Abstract
Immune cells express multiple Toll-like receptors (TLRs) that are concomitantly activated by a variety of pathogen products. Although there is presumably a need to coordinate the expression and function of TLRs in individual cells, little is known about the mechanisms governing this process. We show that a protein associated with TLR4 (PRAT4A) is required for multiple TLR responses. PRAT4A resides in the endoplasmic reticulum, and PRAT4A knockdown inhibited trafficking of TLR1 and TLR4 to the cell surface and ligand-induced trafficking of TLR9 to lysosomes. Other cell-surface molecules were expressed normally on immunocytes from PRAT4A−/− mice. There was impaired cytokine production to TLR ligands, except to the TLR3 ligand poly(I:C), and to whole bacteria. Activation of antigen-specific T helper type 1 responses were also defective. Moreover, PRAT4A−/− bone marrow chimeric mice were resistant to lipopolysaccharide-induced sepsis. These results suggest that PRAT4A regulates the subcellular distribution and response of multiple TLRs and is required for both innate and adaptive immune responses.
Toll-like receptors (TLRs) can sense a variety of microbial products, such as microbial membrane lipids or nucleic acids. Cell-surface TLRs, including TLR4/MD-2, TLR1/TLR2, and TLR6/TLR2, recognize microbial membrane lipids, whereas TLR3, TLR7, TLR8, and TLR9 reside in intracellular organelles and recognize microbial nucleic acids (1, 2). Immune cells such as DCs or macrophages express multiple TLRs, which are concomitantly activated in response to pathogens, because single microbes or viruses express a variety of TLR ligands. Given that multiple TLRs simultaneously respond to pathogens, their distribution and activation need to be orchestrated for optimal immune responses. Indeed, a synergistic relationship between TLR4/MD-2 and TLR7/9 has been recently reported in the triggering of IL-12 and other Th1-promoting cytokines by DCs (3, 4). Dual recognition of Mycobacterium tuberculosis by TLR2 and TLR9 is required for efficient responses (5). On the other hand, there is evidence that the collective utilization of TLRs must be limited to avoid excessive immune responses. For example, overexpression of TLRs causes autoimmunity (6, 7). B cells containing the Y-linked autoimmune accelerator locus are intrinsically biased toward nucleolar antigens because of gene duplication and increased expression of TLR7. Further, autoimmunity was evident in mice that encode multiple copies of the TLR4 gene (6).
It is also of note that dedicated chaperones regulate the activity of specific receptors (8–10). Macrophages lacking the gp96 chaperone were hyporesponsive to a variety of TLR ligands (11), and conversely, TLR4 signaling was positively enforced by artificially expressing gp96 on the cell surface (6). TLR4 as well as commensal flora cause production of anti–double-stranded DNA antibody and immune complex–mediated glomerulonephritis in transgenic mice overexpressing gp96. Despite its role as a general housekeeping chaperone, gp96 is unexpectedly specific for the Toll family of receptors in macrophages. Thus, the total density of TLRs on immune cells can depend on a unique protein maturation pathway in the endoplasmic reticulum (ER). This would in turn limit overall TLR responsiveness against a pathogen and avoid hazardous immune responses. We now describe another global regulator of TLR availability and show that it is rate limiting for innate and adaptive immune responses.
RESULTS
Generation of mice lacking a protein associated with TLR4 (PRAT4A)
We previously described the discovery of PRAT4A, which regulates TLR4's surface expression and responsiveness to LPS (12). Mice lacking PRAT4A have now been generated to allow rigorous assessment of its functions. A targeting vector was constructed to replace the first exon containing the initiation codon with the neomycin resistance gene (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20071132/DC1). PRAT4A−/− mice were screened by Southern hybridization and genomic PCR (Fig. S1 B). The PRAT4A messenger RNA (mRNA) and protein were not detectable in mutant mice homozygous for the disrupted allele (Fig. S1, C and D). PRAT4A−/− mice were born in less than the expected Mendelian ratio (17 out of 156). PRAT4A−/− mice appeared normal when they were born but their growth thereafter was runted (Fig. S1 E), and about half of the mice (10 out of 17) died by the end of the weaning period. The cause of lethality and grow retardation was not unknown. Adjacent genes might have been affected by the Neo promoter in the PRAT4A targeting vector, leading to these phenotypes. We, however, could not find any potential genes potentially causing growth retardation in the adjacent region. Given that PRAT4A has a chaperone activity, PRAT4A might be required for protein maturation of genes regulating growth such as growth hormone, insulin-like growth factors, or their receptors. This possibility will be addressed in further study.
Despite impaired growth and premature death, the phenotype was much milder than in gp96-deficient mice, whose embryos do not survive beyond 5.5 d (11). Thus, PRAT4A is unlikely to have a general housekeeping role like gp96. PRAT4A−/− mice at 4–5 wk were used for further analysis. We also studied BM chimeric mice to exclude the possibility that impaired growth influenced the immune system. Mice reconstituted with PRAT4A−/− BM cells were not impaired in growth and survival. No differences were found between PRAT4A−/− mice and BM chimeric mice regarding immune cell function. Despite the ubiquitous expression of PRAT4A (12), PRAT4A from recipient mice did not rescue PRAT4A−/− hematopoietic cells, demonstrating that PRAT4A works in a cell-autonomous way.
DCs lacking PRAT4A have impaired responses to a variety of TLR ligands
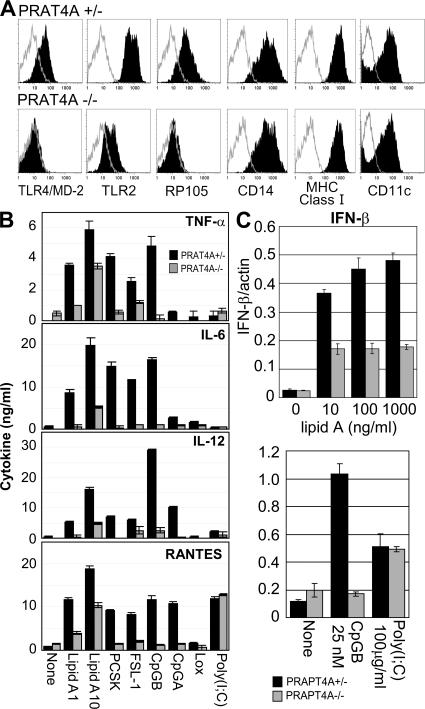
PRAT4A gene silencing in BM-derived DCs (BM-DCs) was previously shown to down-regulate cell-surface expression of TLR4/MD-2 and partially reduce TLR2 (12). BM cells from PRAT4A−/− mice were indistinguishable from wild-type mice with regard to proliferation in the presence of GM-CSF and the expression of cell-surface markers such as CD11c, CD14, and MHC class I antigen (Fig. 1 A). In keeping with the previous results, cell-surface TLR4/MD-2 was down-regulated in PRAT4A−/− DCs (Fig. 1 A). Partial or complete down-regulation was observed in cell-surface TLR2 or the TLR-related molecule radioprotective 105 (RP105) (13), respectively. DCs were then stimulated with a variety of TLR ligands, and production of TNF-α, IL-6, IL-12, and regulated on activation, normal T cell expressed and secreted (RANTES) was measured by ELISA. Partial, but substantial, reduction was observed not only in response to a TLR4/MD-2 ligand (lipid A) but also to a TLR1/TLR2 ligand (Pam3CSK4 [PCSK]), a TLR2/TLR6 ligand (FSL-1), and TLR9 ligands (CpG-B or CpG-A). In contrast, RANTES production in response to poly(I:C) was not altered in PRAT4A−/− DCs (Fig. 1 B). Up-regulation of mRNA encoding IFN-β was partially impaired in response to lipid A and completely impaired in response to CpG-B. No impairment was observed in response to poly(I:C) (Fig. 1 C).
Figure 1.
BM-DCs lacking PRAT4A are impaired in TLR-dependent responses. (A) Shaded histograms show the expression of TLRs and cell-surface markers on BM-DCs from PRAT4A+/− or PRAT4A−/− mice. Open histograms show control staining with the second reagent alone. (B) Bar graphs show cytokine production by BM-DCs in response to a variety of TLR ligands (1 and 10 ng/ml lipid A, 100 ng/ml PCSK, 100 ng/ml FSL-1, 100 nM CpG-B, 1.5 μM CpG-A, 150 μM loxoribine, and 100 μg/ml poly(I:C)). (C) Bar graphs show real-time RT-PCR analyses for mRNA encoding IFN-β, the amount of which was normalized by that of mRNA for β-actin. BM-DCs from BM chimeric mice were stimulated with the indicated TLR ligands, and RNA was collected 2 (lipid A) or 5 (CpG-B and poly(I:C)) h after stimulation, respectively. Data in B and C represent the SD.
PRAT4A is required for TLR-mediated responses in macrophages and B cells
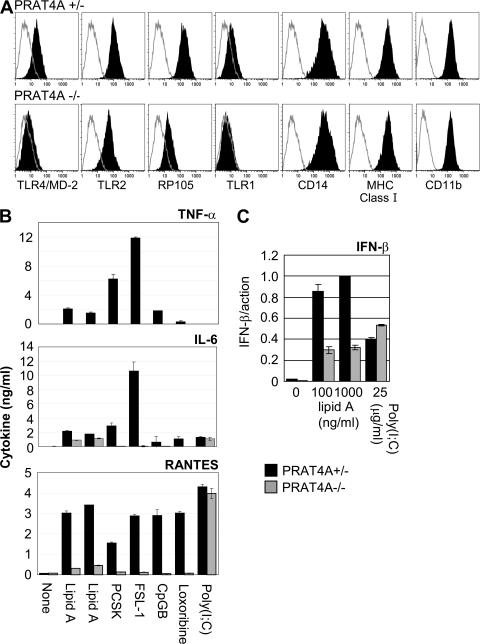
We next studied BM-derived macrophages (BM-macrophages), which were not altered with regard to cell-surface expression of CD14, MHC class I, or CD11b (Fig. 2 A). In contrast, densities of TLR4/MD-2, RP105, and TLR1 were all reduced (Fig. 2 A). TLR2 was only slightly diminished on PRAT4A−/− BM-macrophages. In contrast, TLR2 was reported to be undetectable on gp96−/− BM-macrophages (11). BM-macrophages were stimulated with a variety of TLR ligands, and cytokine production was determined by ELISA (Fig. 2 B). When compared with BM-DCs, more apparent reduction in the production of TNF-α, IL-6, and RANTES was observed in response to TLR ligands. The TLR7 ligand loxoribine induced production of RANTES, a response completely abolished in PRAT4A−/− BM-macrophages. BM-macrophages were similar to BM-DCs in that no impairment was seen in the poly(I:C)-induced production of RANTES. IFN-β mRNA induction in response to lipid A was partially impaired in PRAT4A−/− macrophages, whereas no alteration was seen in response to poly(I:C) (Fig. 2 C).
Figure 2.
PRAT4A is required for TLR responses in macrophages. (A) Shaded histograms show cell-surface expression of the indicated markers on BM-macrophages from PRAT4A+/− or PRAT4A−/− mice. Open histograms show control staining with the second reagent alone. (B) Bar graphs show cytokine production by BM-macrophages in response to a variety of TLR ligands (1 and 10 ng/ml lipid A, 100 ng/ml PCSK, 100 ng/ml FSL-1, 100 nM CpG-B, 150 μM loxoribine, and 100 μg/ml poly(I:C)). (C) Bar graphs show real-time RT-PCR analyses for mRNA encoding IFN-β, the amount of which was normalized by that of mRNA for β-actin. BM-macrophages were stimulated with the indicated TLR ligands and RNA was collected 2 (lipid A) or 5 (poly(I:C)) h after stimulation, respectively. Data in B and C represent the SD.
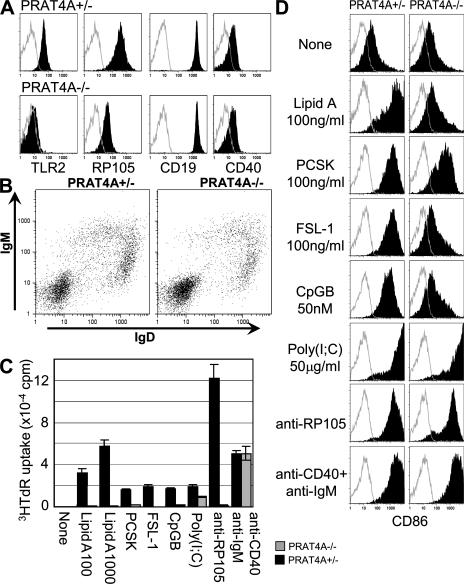
Splenic B cells lacking PRAT4A were normal with regard to cell-surface expression of CD19, CD40, IgM, and IgD (Fig. 3, A and B). The absence of abnormalities in cell-surface IgM and IgD suggests that B cell development is normal in the absence of PRAT4A (Fig. 3 B). In contrast, TLR2 or RP105 were completely or partially down-regulated, respectively (Fig. 3 A). Splenic B cells were enriched from BM chimeric mice and stimulated with a variety of reagents. B cell proliferation was completely abolished in response to all TLR ligands with the exception of poly(I:C) (Fig. 3 C). Despite expression of RP105 on the cell surface, PRAT4A−/− B cells did not proliferate in response to antibody-mediated RP105 ligation. In contrast, no impairment was observed in proliferation when secretory IgM and CD40 were coligated with mAbs. B cell proliferation in response to poly(I:C) was only marginally impaired. Up-regulation of the co-stimulatory molecule CD86 was also impaired in response to lipid A, FSL-1, and CpG-B, and slightly impaired in response to PCSK (Fig. 3 D). No impairment in CD86 up-regulation was seen in B cell responses to poly(I:C), anti-RP105, or anti-CD40 plus anti-IgM (Fig. 3 D). Residual expression of RP105 seemed to be sufficient for partial activation of B cells, as revealed by CD86 up-regulation without proliferation. These results demonstrated that PRAT4A is specifically required for TLR-dependent responses in BM-DCs, BM-macrophages, and B cells.
Figure 3.
PRAT4A is required for TLR responses in B lymphocytes. (A) Shaded histograms show cell-surface expression of the indicated markers on B220-positive splenic B cells. Open histograms show control staining with the second reagent alone. (B) Dot plots show cell-surface expression of IgM and IgD on splenocytes. (C) Proliferation of enriched B cells stimulated with a variety of stimulation (100 and 1,000 ng/ml lipid A, 1,000 ng/ml PCSK, 1,000 ng/ml FSL-1, 500 nM CpG-B, 100 μg/ml poly(I:C), 1 μg/ml anti-RP105 antibody, 1 μg/ml anti-IgM antibody plus 2.25 μg/ml anti-CD40 antibody). Splenic B cells were enriched from BM chimeric mice and stimulated for 3 d. 1 μCi [3H]thymidine was pulsed for the last 6 h, and the incorporated thymidine was counted by a scintillation counter. (D) Shaded histograms show CD86 up-regulation on B cells stimulated with the indicated reagents. The concentration used was as described in C. Open histograms show control staining with the second reagent alone.
N-linked glycosylation of TLR4 is arrested in the ER of PRAT4A knockdown cells
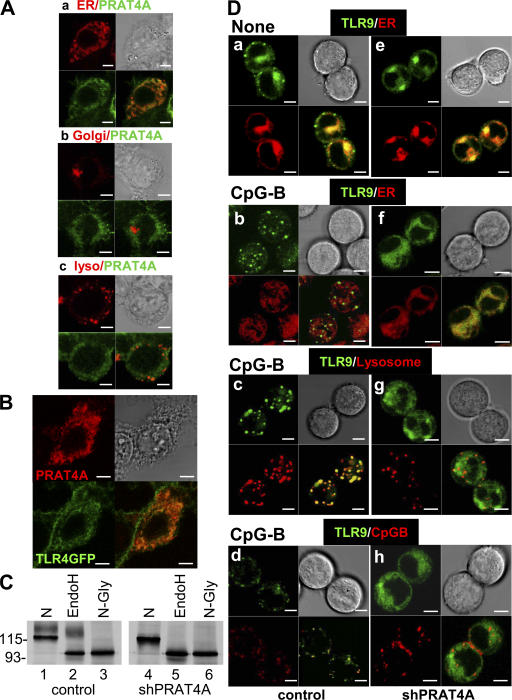
We previously reported that PRAT4A is associated with and regulates cell-surface expression of TLR4 (12). To explore the underlying mechanisms, we determined the subcellular distribution of PRAT4A. The aa sequence of PRAT4A begins with a signal peptide and ends with a sequence similar to the ER retention signal (PDEL), suggesting its localization in the ER (12). Mouse PRAT4A was expressed in HEK293 cells with a Flag epitope at the C terminus and stained with an epitope-specific mAb. PRAT4A was precisely colocalized with an ER marker but not with markers for the Golgi apparatus or lysosomes (Fig. 4 A). When expressed in HEK293 cells, PRAT4A was colocalized with TLR4-GFP (Fig. 4 B), suggesting the physical association of these two molecules in the ER. Given that the ER is a site for protein folding and glycosylation, PRAT4A might have a role in the maturation/glycosylation of TLRs. To address this issue, we conducted further analyses with PRAT4A knockdown cells in which PRAT4A mRNA was silenced by ∼85–90% (12). As published previously (12), the larger species of TLR4 was decreased by PRAT4A gene silencing (Fig. 4 C, compare lane 1 with 4). We suspected that maturation of N-linked oligosaccharides might be impaired by PRAT4A knockdown. Immunoprecipitated TLR4 was treated with endoglycosydase H (endoH), which cleaves high-mannose type N-linked oligosaccharides in the ER but is unable to cleave more mature oligosaccharides on proteins in the Golgi apparatus or on the cell surface. In control cells, the larger TLR4 was resistant to endoH, whereas the smaller TLR4 was sensitive to endoH treatment (Fig. 4 C, lane 2). In PRAT4A knockdown cells, all of the TLR4 was sensitive to endoH treatment (Fig. 4 C, lane 5), indicating that N-linked oligosaccharides remained immature in PRAT4A knockdown cells. Thus, the maturation of TLR4 is likely to be arrested in PRAT4A knockdown cells.
Figure 4.
PRAT4A resides in the ER and is required for TLR4 protein maturation and TLR9 relocation. (A) PRAT4A-GFP was expressed in HEK293 cells together with a marker locating ER (a), the Golgi apparatus (b), or lysosome (c). PRAT4A-GFP (green) and the markers (red) were visualized by confocal microscopy. (B) TLR4-GFP and PRAT4A-FlagHis were expressed in HEK293 cells, and PRAT4A was stained with anti-Flag mAb. TLR4-GFP (green) and PRAT4A-FlagHis (red) were visualized by confocal microscopy. (C) Ba/F3 cells expressing TLR4–Flag/MD-2 were transduced with a control vector (lanes 1–3) or with a vector encoding shPRAT4A (lanes 4–6). Cells were subjected to immunoprecipitation with anti-Flag antibody, followed by treatment with buffer (N), endoH, or N-glycanase (N-Gly), as indicated in the figure. Samples were further subjected to SDS-PAGE and immunoprobing with anti-TLR4 mAb. Molecular masses are shown in kilodaltons. (D) B cell lymphoma M12 cells expressing TLR9-GFP plus a control vector (left) or TLR9-GFP plus shPRAT4A (right) were stimulated with medium (a and e) or 1 μM CpG-B for 2 h (b–d and f–h). Cells were counterstained with markers for ER (a, b, e, and f) or lysosome (c and g) and visualized by confocal microscopy. (bottom) CpG-B conjugated with 1 μM rhodamine was used for stimulation, and its distribution was determined in d and h. Bars, 5 μm.
PRAT4A is associated with TLR1 and required for its cell-surface expression
Our functional studies revealed that PRAT4A is required not only for TLR4-dependent responses but also for TLR2 responses (Figs. 1–3 ). Cell-surface expression of TLR2 was, however, not completely abolished. TLR1, together with TLR2, responds to a lipopeptide, PCSK (14). Cell-surface expression of TLR1 was very low but apparently down-regulated in PRAT4A−/− BM-macrophages (Fig. 2 A). We suspected that cell-surface expression of TLR1 might be dependent on PRAT4A. With Ba/F3 transfectants overexpressing TLR1 and PRAT4A, PRAT4A was found to be associated with TLR1 (Fig. S2 A, available at http://www.jem.org/cgi/content/full/jem.20071132/DC1). As is the case with TLR4 (12), the smaller species of TLR1 was preferentially coprecipitated with PRAT4A (Fig. S2 A, lane 3). The A20 B cell lymphoma expresses TLR1 and TLR2 on the cell surface. The PRAT4A knockdown reduced cell-surface expression of TLR1 more effectively than TLR2 (Fig. 2 B). Additionally, PRAT4A knockdown reduced the completely glycosylated TLR1 (Fig. S2 C, compare lane 1 with 4), and all of the TLR1 in PRAT4A knockdown cells was sensitive to endoH glycosidase (Fig. S2 C, lane 5). Therefore, TLR1, like TLR4, is unable to mature in the ER in the absence of PRAT4A. Hyporesponsiveness of PRAT4A−/− cells to the PCSK TLR1/TLR2 ligand is likely to be explained by down-regulation of cell-surface TLR1. We are currently studying the physical and functional association between PRAT4A and TLR6. In the previous study (12), we were unable to detect physical association between PRAT4A and TLR2. Nonetheless, cell-surface TLR2 was diminished on PRAT4A−/− cells (Figs. 1–3 ). We cannot exclude the possibility that PRAT4A is associated with TLR2. TLR2 is, however, less dependent on PRAT4A than TLR1, because PRAT4A gene silencing in the A20 B cell lymphoma had more apparent effect on TLR1 than on TLR2 (Fig. S2 B, compare the middle and right columns).
TLR9 is unable to traffic to lysosomes upon ligand stimulation in PRAT4A knockdown cells
All TLR9-dependent responses were completely abolished in BM-DCs, BM-macrophages, and B cells from mice lacking PRAT4A (Figs. 1–3 ). Because TLR9 resides in the ER, N-linked oligosaccharides remain immature and are all cleaved by endoH treatment (15, 16). TLR9 may differ from cell-surface TLRs in a way requiring PRAT4A. To address a molecular mechanism by which PRAT4A controls TLR9, we first studied physical association between these two molecules. We were able to detect PRAT4A coprecipitation with TLR9, whereas coprecipitation of TLR9-GFP with PRAT4A-hemagglutinin (HA) was very weak (Fig. S3 A, available at http://www.jem.org/cgi/content/full/jem.20071132/DC1). TLR9-GFP and short hairpin RNA (shRNA) targeting PRAT4A (shPRAT4A) were both expressed in the M12 B cell line. A 75% silencing effect was confirmed by real-time RT-PCR (Fig. S3 B). The TLR9-GFP protein in cells expressing shPRAT4A was still detectable by flow cytometry (Fig. S3 C). We studied the subcellular distribution of TLR9 in the absence of CpG-B stimulation. Given that TLR9 is an ER-resident protein, TLR9-GFP was compared with an ER marker. Colocalization of TLR9-GFP with an ER marker was not affected by shPRAT4A (Fig. 4 D, a and e). Upon CpG-B stimulation, TLR9 was reported to redistribute from the ER to CpG-B DNA–containing lysosomes (15, 16). To see if PRAT4A was required for ligand-induced TLR9 trafficking, M12 cells expressing TLR9-GFP were stimulated with CpG-B, and the subcellular distribution of TLR9-GFP was studied (Fig. 4 D). TLR9-GFP in control cells relocated out of the ER after 2 h of stimulation with CpG-B, whereas TLR9-GFP in M12 cells expressing shPRAT4A remained colocalized with an ER marker (Fig. 4 D, compare b with f). Relocated TLR9 in control M12 cells was colocalized with a lysosome marker and with internalized CpG-B conjugated with rhodamine (Fig. 4 D, c and d), indicating the egress of TLR9 from the ER and its ligand–receptor interaction in lysosomes. In sharp contrast, TLR9-GFP in M12 cells expressing shPRAT4A remained colocalized with an ER marker but not with a lysosome marker or CpG-B–rhodamine (Fig. 4 D, f–h). shPRAT4A did not influence internalization of CpG-B or its trafficking to lyososmes (Fig. 4 D, d and h). These results clearly demonstrate a requirement for PRAT4A in ligand-induced relocation of TLR9 from the ER to lysosomes. Collectively with the results regarding TLR4 and TLR1, TLRs seem to be unable to mature in the ER or to egress from the ER in the absence of PRAT4A.
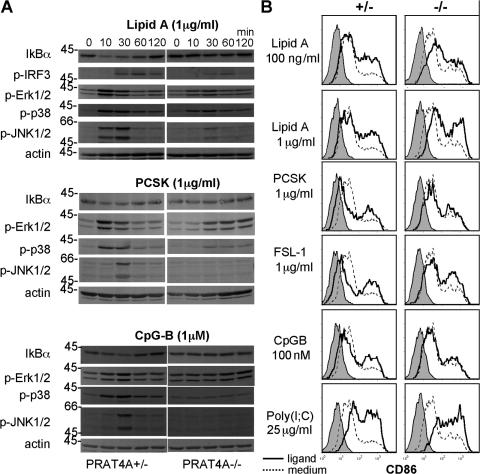
PRAT4A-independent signaling in response to lipid A or a lipopeptide
Some PRAT4A-independent responses were seen in response to LPS or lipopeptides, and this was particularly the case with BM-DCs (Fig. 1). Therefore, we studied signaling events upon stimulation with lipid A, PCSK, or CpG-B (Fig. 5 A). Lipid A–induced IκBα degradation was apparently inhibited, whereas IFN regulatory factor (IRF) 3 and mitogen-activated protein kinase phosphorylation were partially impaired in PRAT4A−/− BM-DCs. After PCSK stimulation, phosphorylation of extracellular signal-regulated kinase 1/2 and p38 was apparently delayed, and c-Jun N-terminal kinase 1/2 phosphorylation was impaired in PRAT4A−/− BM-DCs. CpG-B was distinct from these two TLR ligands in that all of the signaling pathways studied were completely abolished in PRAT4A−/− BM-DCs. TLR9 was completely dependent on PRAT4A, whereas TLR4 and TLR2 were still able to signal in the absence of PRAT4A. PRAT4A-independent lipid A responses were most obvious in up-regulation of the CD86 co-stimulatory molecule. BM-DCs were induced from BM chimeric mice and stimulated with lipid A, lipopeptides, CpG-B, and poly(I:C). Poly(I:C) and lipid A were most potent in inducing CD86 up-regulation, and no alteration was seen between PRAT4A+/− or PRAT4A−/− cells (Fig. 5 B). A lipopeptide (FSL-1) weakly induced CD86 up-regulation in both PRAT4A+/− and PRAT4A−/− mice. The other lipopeptides, PCSK and CpG, weakly induced CD86 only in PRAT4A+/− cells.
Figure 5.
PRAT4A-independent responses in BM-DCs. (A) BM-DCs from BM chimeric mice were stimulated with lipid A, PCSK, or CpG-B, as indicated in the figure. After the indicated periods of time, cells were lysed and subjected to SDS-PAGE, Western blotting, and immunoprobing with the indicated antibodies. (B) CD86 on BM-DCs from BM chimeric mice was stained 48 h after stimulation with the indicated TLR ligands. Shaded histograms show staining with the second reagent alone. Histograms with continuous or dashed lines show CD86 staining of BM-DCs cultured with TLR ligands or medium, respectively.
Complementation with overexpressed PRAT4A
To confirm the critical role for PRAT4A in multiple TLR responses, we tried to complement PRAT4A knockdown cells and PRAT4A−/− cells with PRAT4A overexpression. PRAT4A expression in RAW264.7 cells was down-regulated with shPRAT4A by >90% (Fig. S4 A, available at http://www.jem.org/cgi/content/full/jem.20071132/DC1). Knockdown cells were further transfected with PRAT4A with silent mutation in the target sequence of shPRAT4A (mutPRAT4A). Cell-surface TLR4 was down-regulated by PRAT4A knockdown and recovered by PRAT4A overexpression (Fig. S4 B). In parallel to the results with BM-macrophages (Fig. 2 A), TLR2 was not much more influenced by PRAT4A knockdown than TLR4 in a macrophage cell line, RAW264.7. RAW264.7 cells were stimulated with TLR ligands and cytokine production was determined by ELISA. TNF-α production in response to TLR2/4/9 ligands was down-regulated by shPRAT4A and complemented by PRAT4A overexpression (Fig. S4, C–E). TLR9 responses seemed more resistant to PRAT4A complementation than TLR2/4 responses.
To determine the requirement for the C-terminal sequence similar to the ER retention signal, we next conducted retrovirus-mediated complementation of PRAT4A−/− DCs. PRAT4A−/− BM cells that had been treated with 5-fluorouracil (5-FU) were transduced with retroviral vector encoding PRAT4A or its deletion mutants (Fig. S5 A, available at http://www.jem.org/cgi/content/full/jem.20071132/DC1). After DC induction with GM-CSF, expression of PRAT4A was examined by immunoprobing of the cell lysate with anti-PRAT4A polyclonal antibody. We were able to detect endogenous PRAT4A but not the transduced PRAT4A (unpublished data). Expression of the transduced PRAT4A seemed to be much lower than that of endogenous PRAT4A. We observed that the flag-tagged PRAT4A was secreted into the culture supernatant (unpublished data). We therefore conducted immunoprecipitation and immunoprobing of the culture supernatant with antibody to the flag epitope and were able to confirm expression of transduced PRAT4A (Fig. S5 B). We then looked at cell-surface expression of TLR2 and TLR4. DCs induced from 5-FU–treated BM were distinct from conventional BM-DCs in that cell-surface TLR4 was too low to detect by flow cytometry (unpublished data). Instead, we were able to detect cell-surface TLR2 on PRAT4+/+ DCs but not on PRAT4A−/− DCs (Fig. S5 C). Overexpression of PRAT4A complementary DNA partially but apparently up-regulated cell-surface TLR2 (Fig. S5 C). Complementation was also seen in cytokine production against TLR ligands (Fig. S5, D and E). Only the partial complementation seen in cell-surface expression of TLR2, TLR4-dependent production of IL-12, and TLR9-dependent IL-6 production was probably caused by low expression of the transduced PRAT4A. PRAT4A mutant lacking the putative ER retention signal PDEL (Del-1) or the C-terminal 19 aa (Del-2) were both as effective as the full-length PRAT4A in complementing cell-surface expression of TLR2 and TLR4/9 responses (Fig. S5, C–E). No apparent difference in complementation with the PRAT4A mutant was observed between TLR4 and TLR9 responses and between MyD88-dependent IL-12 production and TRIF-dependent RANTES production.
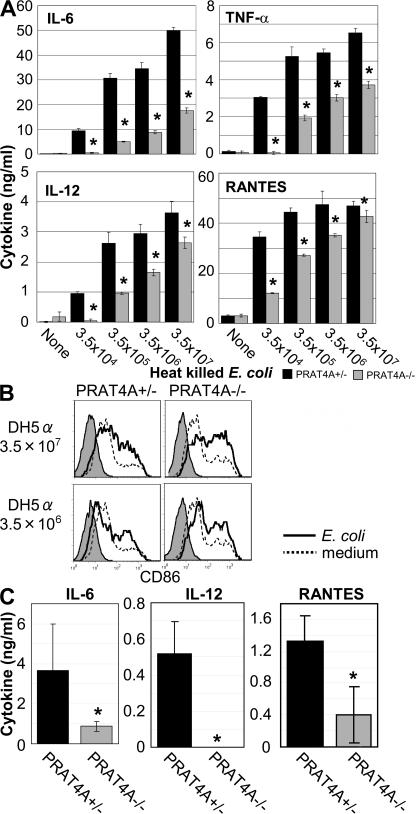
PRAT4A is required for cytokine production against bacteria
The results above (Figs. 1–5 ) have revealed that PRAT4A influences multiple TLR responses in vitro by regulating TLR maturation in the ER and/or the egress from the ER. Bacteria such as Escherichia coli have many TLR ligands, and immune cells such as BM-DCs, macrophages, or B cells express multiple TLRs. Given the apparent importance of PRAT4A for controlling the positioning and densities of multiple TLRs, PRAT4A−/− immune cells are likely to be compromised with respect to bacterial responses. To address this issue, heat-killed E.coli or M. tuberculosis was used for stimulating immune cells in vitro and in vivo. Heat-killed E. coli was effective in inducing the production of IL-6, TNF-α, IL-12, and RANTES in wild-type BM-DCs. All cytokine production was significantly impaired in PRAT4A−/− BM-DCs (Fig. 6 A). In sharp contrast to cytokine production, up-regulation of the CD86 co-stimulatory molecule was unaffected in PRAT4A−/− DCs (Fig. 6 B). In response to M. tuberculosis, cytokine production, particularly of IL-6 and TNF-α, was impaired in PRAT4A−/− BM-DCs (Fig. S6 A, available at http://www.jem.org/cgi/content/full/jem.20071132/DC1), whereas CD86 up-regulation was normal (Fig. S6 B). BM chimeric mice were next stimulated with heat-killed E. coli, and cytokine concentrations in the circulation were measured 3 h after injection. Heat-killed E. coli induced production of IL-12, IL-6, and RANTES in vivo, and all of the production was significantly impaired in PRAT4A−/− mice (Fig. 6 C). Interestingly, IL-12 production against heat-killed bacteria in vivo was more severely impaired than that by BM-DCs (Fig. 6, A and C). An important role for macrophages was recently reported in in vivo IL-12 production in response bacteria (11). Given that PRAT4A−/− macrophages were more severely impaired than PRAT4A−/− BM-DCs in cytokine production against TLR ligands (Figs. 1 and Figs.2), in vivo IL-12 production in response to heat-killed E. coli might be mainly contributed by macrophages.
Figure 6.
PRAT4A is required for cytokine production in response to heat-killed E. coli. (A) BM-DCs from BM chimeric mice were stimulated with heat-killed E. coli, as indicated in the figure. After 24 h of culture, concentrations of cytokines in the supernatant were determined by ELISA. Data represent the SD. *, P < 0.05. (B) CD86 on BM-DCs from BM chimeric mice was stained 48 h after stimulation with heat-killed E. coli. Shaded histograms show staining with the second reagent alone. Histograms with solid or dashed lines show CD86 staining of BM-DCs cultured with heat-killed E. coli or medium, respectively. (C) BM chimeric mice (n = 5) were injected with heat-killed E. coli (3.5 × 108 cells per mouse) and bled 3 h after injection. Serum cytokines were measured by ELISA. Data represent the SD. *, P < 0.05.
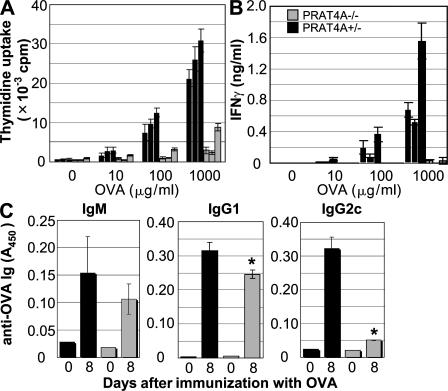
A profound defect in Th1 responses in PRAT4A−/− mice
Impaired production of IL-12 in response to heat-killed bacteria suggested a defect in antigen-specific adaptive immune responses. To address this possibility, mice were immunized (hind legs) with OVA mixed with CFA, which contained heat-killed M. tuberculosis. Immune responses were analyzed 8 d after immunization. Recovered draining lymph node cells from PRAT4A−/− cells poorly proliferated and produced IFN-γ in response to OVA (Fig. 7, A and B). OVA-specific antibodies were next measured by ELISA. IgG2a/c production was profoundly impaired, whereas IgM and IgG1 were not impaired or only weakly impaired, respectively. These results demonstrated a profound defect in Th1 responses in PRAT4A−/− mice.
Figure 7.
Th1 responses are impaired in PRAT4A−/− mice. Cells from draining popliteal lymph nodes were collected from mice 8 d after immunization (hind legs) with OVA in CFA. (A) The proliferation of antigen-specific lymph node cells from PRAT4A+/− or PRAPT4A−/− BM chimeric mice was measured by culturing these cells with OVA for 72 h. (B) IFN-γ production was determined by ELISA from supernatants of antigen-stimulated lymph node cells from PRAT4A+/− or PRAT4A−/− BM chimeric mice. Data are means + SD from each mouse, and the results of three independent mice per group are shown. Similar results were obtained in two independent experiments. (C) Serum was collected before (day 0) and after (day 8) immunization with OVA plus CFA, and the titers of OVA-specific IgM, IgG2a/c, and IgG2b were determined by ELISA. Data are means + SD from three mice per group. Similar results were obtained in two independent experiments. *, P < 0.01.
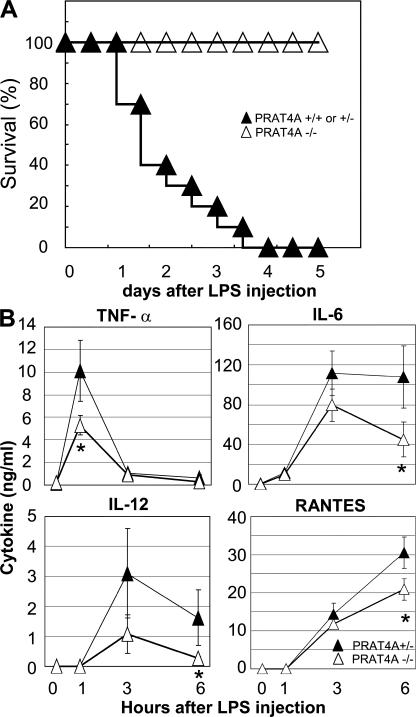
PRAT4A−/− mice are resistant to LPS-induced sepsis
To further confirm the physiological relevance of PRAT4A in in vivo innate immune responses, BM chimeric mice were administered a lethal dose of LPS, and the mortality was monitored. Although control BM chimeric mice all died by 4 d after LPS injection, all of the PRAT4A−/− BM chimeric mice survived the LPS challenge (Fig. 8 A). Sera after LPS injection were collected from LPS-injected mice, and the concentration of cytokines was determined by ELISA. PRAT4A−/− BM chimeric mice showed significantly lower production of cytokines such as TNF-α, IL-6, IL-12, and RANTES (Fig. 8 B).
Figure 8.
PRAT4A−/− BM chimeric mice are resistant to LPS-induced sepsis. (A) PRAT4A−/− (n = 10), PRAT4A+/− (n = 5), and PRAT4A+/+ (n = 5) BM chimeric mice were administered with 400–500 μg LPS intraperitoneally, and the mortality was shown as the percentage of survival. (B) Blood samples from PRAT4A+/− (n = 6) and PRAT4A−/− (n = 6) mice were collected from the vena cava at 0 (no treatment), 1, 3, and 6 h after injection of a lethal dose of LPS (500 μg per mouse). The concentrations of TNF-α, IL-6, IL-12, and RANTES were determined by ELISA. Data represent the SD. *, P < 0.01.
Given the critical role for PRAT4A in innate and acquired immune responses, we finally addressed a possibility that PRAT4A might have a role in limiting excessive TLR responses by examining PRAT4A mRNA expression in BM-DCs after stimulation with a variety of TLR ligands. We found significant down-regulation of PRAT4A mRNA upon stimulation with TLR ligands, whereas gp96, another chaperone required for multiple TLRs, was not down-regulated at all (Fig. S7, available at http://www.jem.org/cgi/content/full/jem.20071132/DC1). These results suggest a regulatory role for PRAT4A in immune responses.
DISCUSSION
Successful targeting of the PRAT4A gene revealed that the corresponding protein is required for many TLR-dependent functions and has the potential for coordinating innate and adaptive immune responses. Characterization of the knockout mice was informative about many aspects of TLR maturation, as well as trafficking within and display on immune cells. Moreover, we were able to categorize particular cellular responses to TLR ligands according to their dependence on PRAT4A.
PRAT4A is required for cell-surface expression of the Toll family of receptors and a related molecule, RP105. No abnormalities were found in cell-surface expression of non-TLR molecules, including CD11c, CD11b, CD14, B220, MHC class I, CD19, IgM, IgD, and others in PRAT4A−/− immune cells. Importantly, PRAT4A was not required for antigen receptor expression and function, because PRAT4A−/− B cells developed normally and responded to BCR-mediated signaling (Fig. 3 B). In this regard, PRAT4A is distinct from gp96, which was reported to chaperone Ig (17). Although gp96 is a general chaperone, PRAT4A seems to be more dedicated to TLRs in immune cells. Immature glycosylation of cell-surface TLRs such as TLR1 and TLR4 in PRAT4A knockdown cells suggests an important role for PRAT4A in TLR protein maturation in the ER (Figs. 4 C and S2 C) (12). Consequently, TLR2/4 responses were impaired in PRAT4A−/− BM-DCs, macrophages, and B cells (Figs. 1–3 ).
PRAT4A is also required for intracellular nucleic acid–sensing TLR7/9. ER-resident TLR9 does not require maturation of N-linked oligosaccharide in the Golgi apparatus. Nonetheless, TLR9 was unable to traffic to the endosome/lysosome during ligand stimulation in PRAT4A knockdown cells (Fig. 4 D). Given that the TLR9 ligand CpG is sequestered to lysosomes, TLR9 is unlikely to interact with its ligand DNA in the absence of PRAT4A. As a result, TLR9 responses and signaling were completely abolished (Figs. 1–5 ). TLR7 responses were impaired as much as TLR9 responses in PRAT4A−/− BM-DCs, BM-macrophages, and splenic B cells (Figs. 1–3 ). Although TLR7 has not been reported to relocate upon ligand stimulation, the roles of PRAT4A for TLR7 are probably similar to those for TLR9. In this regard, TLR3 seemed to be different from TLR7/9. Although poly(I:C) stimulates both TLR3 and cytoplasmic sensors such as melanoma differentiation-associated gene 5 or retinoic acid–inducible gene I (18), mice lacking TLR3 were shown to be impaired in poly(I:C)-induced production of RANTES and IFN-β by macrophages and in CD86 up-regulation on B cells (19, 20). Given that mice lacking PRAT4A were not impaired in these responses to poly(I:C), PRAT4A is unlikely to be required for TLR3.
Complementation with PRAT4A deletion mutants revealed that the C-terminal 19 aa including the putative ER retention signal (PDEL) was not required for TLRs. We previously reported that TLR4 was still associated with PRAT4A mutants lacking PDEL or the C-terminal 19 aa (Del-1 and -2) (21). As long as PRAT4A is associated with TLRs, the ER retention signal may not be so important. Further study with more deletion mutants is needed to understand the mechanisms by which PRAT4A interacts with TLRs.
Dependency on PRAT4A seems to vary among TLRs and cell types. Although TLR7/9 responses were completely dependent on PRAT4A, TLR2/4 responses were still detectable in PRAT4A−/− immune cells. PRAT4A-independent TLR2/4 signaling and responses were more evident in BM-DCs than in macrophages or splenic B cells (Figs. 1–3 and 5 A). We previously reported PRAT4B as a molecule similar to PRAT4A in aa sequence and in physical association with TLR4 (21). PRAT4B might be responsible for PRAT4A-independent TLR2/4 responses, particularly in BM-DCs. This possibility is currently being studied.
Despite that cell-surface TLR4 was not detectable on PRAT4A−/− BM-DCs (Fig. 1 A), there were some responses to lipid A in PRAT4A−/− mice, and this was most evident in up-regulation of CD86 on BM-DCs (Fig. 5 B), suggesting that intracellular TLR4 may still be able to sense LPS. TLR4 was reported to reside in the Golgi apparatus in intestinal epithelial cells and respond to internalized LPS (22). LPS internalization is dependent on cell-surface CD14 (23). If PRAT4A-independent TLR4 responses are dependent on LPS internalization, PRAT4A-independent lipid A responses should not be detected in CD14-negative cells. Indeed, LPS-induced up-regulation of CD86 was seen in PRAT4A−/− BM-DCs but not in PRAT4A−/− B cells (Figs. 3 and Figs.5). LPS-induced up-regulation of CD86 in DCs or macrophages was shown to be dependent on MyD88-independent signaling (24), which induces IRF3 phosphorylation and IFN-β production. Impaired but detectable phosphorylation of IRF3 and IFN-β production was observed with PRAT4A−/− BM-DCs (Figs. 1 and Figs.5). This was apparently sufficient for up-regulation of a co-stimulatory molecule but not completely sufficient for production of MyD88-independent cytokines such as RANTES and IFN-β. As is the case with TLR9 (25), the subcellular distribution of TLR4/MD-2 might influence LPS responses. Further studies are needed to determine the site of LPS recognition in PRAT4A−/− cells.
Although TLR1 was not detectable on PRAT4A−/− BM-DCs, TLR2 was still detectable (Fig. 1 A and not depicted). PCSK was shown to stimulate TLR2 in the absence of TLR1 (26). Interestingly, PCSK-dependent activation of extracellular signal-regulated kinase 1/2 and p38 was delayed in PRAT4A−/− BM-DCs (Fig. 5 A), suggesting a qualitative difference between PRAT4A-dependent and -independent responses to PCSK. It is possible that only the TLR1-independent signaling pathway was activated in PRAT4A−/− BM-DCs. PRAT4A−/− cells might reveal an as yet uncharacterized lipopeptide recognition that is dependent on TLR2 but not TLR1.
Alteration in multiple TLR responses in PRAT4A−/− cells led to impaired cytokine production against bacteria in vitro and in vivo (Fig. 6, A and C). Of particular interest, IL-12 production in response to heat-killed bacteria was profoundly down-regulated in mice lacking PRAT4A−/−. In contrast, up-regulation of CD86 in response to heat-killed bacteria was normal in PRAT4A−/− BM-DCs (Fig. 6 B). Profound defects in multiple TLR responses resulted in a change in adaptive immunity. Upon immunization with OVA mixed with bacteria-containing CFA, PRAT4A−/− BM chimeric mice failed to mount Th1 responses, as revealed by impaired production of IFN-γ and OVA-specific IgG2a/c antibody (Fig. 7). Impaired Th1-dependent responses were reported in mice lacking MyD88 (27, 28). PRAT4A−/− BM-DCs were normal with respect to CD86 up-regulation upon lipid A stimulation and TLR3-dependent responses, both of which are MyD88 independent. Collectively, TLR signaling without PRAT4A may be shifted to MyD88-independent responses, suggesting an unexpected link between the subcellular distribution of TLRs and their downstream signaling pathways. PRAT4A may have a role in determining the balance between MyD88 and MyD88-independent immune responses against a pathogen.
Another change in in vivo immune responses was revealed in the LPS-induced sepsis model. PRAT4A−/− mice were resistant to the LPS-induced sepsis model (Fig. 8 A). LPS-induced cytokine production in vivo was partially but significantly reduced (Fig. 8 B). It is not clear whether such partial impairment in cytokine production was responsible for resistance against LPS-induced sepsis. Mice treated with lethal doses of LPS succumbed at latencies of up to 4 d (Fig. 8 A), long after serum TNF-α and IL-12 returned to basal levels (Fig. 8 B), suggesting that mediators other than these cytokines might contribute to LPS-induced sepsis. In this regard, high-mobility group box 1 (HMGB-1) has been shown to serve as a late mediator in sepsis (29). HMGB-1 has been shown to act on TLRs (30). It is interesting to study whether PRAT4A−/− cells are impaired in their secretion of or responsiveness to HMGB-1. PRAT4A might have a role in detrimental immune responses in LPS-induced sepsis. In this regard, it is of note that PRAT4A mRNA was down-regulated after responses to a variety of TLR ligands (Fig. S7). PRAT4A mRNA down-regulation might have a role in limiting innate immune responses.
Molecular pathways for responses to highly purified microbial/viral products have been extensively studied, and there is clearly more to learn from that type of experimentation. However, a given immunocyte utilizes combinations of receptors to simultaneously recognize multiple ligands, and there must be mechanisms to ensure that the corresponding response is appropriate. Further studies on PRAT4A would cast light on such mechanisms.
MATERIALS AND METHODS
Generation of PRAT4A−/− mice and immunizations.
PRAT4A−/− mice were generated at Unitech. In brief, the targeting vector was constructed to replace the exon 1 with the neomycin resistance gene (Fig. S1). The targeting vector was transfected by electroporation of C57BL/6 embryonic stem (ES) cells. After selection with G418, ES clones were subject to PCR analyses and Southern blotting to identify homologous recombinant clones. Recombinant ES cells were microinjected into C57BL/6 blastocysts to obtain chimeric mice, which were then mated to generate heterozygous and homozygous mice. BM chimeric mice were generated by transferring BM cells from PRAT4A+/− or PRAT4A−/− mice into C57BL/6 mice (6–7 wk of age) that had been irradiated at 950 rad. BM chimeric mice were used 6–10 wk after transfer. Chimerism was confirmed by down-regulated TLR2 and RP105 on splenic B cells from PRAT4A−/− cells.
Chimeric mice were immunized via injections into each hind footpad with OVA emulsified 1:1 in CFA (Difco Laboratories). 8 d after immunization, the draining popliteal and inguinal lymph nodes were isolated, and serum was collected from the mice before (day 0) and after immunization. All animal experiments were conducted with the approval of the Animal Research Committee at the Institute of Medical Science at the University of Tokyo.
Reagents, cells, and mice.
Anti-Flag antibody and anti-Flag–agarose were purchased from Sigma-Aldrich. Anti-HA antibody was purchased from Roche. Anti-GFP antibody was purchased from Invitrogen. The antibodies for flow cytometry (anti-TLR2, anti-CD14, anti-CD11c, and anti-CD86) were purchased from eBioscience. Rat anti–mouse TLR1 mAb (TR23, IgG2a/κ) was established in our laboratory by immunizing a rat with Ba/F3 cells expressing TLR1. The mAb was chosen that stained the line used for immunization but not the original Ba/F3 cells. pAb to mouse PRAT4A was established in our laboratory by immunizing a rabbit with the fusion protein in which mature mouse PRAT4A (aa 38–276) was fused with glutathione S-transferase.
Lipid A purified from Salmonella minnesota (Re-595) and LPS from E. coli (O55:B5) were purchased from Sigma-Aldrich. PCSK and FSL-1 were purchased from EMC Microcollections. CpG-B (TCCATGACGTTCCTGATGCT) and CpG-A (GGGGTCAACGTTGAGGGGGG) DNA were synthesized by Hokkaido System Science. Poly(I:C) and loxoribine (7-allyl-7,8-dihydro-8-oxo-guanosine) were purchased from InvivoGen.
RAW264, Ba/F3, M12, A20, HEK293, and NIH3T3 cells were cultured as previously described (31). C57BL/6 mice were purchased from Japan SLC Inc. and maintained in the animal facility at the Institute of Medical Science at the University of Tokyo. BM-DCs were prepared as previously described (12). In brief, BM cells were plated at 106 cells per milliliter in 24-well plates with 10% FCS–RPMI 1640 supplemented with 10 ng/ml of recombinant mouse GM-CSF (Genzyme). At day 7, cells were harvested and used for flow cytometry. To enrich PRAT4A−/− DCs expressing PRAT4A, we used a retroviral vector with the neomycin resistance gene. BM was isolated from PRAT4A−/− or PRAT4A+/+ mice that had been injected intraperitoneally 4 d before with 5 mg 5-FU. After 48 h of culture, cells were transduced with retroviral supernatant on two successive days. After the second transduction, cells were washed and cultured in the presence of GM-CSF. During DC induction, 400 μg/ml neomycin was included to enrich DCs expressing shRNA.
Flow cytometry.
Cell-surface staining and analyses on a FACSCalibur (BD Biosciences) were previously described (12).
Immunoprecipitation and immunoprobing.
Immunoprecipitation and immunoprobing were previously described (12). The antibodies used for immunoprobing were anti-Flag (M2), anti-HA, and anti-GFP. The second antibodies were goat anti–mouse IgG–alkaline phosphatase conjugate, goat anti–rat IgG–alkaline phosphatase conjugate (American Qualex), and goat anti–rabbit IgG–alkaline phosphatase conjugate (Bio-Rad Laboratories).
endoH and N-glycosidase assay.
Cells were lysed in 1% Triton X-100 lysis buffer. Lysates were incubated with anti-Flag (M2) or anti-GFP antibody-conjugated beads at 4°C for 2 h. Proteins were eluted and treated with endoH or N-glycosidase according to the manufacturer's instructions (New England Biolabs, Inc.). Samples were subjected to SDS-PAGE and immunoprobing with anti-TLR4 or anti-GFP antibodies.
siRNA.
To inhibit mouse PRAT4A expression, we produced a retroviral vector (provided by H. Iba and T. Mizutani, The University of Tokyo, Tokyo, Japan) expressing shRNA, as previously described (12). The target sequence was 5′-gagtttgaagaggtgattgag-3′. As a control, we used cells expressing an empty vector. For complementation of PRAT4A gene silencing, four silent mutations were introduced in the target sequence, as indicated by underlining (5′-gagttcgaggaagtaattgag-3′). This PRAT4A mutant was cloned into a retroviral vector and transduced into RAW264 cells expressing shPRAT4A.
Quantitative RT-PCR.
Quantitative PCR was conducted as previously described (12). In brief, 1 μg of total RNA was reverse transcribed into complementary DNA using ReverTra Ace -α- (TOYOBO), according to the manufacturer's instructions. Quantitative PCR analyses were performed using a PCR system (7300 Fast Real-Time PCR System; Applied Biosystems) with gene expression assays (TaqMan; Applied Biosystems) for mouse PRAT4A (Mm00511161) and IFN-β (Mm00439546). Each sample was normalized using TaqMan gene expression assays for mouse β-actin (Mm00607939).
Immunofluorescence.
Cells were plated on a glass-bottom dish (Matsunami) and used for staining. In some experiments, CpG-rhodamine (Hokkaido System Science) was used to stimulate TLR9-GFP. Cells were fixed by 4% formaldehyde–BS for 10 min at room temperature and washed three times with PBS. To analyze the location of the TLR-GFP fusion proteins, cells were stained with biotinylated cholera toxin subunit B (Sigma-Aldrich), followed by streptavidin–Texas red (Invitrogen), LysoTracker green, or ER tracker red (Invitrogen). DsRed2-ER vector and DsRed-monomer-Golgi vector were purchased from Clontech Laboratories, Inc. TLR-GFP and markers were visualized with confocal microscopy (LSM5 PASCAL; Carl Zeiss, Inc.).
ELISA.
Cells were plated at 105 cells per well in 96-well plates and stimulated with a variety of TLR ligands. After 24 h, supernatant was collected, and concentrations of TNF-α, IL-6, IL-12, and RANTES were determined by ELISA kits (R&D Systems). To measure OVA-specific antibodies, 10 μg/ml OVA was used to coat a microtiterplate, and bound Ig isotypes were detected with specific secondary antibodies (horseradish peroxidase–conjugated goat anti–mouse IgM, IgG1, and IgG2c; SouthernBiotech).
Statistical analysis.
Data from triplicate samples were used for statistical analysis. Statistical significance was calculated by the Student's t-test. P < 0.05 was considered significant.
Online supplemental material.
Fig. S1 shows the strategy for the targeted disruption of the mouse PRAT4A gene. Fig. S2 shows that PRAT4A is associated with TLR1 and is required for its cell-surface expression. Fig. S3 shows the physical association between PRAT4A and TLR9. Fig. S4 shows the complementation of PRAT4A knockdown by PRAT4A overexpression in the RAW264.7 macrophage cell line. Fig. S5 shows the complementation of PRAT4A−/− DCs by retrovirus-mediated overexpression of PRAT4A. Fig. S6 shows the requirement for PRAT4A in cytokine production by BM-DCs against heat-killed M. tuberculosis. Fig. S7 shows down-regulation of PRAT4A mRNA upon stimulation by TLR ligands. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071132/DC1.
Supplemental Material
Acknowledgments
We thank Drs. Hideo Iba and Taketoshi Mizutani for vectors expressing shRNA, and Drs. P.W. Kincade and M. Ogata and Mr. Jennings for critically reviewing the manuscript.
This study was supported by strategic cooperation to control emerging and reemerging infections funded by the Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), a contract research fund from MEXT for the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, the Uehara Memorial Foundation, the Takeda Foundation, and the Naito Foundation.
The authors have no conflicting financial interests.
Abbreviations used: 5-FU, 5-fluorouracil; BM-DC, BM-derived DC; BM-macrophage, BM-derived macrophage; endoH, endoglycosydase H; ER, endoplasmic reticulum; ES, embryonic stem; HA, hemagglutinin; HMGB-1, high-mobility group box 1; IRF, IFN regulatory factor; mRNA, messenger RNA; PCSK, Pam3CSK4; PRAT4A, protein associated with TLR4; RANTES, regulated on activation, normal T cell expressed and secreted; RP105, radioprotective 105; shPRAT4A, shRNA targeting PRAT4A; shRNA, short hairpin RNA; TLR, Toll-like receptor.
K. Takahashi, T. Shibata, and S. Akashi-Takamura contributed equally to this study.
T. Kouro, Y. Nagai, and K. Takatsu's present address is Dept. of Immunobiology and Pharmacological Genetics, Graduate School of Medicine and Pharmaceutical Science for Research, University of Toyama, Toyama-shi, Toyama 930-0194, Japan.
References
- 1.Beutler, B., Z. Jiang, P. Georgel, K. Crozat, B. Croker, S. Rutschmann, X. Du, and K. Hoebe. 2006. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 24:353–389. [DOI] [PubMed] [Google Scholar]
- 2.Kaisho, T., and S. Akira. 2006. Toll-like receptor function and signaling. J. Allergy Clin. Immunol. 117:979–987. [DOI] [PubMed] [Google Scholar]
- 3.Gautier, G., M. Humbert, F. Deauvieau, M. Scuiller, J. Hiscott, E.E. Bates, G. Trinchieri, C. Caux, and P. Garrone. 2005. A type I interferon autocrine–paracrine loop is involved in Toll-like receptor–induced interleukin-12p70 secretion by dendritic cells. J. Exp. Med. 201:1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Napolitani, G., A. Rinaldi, F. Bertoni, F. Sallusto, and A. Lanzavecchia. 2005. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 6:769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bafica, A., C.A. Scanga, C.G. Feng, C. Leifer, A. Cheever, and A. Sher. 2005. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J. Exp. Med. 202:1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, B., Y. Yang, J. Dai, R. Medzhitov, M.A. Freudenberg, P.L. Zhang, and Z. Li. 2006. TLR4 up-regulation at protein or gene level is pathogenic for lupus-like autoimmune disease. J. Immunol. 177:6880–6888. [DOI] [PubMed] [Google Scholar]
- 7.Pisitkun, P., J.A. Deane, M.J. Difilippantonio, T. Tarasenko, A.B. Satterthwaite, and S. Bolland. 2006. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 312:1669–1672. [DOI] [PubMed] [Google Scholar]
- 8.Culi, J., and R.S. Mann. 2003. Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell. 112:343–354. [DOI] [PubMed] [Google Scholar]
- 9.Okajima, T., A. Xu, L. Lei, and K.D. Irvine. 2005. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 307:1599–1603. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J.R., S. Urban, C.F. Garvey, and M. Freeman. 2001. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 107:161–171. [DOI] [PubMed] [Google Scholar]
- 11.Yang, Y., B. Liu, J. Dai, P.K. Srivastava, D.J. Zammit, L. Lefrancois, and Z. Li. 2007. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 26:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakabayashi, Y., M. Kobayashi, S. Akashi-Takamura, N. Tanimura, K. Konno, K. Takahashi, T. Ishii, T. Mizutani, H. Iba, T. Kouro, et al. 2006. A protein associated with Toll-like receptor 4 (PRAT4A) regulates cell surface expression of TLR4. J. Immunol. 177:1772–1779. [DOI] [PubMed] [Google Scholar]
- 13.Nagai, Y., T. Kobayashi, Y. Motoi, K. Ishiguro, S. Akashi, S. Saitoh, Y. Kusumoto, T. Kaisho, S. Akira, M. Matsumoto, et al. 2005. The radioprotective 105/MD-1 complex links TLR2 and TLR4/MD-2 in antibody response to microbial membranes. J. Immunol. 174:7043–7049. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi, O., S. Sato, T. Horiuchi, K. Hoshino, K. Takeda, Z. Dong, R.L. Modlin, and S. Akira. 2002. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169:10–14. [DOI] [PubMed] [Google Scholar]
- 15.Latz, E., A. Schoenemeyer, A. Visintin, K.A. Fitzgerald, B.G. Monks, C.F. Knetter, E. Lien, N.J. Nilsen, T. Espevik, and D.T. Golenbock. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5:190–198. [DOI] [PubMed] [Google Scholar]
- 16.Leifer, C.A., M.N. Kennedy, A. Mazzoni, C. Lee, M.J. Kruhlak, and D.M. Segal. 2004. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J. Immunol. 173:1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melnick, J., J.L. Dul, and Y. Argon. 1994. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 370:373–375. [DOI] [PubMed] [Google Scholar]
- 18.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K.J. Ishii, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science. 301:640–643. [DOI] [PubMed] [Google Scholar]
- 20.Alexopoulou, L., A.C. Holt, R. Medzhitov, and R.A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738. [DOI] [PubMed] [Google Scholar]
- 21.Konno, K., Y. Wakabayashi, S. Akashi-Takamura, T. Ishii, M. Kobayashi, K. Takahashi, Y. Kusumoto, S. Saitoh, Y. Yoshizawa, and K. Miyake. 2006. A molecule that is associated with Toll-like receptor 4 and regulates its cell surface expression. Biochem. Biophys. Res. Commun. 339:1076–1082. [DOI] [PubMed] [Google Scholar]
- 22.Hornef, M.W., B.H. Normark, A. Vandewalle, and S. Normark. 2003. Intracellular recognition of lipopolysaccharide by Toll-like receptor 4 in intestinal epithelial cells. J. Exp. Med. 198:1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitchens, R.L., P. Wang, and R.S. Munford. 1998. Bacterial lipopolysaccharide can enter monocytes via two CD14-dependent pathways. J. Immunol. 161:5534–5545. [PubMed] [Google Scholar]
- 24.Kaisho, T., O. Takeuchi, T. Kawai, K. Hoshino, and S. Akira. 2001. Endotoxin-induced maturation of MyD88-deficient dendritic cells. J. Immunol. 166:5688–5694. [DOI] [PubMed] [Google Scholar]
- 25.Honda, K., Y. Ohba, H. Yanai, H. Negishi, T. Mizutani, A. Takaoka, C. Taya, and T. Taniguchi. 2005. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 434:1035–1040. [DOI] [PubMed] [Google Scholar]
- 26.Buwitt-Beckmann, U., H. Heine, K.-H. Wiesmüller, G. Jung, R. Brock, S. Akira, and A.J. Ulmer. 2006. TLR1- and TLR6-independent recognition of bacterial lipopeptides. J. Biol. Chem. 281:9049–9057. [DOI] [PubMed] [Google Scholar]
- 27.Schnare, M., G.M. Barton, A.C. Holt, K. Takeda, S. Akira, and R. Medzhitov. 2001. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2:947–950. [DOI] [PubMed] [Google Scholar]
- 28.Gavin, A.L., K. Hoebe, B. Duong, T. Ota, C. Martin, B. Beutler, and D. Nemazee. 2006. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 314:1936–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, H., O. Bloom, M. Zhang, J.M. Vishnubhakat, M. Ombrellino, J. Che, A. Frazier, H. Yang, S. Ivanova, L. Borovikova, et al. 1999. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 285:248–251. [DOI] [PubMed] [Google Scholar]
- 30.Tsung, A., R. Sahai, H. Tanaka, A. Nakao, M.P. Fink, M.T. Lotze, H. Yang, J. Li, K.J. Tracey, D.A. Geller, and T.R. Billiar. 2005. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J. Exp. Med. 201:1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akashi-Takamura, S., T. Furuta, K. Takahashi, N. Tanimura, Y. Kusumoto, T. Kobayashi, S. Saitoh, Y. Adachi, T. Doi, and K. Miyake. 2006. Agonistic antibody to TLR4/MD-2 protects mice from acute lethal hepatitis induced by TNF-alpha. J. Immunol. 176:4244–4251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.