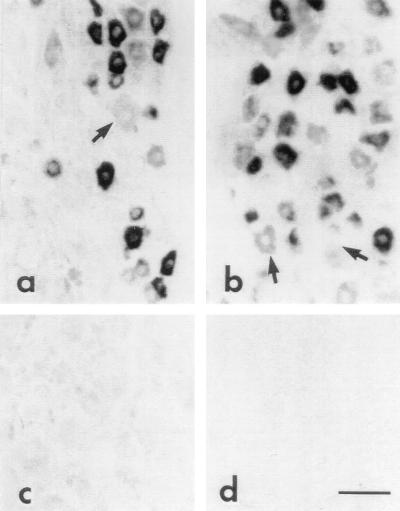
In situ hybridization revealed intense (≥5-fold above background levels) NaN hybridization signals in 82.9% (n = 241) of small (<30-μm diameter) neurons in DRG (Fig. 4b). In contrast, only 18.8% (n = 64) of larger (>30 μm) neurons displayed significant NaN mRNA hybridization signals. Trigeminal ganglion neurons also expressed high levels of NaN mRNA (Fig. 4a). No hybridization signal was detected in cerebellum (Fig. 4d), spinal cord, liver, heart, kidney, or adrenal gland (data not shown) or in DRG hybridized with NaN sense riboprobe (Fig. 4c).
Figure 4.
NaN mRNA expression in adult rat tissue by in situ hybridization. Strong hybridization signal for NaN mRNA is present in many small-diameter neurons within trigeminal ganglia (a) and DRG (b). Large-diameter neurons (arrow) generally lack NaN mRNA hybridization signal. DRG neurons hybridized with NaN sense riboprobe do not show signal above background (c). Hybridization signal for NaN mRNA is not present in cerebellum (d) or in liver, spinal cord, heart, kidney, and adrenal (not shown). (Bar = 50 μm.)
NaN Transcript Levels Decrease After Axotomy of DRG Neurons.
Because TTX-R Na currents are known to decrease in DRG neurons after axonal transection (21, 26), DRG neurons were axotomized via sciatic nerve transection, and NaN transcript levels were examined by quantitative RT-PCR and in situ hybridization at 7 dpa, a time when TTX-R Na currents are reduced substantially (21, 26).
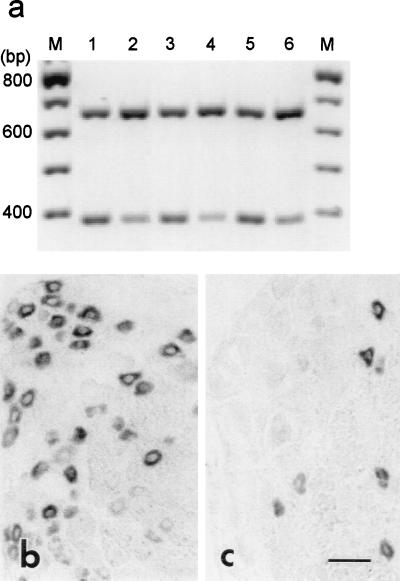
Amplification products of NaN and GAPDH migrated in the gel consistent with their predicted sizes of 392 and 666 bp, respectively (Fig. 5a). A significant reduction in NaN amplification product is evident in the ipsilateral side after axotomy (Fig. 5a, lanes 2, 4, and 6). RT-PCR showed a decrease to approximately 40% of control NaN levels in axotomized DRG (mean ratios ± SD for NaN/GAPDH products, from seven independent amplifications, for uninjured and axotomized DRG neurons = 0.8200 ± 0.0857 and 0.3054 ± 0.0313, respectively; P < 0.001). Consistent with the RT-PCR results, the hybridization signal for NaN mRNA was attenuated (to approximately 60% of control levels) in DRG neurons at 7 dpa (OD = 0.111 ± 0.032 ipsilateral, n = 138, vs. 0.183 ± 0.04 contralateral, n = 240; P < 0.001) (Fig. 5 b and c). A few, small DRG neurons continued to display high levels of NaN mRNA; these are likely to represent those DRG neurons not axotomized by midthigh sciatic nerve transection.
Figure 5.
Analysis of NaN expression in control and axotomized DRG neurons by quantitative RT-PCR (a) and in situ hybridization (b and c). (a) A representative gel analysis of three independent coamplifications of NaN (392 bp) and GAPDH (666 bp) products from control (lanes 1, 3, and 5) and axotomized (lanes 2, 4, and 6) DRG. (b) NaN mRNA is expressed in many small neurons in control DRG. (c) At 7 dpa, NaN hybridization signal is attenuated, with only a few small neurons displaying hybridization signal. (Bar = 50 μm.)
DISCUSSION
Our major findings are as follows. (i) We have identified and sequenced a previously unidentified Na channel, which we have termed NaN. (ii) Based on its predicted amino acid sequence, NaN is expected to produce a TTX-R current and may have altered voltage-dependent properties compared with SNS/PN3 channels. (iii) NaN is expressed preferentially in DRG and trigeminal ganglia neurons, particularly C type DRG neurons. (iv) NaN transcripts are down-regulated in DRG neurons after transection of their axons within the sciatic nerve.
Previously cloned Na channels, expressed within the nervous system, include αI, -II, -III (42–44), and Na6 (45); of these, αI, -II, and -III now have been characterized in heterologous expression systems (46–48) and are known to be TTX-S. NaG, originally thought to be glial-specific (49), also is expressed in sensory neurons of neural crest origin (38). PN1, cloned from PC-12 cells, is expressed preferentially in peripheral nervous system tissues (50). SNS/PN3, preferentially expressed in DRG and trigeminal neurons, produces a TTX-R current (4, 6).
NaN is distinct from all of the known rat Na channels, exhibiting ≤50% similarity with them (similarity highest to μI/SkM1; 50%) as determined by the clustal method. However, NaN′s 1,765 aa can be aligned precisely with all known Na channels and contains all the relevant landmark sequences at the predicted positions. Surprisingly, NaN is only 47% similar to the other TTX-R channel expressed in DRG and trigeminal neurons (SNS/PN3) and 49% similar to the TTX-R cardiac muscle Na channel (rH1/SkM2). Indeed, NaN appears to be equally distant from all previously identified mammalian Na channels (data not shown). Coupled with the fact that NaN shares structural features characteristic of both subfamilies 1 and 2, this may suggest an ancestral relationship between NaN and these Na channels. Alternatively, NaN may represent a third Na channel subfamily, with properties intermediate between the existing two subfamilies.
NaN demonstrates features in common with both subfamilies 1 and 2 (51). NaN transcripts lack the out-of-frame ATG at position −8 to −6 that is present in all previously cloned subfamily 1 cDNAs (data not shown), but is absent in transcripts of subfamily 2 members (5, 51). The inactivation IFM tripeptide (40) is present in NaN; the sequence of L3 including IFM is more like subfamily 1 than it is like subfamily 2. Notably, a serine residue is located in DI-SS2 of NaN at a position analogous to that of SNS/PN3 and to the cysteine residue in rH1 where they confer TTX-R phenotype (52, 53), which suggests that NaN encodes a TTX-R channel. The linker joining S3 and S4 of D4 of NaN is longer, as in SNS/PN3, than those of the other channels; site-directed mutagenesis suggests that this may contribute to faster recovery from inactivation (54).
NaN has distinct structural features, compared with SNS/PN3 and rH1, that suggest different properties and/or modulation. Because NaN has a reduced number [intermediate between that of subfamilies 1 and 2 (5, 51)] of positively charged residues in DII- and DIII-S4 segments, it is not unreasonable to predict that NaN may have altered voltage-dependence or kinetic properties compared with SNS/PN3 and rH1. The putative intracellular L1 and L2 loops of NaN show the least similarity to the other channels. L2 of NaN shows only 18% similarity to L2 of SNS/PN3. These loops contain predicted phosphorylation sites that have been shown to modulate Na currents (55, 56). The different sequence of these loops in NaN and SNS/PN3 suggests that NaN may be regulated/modulated differently than SNS/PN3 in vivo.
Our RT-PCR and Northern analysis results indicate that NaN is expressed preferentially in peripheral sensory ganglia, i.e., DRG and trigeminal ganglia, but is not detectable by these assays in other neural tissues, nor is it detectable in other nonneuronal tissues that possess excitable membranes. Very low levels of NaN expression were detected in the retina and cerebral hemisphere. Retinal ganglion neurons of the newt have been shown recently to produce a TTX-R current (57) that could arise from a NaN-like channel; rat retina lacks transcripts of SNS/PN3 and displays only very low levels of rH1, the two previously identified TTX-R channels, as determined by RT-PCR and in situ hybridization (35).
In situ hybridization confirms the results of RT-PCR and Northern blot analysis and indicates that NaN is expressed preferentially in small DRG neurons, which include C type nociceptive cells (58). Consistent with this result, small, C type DRG neurons preferentially express TTX-R currents (12–15, 21). The presence of transcripts of NaN, in addition to those of SNS/PN3 in these cells, suggests that they express two different TTX-R Na channels. Electrophysiological and pharmacological studies have demonstrated heterogeneity in the TTX-R currents in DRG and nodose ganglia neurons (15, 31) and, in fact, led Rush and Elliott (32) to suggest that there are at least two distinct TTX-R Na channels in these cells.
TTX-R Na currents contribute to the encoding and/or transmission of nociceptive information in DRG neurons and appear to participate in the generation of abnormal burst activity underlying paresthesias and chronic pain (18, 21, 26, 28, 30). Previous electrophysiological studies have shown that after transection of their axons within peripheral nerve, TTX-R currents are attenuated in DRG neurons (21, 26). The present observations, of a significant reduction in NaN transcripts in DRG neurons after axotomy, are consistent with the idea that NaN encodes a TTX-R Na channel in these cells.
In summary, the present results demonstrate the presence of a previously unidentified Na channel, NaN, which may represent a prototype of a third class of Na channels in C type DRG and trigeminal neurons. The preferential expression of NaN in these sensory neurons and the plasticity of NaN transcript levels after axotomy suggest that NaN produces TTX-R currents and influences the generation of electrical activity in these cells.
Acknowledgments
We thank B. R. Toftness for technical assistance. This work was supported in part by a grant from the National Multiple Sclerosis Society.
ABBREVIATIONS
- DIG
digoxigenin
- DRG
dorsal root ganglia
- dpa
days post axotomy
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- RT-PCR
reverse transcription–PCR
- RACE
rapid amplification of cDNA ends
- TTX
tetrodotoxin
- TTX-R and -S
TTX-resistant and -sensitive
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF059030).
References
- 1.Barchi R L. Annu Rev Physiol. 1995;57:355–385. doi: 10.1146/annurev.ph.57.030195.002035. [DOI] [PubMed] [Google Scholar]
- 2.Catterall W A. Trends Neurosci. 1993;16:500–506. doi: 10.1016/0166-2236(93)90193-p. [DOI] [PubMed] [Google Scholar]
- 3.Goldin A L. In: Handbook of Receptors and Channels. North R A, editor. Boca Raton, FL: CRC; 1995. pp. 73–100. [Google Scholar]
- 4.Akopian A N, Sivilotti L, Wood J N. Nature (London) 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- 5.Akopian A N, Souslova V, Sivilotti L, Wood J N. FEBS Lett. 1997;400:183–187. doi: 10.1016/s0014-5793(96)01389-0. [DOI] [PubMed] [Google Scholar]
- 6.Sangameswaran L, Delgado S G, Fish L M, Koch B D, Jakeman L B, Stewart G R, Sze P, Hunter J C, Eglen R M, Herman R C. J Biol Chem. 1996;271:5953–5956. doi: 10.1074/jbc.271.11.5953. [DOI] [PubMed] [Google Scholar]
- 7.Beckh S, Noda M, Lubbert H, Numa S. EMBO J. 1989;8:3611–3616. doi: 10.1002/j.1460-2075.1989.tb08534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizzo M A, Kocsis J D, Waxman S G. Eur Neurol. 1996;36:3–12. doi: 10.1159/000117192. [DOI] [PubMed] [Google Scholar]
- 9.Dumaine R, Wang Q, Keating M T, Hartmann H A, Schwartz P J, Brown A M, Kirsch G E. Circ Res. 1996;78:916–924. doi: 10.1161/01.res.78.5.916. [DOI] [PubMed] [Google Scholar]
- 10.Ptacek L J. Neuromuscular Dis. 1997;7:250–255. doi: 10.1016/s0960-8966(97)00046-1. [DOI] [PubMed] [Google Scholar]
- 11.Cannon S C. Neuromuscular Dis. 1997;7:241–249. doi: 10.1016/s0960-8966(97)00430-6. [DOI] [PubMed] [Google Scholar]
- 12.Roy M L, Narahashi T. J Neurosci. 1992;12:2104–2111. doi: 10.1523/JNEUROSCI.12-06-02104.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott A A, Elliott J R. J Physiol. 1993;463:39–56. doi: 10.1113/jphysiol.1993.sp019583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caffrey J M, Eng D L, Black J A, Waxman S G, Kocsis J D. Brain Res. 1992;592:283–297. doi: 10.1016/0006-8993(92)91687-a. [DOI] [PubMed] [Google Scholar]
- 15.Rizzo M A, Kocsis J D, Waxman S G. J Neurophysiol. 1994;72:2796–2815. doi: 10.1152/jn.1994.72.6.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black J A, Dib-Hajj S, McNabola K, Jeste S, Rizzo M A, Kocsis J D, Waxman S G. Mol Brain Res. 1996;43:117–131. doi: 10.1016/s0169-328x(96)00163-5. [DOI] [PubMed] [Google Scholar]
- 17.Honmou O, Utzschneider D A, Rizzo M A, Bowe C M, Waxman S G, Kocsis J D. J Neurophysiol. 1994;71:1627–1637. doi: 10.1152/jn.1994.71.5.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeftinija S. Brain Res. 1994;639:125–134. doi: 10.1016/0006-8993(94)91772-8. [DOI] [PubMed] [Google Scholar]
- 19.Schild J H, Clark J W, Hay M, Mendelowitz D, Andresen M C, Kunze D L. J Neurophysiol. 1994;71:2338–2358. doi: 10.1152/jn.1994.71.6.2338. [DOI] [PubMed] [Google Scholar]
- 20.Dib-Hajj S, Black J A, Felts P, Waxman S G. Proc Natl Acad Sci USA. 1996;93:14950–14954. doi: 10.1073/pnas.93.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cummins T R, Waxman S G. J Neurosci. 1997;17:3503–3514. doi: 10.1523/JNEUROSCI.17-10-03503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallego R, Ivorra I, Morales A. J Physiol (London) 1987;391:39–56. doi: 10.1113/jphysiol.1987.sp016724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuno M, Llinas R. J Physiol (London) 1970;210:807–821. doi: 10.1113/jphysiol.1970.sp009243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Titmus M J, Faber D S. J Neurophysiol. 1986;55:1440–1454. doi: 10.1152/jn.1986.55.6.1440. [DOI] [PubMed] [Google Scholar]
- 25.Sernagor E, Yarom Y, Werman R. Proc Natl Acad Sci USA. 1986;83:7966–7970. doi: 10.1073/pnas.83.20.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizzo M A, Kocsis J D, Waxman S G. Neurobiol Dis. 1995;2:87–96. doi: 10.1006/nbdi.1995.0009. [DOI] [PubMed] [Google Scholar]
- 27.Waxman S G, Kocsis J D, Black J A. J Neurophysiol. 1994;72:466–470. doi: 10.1152/jn.1994.72.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gold M S, Reichling D B, Shuster M J, Levine J D. Proc Natl Acad Sci USA. 1996;93:1108–1112. doi: 10.1073/pnas.93.3.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.England S, Bevan S, Docherty R J. J Physiol (London) 1996;495:429–440. doi: 10.1113/jphysiol.1996.sp021604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka M, Cummins T R, Ishikawa K, Dib-Hajj S D, Black J A, Waxman S G. NeuroReport. 1998;9:967–972. doi: 10.1097/00001756-199804200-00003. [DOI] [PubMed] [Google Scholar]
- 31.Schild J H, Kunze D L. J Neurophysiol. 1997;78:3198–3209. doi: 10.1152/jn.1997.78.6.3198. [DOI] [PubMed] [Google Scholar]
- 32.Rush A M, Elliott J R. Neurosci Lett. 1997;226:95–98. doi: 10.1016/s0304-3940(97)00258-9. [DOI] [PubMed] [Google Scholar]
- 33.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. In: Current Protocols. Janssen K, editor. New York: Wiley; 1994. [Google Scholar]
- 34.Dib-Hajj S D, Hinson A W, Black J A, Waxman S G. FEBS Lett. 1996;384:78–82. doi: 10.1016/0014-5793(96)00273-6. [DOI] [PubMed] [Google Scholar]
- 35.Fjell J, Dib-Hajj S, Fried K, Black J A, Waxman S G. Mol Brain Res. 1997;50:197–204. doi: 10.1016/s0169-328x(97)00187-3. [DOI] [PubMed] [Google Scholar]
- 36.Dib-Hajj S D, Black J A, Cummins T R, Kenney A M, Kocsis J D, Waxman S G. J Neurophysiol. 1998;79:2668–2676. doi: 10.1152/jn.1998.79.5.2668. [DOI] [PubMed] [Google Scholar]
- 37.Kinoshita T, Imamura J, Nagai H, Shimotohno K. Anal Biochem. 1992;206:231–235. doi: 10.1016/0003-2697(92)90358-e. [DOI] [PubMed] [Google Scholar]
- 38.Felts P A, Black J A, Dib-Hajj S D, Waxman S G. Glia. 1997;21:269–276. doi: 10.1002/(sici)1098-1136(199711)21:3<269::aid-glia2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 39.Kozak M. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 40.West J W, Patton D E, Scheuer T, Wang Y, Goldin A L, Catterall W A. Proc Natl Acad Sci USA. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang N, George A L, Jr, Horn R. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 42.Noda M, Ikeda T, Kayano T, Suzuki H, Takeshima H, Kurasaki M, Takahashi H, Numa S. Nature (London) 1986;320:188–192. doi: 10.1038/320188a0. [DOI] [PubMed] [Google Scholar]
- 43.Auld V J, Goldin A L, Krafte D S, Marshall J, Dunn J M, Catterall W A, Lester H A, Davidson N, Dunn R J. Neuron. 1988;1:449–461. doi: 10.1016/0896-6273(88)90176-6. [DOI] [PubMed] [Google Scholar]
- 44.Kayano T, Noda M, Flockerzi V, Takahashi H, Numa S. FEBS Lett. 1988;228:187–194. doi: 10.1016/0014-5793(88)80614-8. [DOI] [PubMed] [Google Scholar]
- 45.Schaller K L, Krzemien D M, Yarowsky P J, Krueger B K, Caldwell J H. J Neurosci. 1995;15:3231–3242. doi: 10.1523/JNEUROSCI.15-05-03231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda M, Ikeda T, Suzuki H, Takeshima H, Takahashi T, Kuno M, Numa S. Nature (London) 1986;322:826–828. doi: 10.1038/322826a0. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki H, Beckh S, Kubo H, Yahagi N, Ishida H, Kayano T, Noda M, Numa S. FEBS Lett. 1988;228:195–200. doi: 10.1016/0014-5793(88)80615-x. [DOI] [PubMed] [Google Scholar]
- 48.Smith R D, Goldin A L. J Neurosci. 1998;18:811–820. doi: 10.1523/JNEUROSCI.18-03-00811.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gautron S, Dos Santos G, Pinto-Henrique D, Koulakoff A, Gros F, Berwald-Netter Y. Proc Natl Acad Sci USA. 1992;89:7272–7276. doi: 10.1073/pnas.89.15.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toledo-Aral J J, Moss B L, He Z J, Koszowski A G, Whisenand T, Levinson S R, Wolf J J, Silossantiago I, Halegoua S, Mandel G. Proc Natl Acad Sci USA. 1997;94:1527–1532. doi: 10.1073/pnas.94.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Felipe A, Knittle T J, Doyle K L, Tamkun M M. J Biol Chem. 1994;269:30125–30131. [PubMed] [Google Scholar]
- 52.Satin J, Kyle J W, Chen M, Bell P, Cribbs L L, Fozzard H A, Rogart R B. Science. 1992;256:1202–1205. doi: 10.1126/science.256.5060.1202. [DOI] [PubMed] [Google Scholar]
- 53.Sivilotti L, Okuse K, Akopian A N, Moss S, Wood J N. FEBS Lett. 1997;409:49–52. doi: 10.1016/s0014-5793(97)00479-1. [DOI] [PubMed] [Google Scholar]
- 54.Dib-Hajj S D, Ishikawa K, Cummins T R, Waxman S G. FEBS Lett. 1997;416:11–14. doi: 10.1016/s0014-5793(97)01154-x. [DOI] [PubMed] [Google Scholar]
- 55.Li M, West J W, Numann R, Murphy B J, Scheuer T, Catterall W A. Science. 1993;261:1439–1442. doi: 10.1126/science.8396273. [DOI] [PubMed] [Google Scholar]
- 56.Smith R D, Goldin A L. J Neurosci. 1997;17:6086–6093. doi: 10.1523/JNEUROSCI.17-16-06086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaneko Y, Matsumoto G, Hanyu Y. Biochem Biophys Res Commun. 1997;240:651–656. doi: 10.1006/bbrc.1997.7696. [DOI] [PubMed] [Google Scholar]
- 58.Harper A A, Lawson S N. J Physiol (London) 1985;359:31–46. doi: 10.1113/jphysiol.1985.sp015573. [DOI] [PMC free article] [PubMed] [Google Scholar]