Abstract
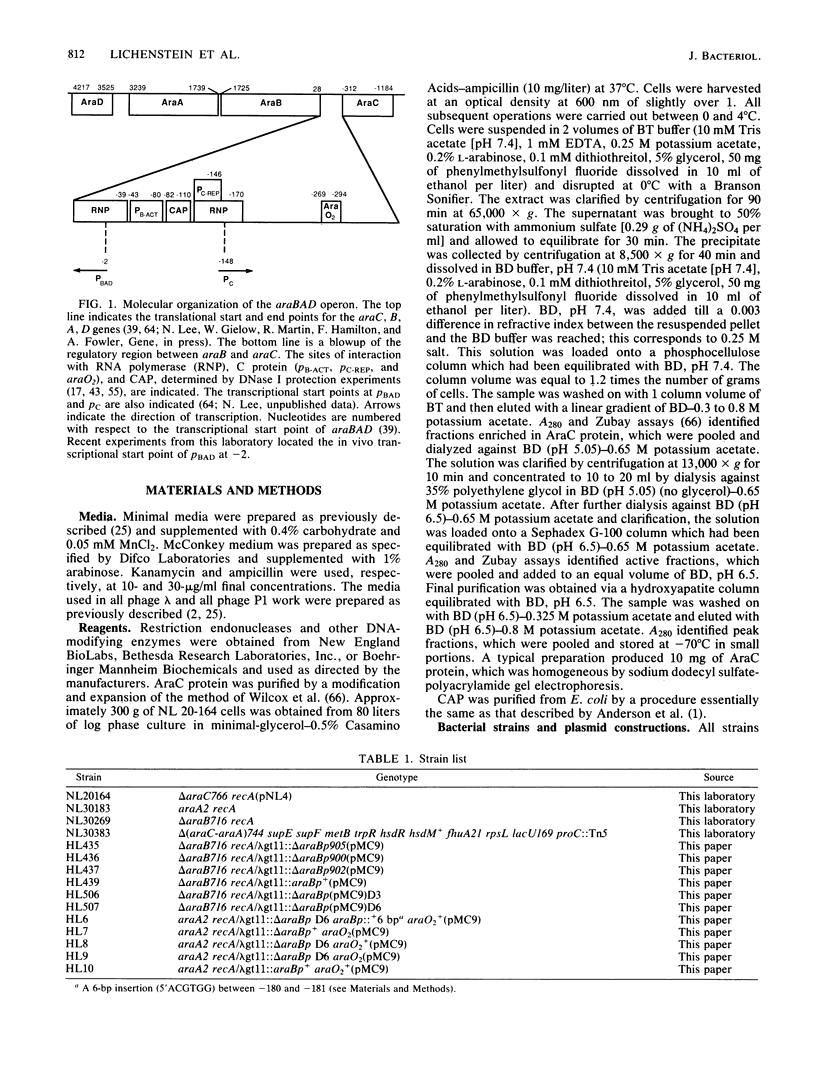
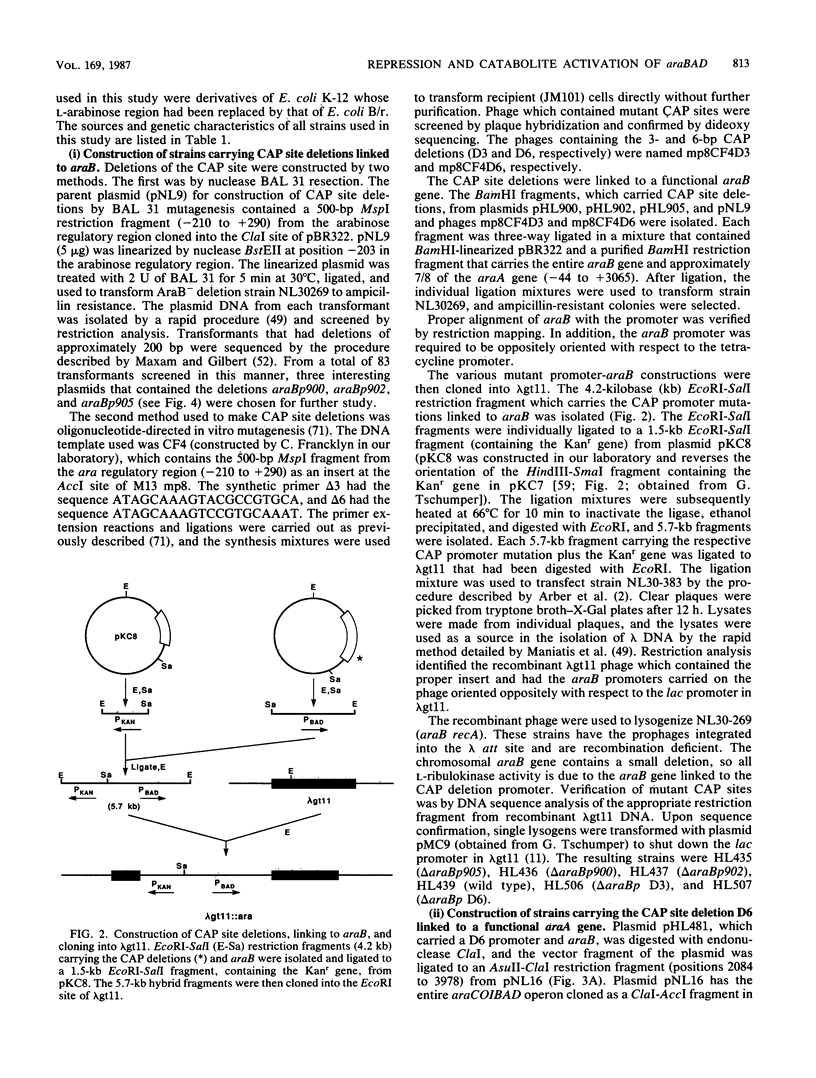
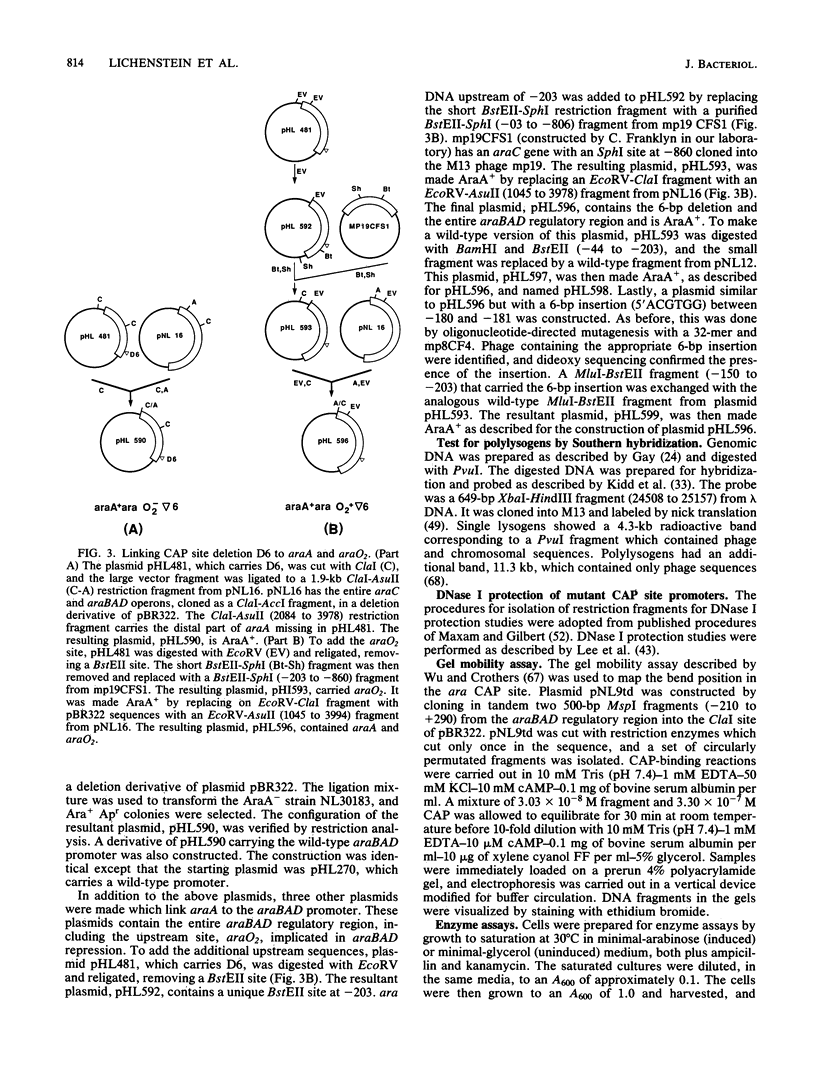
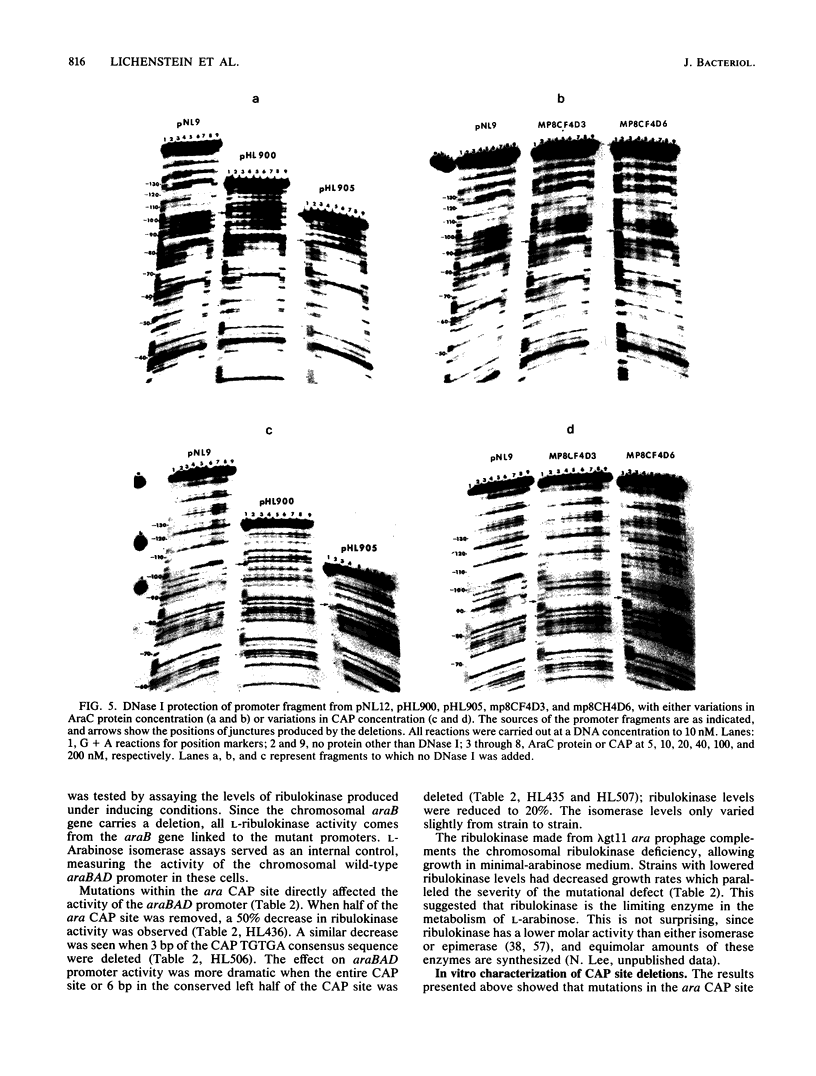
Catabolite gene activation of the araBAD operon was examined by using catabolite gene activator protein (CAP) site deletion mutants. A high-affinity CAP-binding site between the divergently orientated araBAD and araC operons has been previously identified by DNase I footprinting techniques. Subsequent experiments disagreed as to whether this site is directly involved in stimulating araBAD expression. In this paper, we present data showing that deletions generated by in vitro mutagenesis of the CAP site led to a five- to sixfold reduction in single-copy araBAD promoter activity in vivo. We concluded that catabolite gene activation of araBAD involves this CAP site. The hypothesis that CAP stimulates the araBAD promoter primarily by relieving repression was then tested. The upstream operator araO2 was required for repression, but we observed that the magnitude of CAP stimulation was unaffected by the presence or absence of araO2. We concluded that CAP plays no role in relieving repression. Other experiments showed that when CAP binds it induces a bend in the ara DNA; similar bending has been reported upon CAP binding to lac DNA. This conformational change in the DNA may be essential to the mechanism of CAP activation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass R., Heffernan L., Sweadner K., Englesberg E. The site for catabolite deactivation in the L-arabinose BAD operon in Escherichia coli B/r. Arch Microbiol. 1976 Oct 11;110(1):135–143. doi: 10.1007/BF00416978. [DOI] [PubMed] [Google Scholar]
- Beckwith J., Rossow P. Analysis of genetic regulatory mechanisms. Annu Rev Genet. 1974;8:1–13. doi: 10.1146/annurev.ge.08.120174.000245. [DOI] [PubMed] [Google Scholar]
- Bingham A. H., Ponnambalam S., Chan B., Busby S. Mutations that reduce expression from the P2 promoter of the Escherichia coli galactose operon. Gene. 1986;41(1):67–74. doi: 10.1016/0378-1119(86)90268-4. [DOI] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Brown C. E., Hogg R. W. A second transport system for L-arabinose in Escherichia coli B-r controlled by the araC gene. J Bacteriol. 1972 Aug;111(2):606–613. doi: 10.1128/jb.111.2.606-613.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S., Aiba H., de Crombrugghe B. Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein. J Mol Biol. 1982 Jan 15;154(2):211–227. doi: 10.1016/0022-2836(82)90061-4. [DOI] [PubMed] [Google Scholar]
- Busby S., Dreyfus M. Segment-specific mutagenesis of the regulatory region in the Escherichia coli galactose operon: isolation of mutations reducing the initiation of transcription and translation. Gene. 1983 Jan-Feb;21(1-2):121–131. doi: 10.1016/0378-1119(83)90154-3. [DOI] [PubMed] [Google Scholar]
- Busby S., Kotlarz D., Buc H. Deletion mutagenesis of the Escherichia coli galactose operon promoter region. J Mol Biol. 1983 Jun 25;167(2):259–274. doi: 10.1016/s0022-2836(83)80335-0. [DOI] [PubMed] [Google Scholar]
- CRIBBS R., ENGLESBERG E. L-ARABINOSE NEGATIVE MUTANTS OF THE L-RIBULOKINASE STRUCTURAL GENE AFFECTING THE LEVELS OF L-ARABINOSE ISOMERASE IN ESCHERICHIA COLI. Genetics. 1964 Jan;49:95–108. doi: 10.1093/genetics/49.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Lebkowski J. S., Botchan M. R. High mutation frequency in DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1983 May;80(10):3015–3019. doi: 10.1073/pnas.80.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Regulation of the regulatory gene for the arabinose pathway, araC. J Mol Biol. 1976 Jul 5;104(3):557–566. doi: 10.1016/0022-2836(76)90120-0. [DOI] [PubMed] [Google Scholar]
- Chapon C. Role of the catabolite activator protein in the maltose regulon of Escherichia coli. J Bacteriol. 1982 May;150(2):722–729. doi: 10.1128/jb.150.2.722-729.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Johnson P. Nucleotide sequence changes produced by mutations in the lac promoter of Escherichia coli. J Mol Biol. 1977 Mar 25;111(1):65–75. doi: 10.1016/s0022-2836(77)80132-0. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Schleif R. Deletion analysis of the Escherichia coli ara PC and PBAD promoters. J Mol Biol. 1984 Nov 25;180(1):201–204. doi: 10.1016/0022-2836(84)90437-6. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Englesberg E., Squires C., Meronk F., Jr The L-arabinose operon in Escherichia coli B-r: a genetic demonstration of two functional states of the product of a regulator gene. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1100–1107. doi: 10.1073/pnas.62.4.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englesberg E., Wilcox G. Regulation: positive control. Annu Rev Genet. 1974;8:219–242. doi: 10.1146/annurev.ge.08.120174.001251. [DOI] [PubMed] [Google Scholar]
- GROSS J., ENGLESBERG E. Determination of the order of mutational sites governing L-arabinose utilization in Escherichia coli B/r bv transduction with phage Plbt. Virology. 1959 Nov;9:314–331. doi: 10.1016/0042-6822(59)90125-4. [DOI] [PubMed] [Google Scholar]
- Gay N. J. Construction and characterization of an Escherichia coli strain with a uncI mutation. J Bacteriol. 1984 Jun;158(3):820–825. doi: 10.1128/jb.158.3.820-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Evidence for the existence of stable curvature of DNA in solution. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4632–4636. doi: 10.1073/pnas.81.15.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Dunn T., Schleif R. Upstream repression and CRP stimulation of the Escherichia coli L-arabinose operon. J Mol Biol. 1984 Nov 25;180(1):61–72. doi: 10.1016/0022-2836(84)90430-3. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hendrickson W., Schleif R. Transcription of Escherichia coli ara in vitro. The cyclic AMP receptor protein requirement for PBAD induction that depends on the presence and orientation of the araO2 site. J Mol Biol. 1986 Apr 5;188(3):355–367. doi: 10.1016/0022-2836(86)90160-9. [DOI] [PubMed] [Google Scholar]
- Hahn S., Schleif R. In vivo regulation of the Escherichia coli araC promoter. J Bacteriol. 1983 Aug;155(2):593–600. doi: 10.1128/jb.155.2.593-600.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan L., Bass R., Englesberg E. Mutations affecting catabolite repression of the L-arabinose regulon in Escherichia coli B/r. J Bacteriol. 1976 Jun;126(3):1119–1131. doi: 10.1128/jb.126.3.1119-1131.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh J., Schleif R. The araC promoter: transcription, mapping and interaction with the araBAD promoter. Cell. 1977 Jul;11(3):545–550. doi: 10.1016/0092-8674(77)90072-1. [DOI] [PubMed] [Google Scholar]
- Katz L., Englesberg E. Hyperinducibility as a result of mutation in structural genes and self-catabolite repression in the ara operon. J Bacteriol. 1971 Jul;107(1):34–52. doi: 10.1128/jb.107.1.34-52.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Busby S., Herbert M., Kotlarz D., Buc H. Comparison of the binding sites for the Escherichia coli cAMP receptor protein at the lactose and galactose promoters. EMBO J. 1983;2(2):217–222. doi: 10.1002/j.1460-2075.1983.tb01408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodrubetz D., Schleif R. L-arabinose transport systems in Escherichia coli K-12. J Bacteriol. 1981 Nov;148(2):472–479. doi: 10.1128/jb.148.2.472-479.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodrubetz D., Schleif R. Regulation of the L-arabinose transport operons in Escherichia coli. J Mol Biol. 1981 Sep 15;151(2):215–227. doi: 10.1016/0022-2836(81)90512-x. [DOI] [PubMed] [Google Scholar]
- LEE N., ENGLESBERG E. COORDINATE VARIATIONS IN INDUCED SYNTHESES OF ENZYMES ASSOCIATED WITH MUTATIONS IN A STRUCTURAL GENE. Proc Natl Acad Sci U S A. 1963 Oct;50:696–702. doi: 10.1073/pnas.50.4.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lee N. L., Gielow W. O., Wallace R. G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Bendet I. Crystalline L-ribulokinase from Escherichia coli. J Biol Chem. 1967 May 10;242(9):2043–2050. [PubMed] [Google Scholar]
- Lee N., Carbon J. Nucleotide sequence of the 5' end of araBAD operon messenger RNA in Escherichia coli B/r. Proc Natl Acad Sci U S A. 1977 Jan;74(1):49–53. doi: 10.1073/pnas.74.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Wilcox G., Gielow W., Arnold J., Cleary P., Englesberg E. In vitro activation of the transcription of araBAD operon by araC activator. Proc Natl Acad Sci U S A. 1974 Mar;71(3):634–638. doi: 10.1073/pnas.71.3.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. T., Schleif R. Different cyclic AMP requirements for induction of the arabinose and lactose operons of Escherichia coli. J Mol Biol. 1973 Sep 5;79(1):149–162. doi: 10.1016/0022-2836(73)90276-3. [DOI] [PubMed] [Google Scholar]
- Lumpkin O. J. Mobility of DNA in gel electrophoresis. Biopolymers. 1982 Nov;21(11):2315–2316. doi: 10.1002/bip.360211116. [DOI] [PubMed] [Google Scholar]
- Majors J. Specific binding of CAP factor to lac promoter DNA. Nature. 1975 Aug 21;256(5519):672–674. doi: 10.1038/256672a0. [DOI] [PubMed] [Google Scholar]
- Malan T. P., Kolb A., Buc H., McClure W. R. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J Mol Biol. 1984 Dec 25;180(4):881–909. doi: 10.1016/0022-2836(84)90262-6. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miyada C. G., Soberón X., Itakura K., Wilcox G. The use of synthetic oligodeoxyribonucleotides to produce specific deletions in the araBAD promoter of Escherichia coli B/r. Gene. 1982 Feb;17(2):167–177. doi: 10.1016/0378-1119(82)90070-1. [DOI] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastan I., Adhya S. Cyclic adenosine 5'-monophosphate in Escherichia coli. Bacteriol Rev. 1976 Sep;40(3):527–551. doi: 10.1128/br.40.3.527-551.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick J. W., Lee N. Purification and properties of an L-arabinose isomerase from Escherichia coli. J Biol Chem. 1968 Aug 25;243(16):4312–4318. [PubMed] [Google Scholar]
- Patrick J. W., Lee N. Subunit structure of L-arabinose isomerase from Escherichia coli. J Biol Chem. 1969 Aug 25;244(16):4277–4283. [PubMed] [Google Scholar]
- Rao R. N., Rogers S. G. Plasmid pKC7: a vector containing ten restriction endonuclease sites suitable for cloning DNA segments. Gene. 1979 Sep;7(1):79–82. doi: 10.1016/0378-1119(79)90044-1. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. II. Mutations induced by bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1973 Oct 25;80(2):297–314. doi: 10.1016/0022-2836(73)90174-5. [DOI] [PubMed] [Google Scholar]
- Stoner C., Schleif R. The araE low affinity L-arabinose transport promoter. Cloning, sequence, transcription start site and DNA binding sites of regulatory proteins. J Mol Biol. 1983 Dec 25;171(4):369–381. doi: 10.1016/0022-2836(83)90035-9. [DOI] [PubMed] [Google Scholar]
- Vidal-Ingigliardi D., Raibaud O. The mac promoters: functional hybrid promoters activated by the malT product and repressed by the lacI product. Nucleic Acids Res. 1985 Feb 25;13(4):1163–1172. doi: 10.1093/nar/13.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. G., Lee N., Fowler A. V. The araC gene of Escherichia coli: transcriptional and translational start-points and complete nucleotide sequence. Gene. 1980 Dec;12(3-4):179–190. doi: 10.1016/0378-1119(80)90100-6. [DOI] [PubMed] [Google Scholar]
- Wilcox G., Meuris P., Bass R., Englesberg E. Regulation of the L-arabinose operon BAD in vitro. J Biol Chem. 1974 May 10;249(9):2946–2952. [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. M., Reznikoff W. S. Deletion analysis of the CAP-cAMP binding site of the Escherichia coli lactose promoter. Nucleic Acids Res. 1984 Jul 11;12(13):5449–5464. doi: 10.1093/nar/12.13.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. M., Reznikoff W. S. Deletion analysis of the Escherichia coli lactose promoter P2. Nucleic Acids Res. 1985 Apr 11;13(7):2457–2468. doi: 10.1093/nar/13.7.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]
- Zubay G., Schwartz D., Beckwith J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc Natl Acad Sci U S A. 1970 May;66(1):104–110. doi: 10.1073/pnas.66.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]





