Abstract
Antibody class switch recombination (CSR) occurs by an intrachromosomal deletion requiring generation of double-stranded breaks (DSBs) in switch-region DNA. The initial steps in DSB formation have been elucidated, involving cytosine deamination by activation-induced cytidine deaminase and generation of abasic sites by uracil DNA glycosylase. However, it is not known how abasic sites are converted into single-stranded breaks and, subsequently, DSBs. Apurinic/apyrimidinic endonuclease (APE) efficiently nicks DNA at abasic sites, but it is unknown whether APE participates in CSR. We address the roles of the two major mammalian APEs, APE1 and APE2, in CSR. APE1 deficiency causes embryonic lethality in mice; we therefore examined CSR and DSBs in mice deficient in APE2 and haploinsufficient for APE1. We show that both APE1 and APE2 function in CSR, resulting in the DSBs necessary for CSR and thereby describing a novel in vivo function for APE2.
Ig class switch recombination (CSR) occurs by intrachromosomal deletion and results in the exchange of antibody constant regions, thereby improving the effector function of the humoral immune response. Recombination between the switch (S) regions located 5′ of each constant region gene (except Cδ) results in CSR (1). DNA double-stranded breaks (DSBs) in the S regions are essential intermediates in the recombination process. In recent years, much progress has been made toward understanding the molecular mechanism of CSR because of the discovery that the enzyme activation-induced cytidine deaminase (AID) is required for CSR (2, 3). Compelling data show that AID initiates the formation of DSBs by deamination of cytosines, generating uracils in S regions (4–7). The base excision repair enzyme uracil DNA glycosylase (UNG) subsequently excises the uracils, resulting in abasic sites, and mice and humans deficient in UNG have greatly reduced CSR and S region DSBs (6–8). How the abasic sites in S regions are converted to DNA breaks during CSR is unknown. In the base excision repair pathway, the enzyme that acts subsequent to UNG is apurinic/apyrimidinic endonuclease (APE), which nicks the DNA backbone at abasic sites to create DNA single-stranded (SS) breaks (SSBs) (9–11). DNA SS nicks that are spaced sufficiently close on opposite DNA strands could form a DSB spontaneously. However, during normal base excision repair, the SSBs introduced by APE do not progress to DSBs but are instead repaired by DNA polymerase β and ligase. During CSR, DNA abasic sites resulting from UNG activity might be cleaved by APE, but no evidence for involvement of APE in CSR has been reported.
In mammals, two homologues of the Escherichia coli exonuclease III have been cloned and characterized, representing the APEs. APE1 is considered to be the main APE: it is essential for early embryonic development in mice and for the viability of human cell lines (12, 13). APE1 has strong endonuclease activity and weak 3′-5′ exonuclease and 3′-phosphodiesterase activities (14, 15). In addition to its function in base excision repair, APE1 also regulates transcription factors such as p53, activator protein 1, Myb, and NF-κB, although the mechanism is not yet clear (16–19). However, the yeast Apn1 protein, which lacks the ability to stimulate transcription factors, is able to restore viability of human cell lines in which APE1 expression was downmodulated using small interfering RNA (12). These results show that the transcription regulatory function of APE1 is not essential for cellular viability.
Much less is known about APE2, which is encoded on the X chromosome. APE2-deficient mice show a slight growth defect and have a twofold reduction of white blood cells in the periphery, mainly affecting T and B cells (20). Furthermore, splenocytes and thymocytes from these mice exhibit a moderate accumulation in the G2/M phase of the cell cycle upon mitogen stimulation (20). Enzyme assays using abasic site–containing oligonucleotide substrates showed that recombinant purified human APE2 has a weaker APE activity than APE1 (21). Its 3′-5′ exonuclease activity, however, is strong compared with APE1 (14). It is not clear to what extent APE2 carries out these activities in vivo, but they are not required for embryonic development or cell viability because they are essentially normal in APE2-deficient mice. APE2 might be important for the repair of oxidative damage in mitochondrial DNA, as it has a putative mitochondrial targeting signal and was detected in mitochondria by electron microscopic immunocytochemistry and Western blotting (22, 23).
In this study, we demonstrate that both APE1 and APE2 are important for CSR, as we find reduced switching to all isotypes in splenic B cells from ape1+/−, ape2Y/−, and ape1+/−/ape2Y/− APE double-deficient (DBL) mice. Because APE2 is located on the X chromosome, we used APE2-deficient (ape2Y/−) male mice in all experiments. Linker ligation-mediated PCR (LM-PCR) analysis shows that DSBs in Sμ are greatly reduced in DBL B cells and, unlike DSBs in WT B cells, do not occur preferentially at the G:C bp or AID hotspots. Our data show that APE2 contributes considerably to the CSR mechanism, describing a new role for this protein.
RESULTS
Splenic B cell subsets in ape1+/−, ape2Y/−, and DBL mice
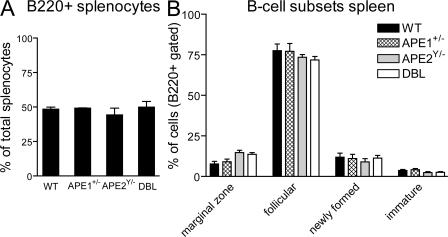
APE1 deficiency causes early embryonic lethality in mice (13); therefore, in this study we used APE1 heterozygous mice (ape1+/−), which have DNA repair defects (24, 25). Adult ape2Y/− mice have reduced white blood cell counts and fewer B and T cells in the bone marrow and thymus, respectively, compared with WT littermates (20). Ape2Y/− and DBL mice have smaller spleens compared with WT littermate controls (unpublished data). However, the proportions of cells in splenic mature B cell subsets from ape1+/−, ape2Y/−, or DBL mice are not different from WT mice, as shown by flow cytometry analysis using anti-B220, -CD23, -CD21, and -CD24 antibodies (Fig. 1). Although the fraction of marginal zone B cells in ape2Y/− and DBL mice appears to be increased, and the percentage of follicular B cells slightly decreased, the differences are not statistically significant.
Figure 1.
Normal splenic B cell subsets in WT, ape1+/−, ape2Y/−, and DBL mice. FACS analysis of ex vivo splenocytes gated on B220+ lymphocytes and stained for CD21, CD23, and CD24. (A) Mean percentages (+SEM) of B220+ splenocytes are shown. (B) Mean percentages (+SEM) of marginal zone B cells (CD21hi/CD23lo), follicular B cells (CD21lo/CD23hi), newly formed B cells (CD21lo/CD23lo), and immature B cells (B220lo/CD24hi) from three to five mice are shown. B cell subsets are shown as a percentage of B220+ splenocytes, except for immature B cells, which are a percentage of total splenocytes.
Expression of APE1 and APE2 proteins in stimulated B cells
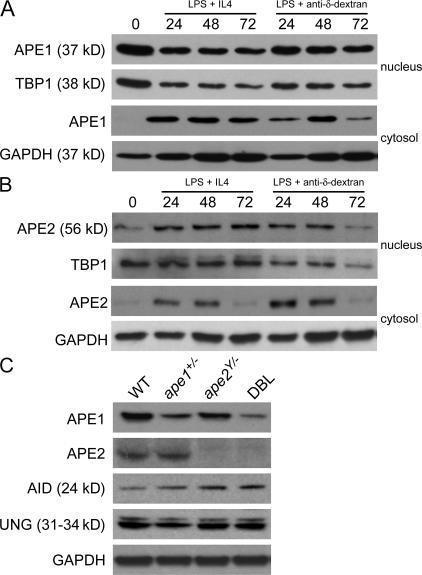
The expression of APE1 and APE2 in unstimulated ex vivo splenic B cells and in B cells stimulated in vitro for CSR was examined by immunoblotting of nuclear and cytosolic extracts. APE1 protein is expressed constitutively in the nuclei of both resting and stimulated B cells (Fig. 2 A). APE1 is detected in the cytosol only after stimulation, suggesting that induction of synthesis and turnover of APE1 protein occurs upon B cell stimulation. A negative regulatory element, which binds the APE1 protein itself, has been identified in the APE1 promoter (26), consistent with the lack of an increase in APE1 in stimulated B cells. In contrast, APE2 is induced in both the nucleus and the cytosol of B cells stimulated to switch (Fig. 2 B). Both APE1 and APE2 are abundant 48 h after stimulation, when AID protein expression and DNA DSBs in Sμ are detectable (7). Cytosolic APE2 expression declines to basal levels after 72 h, consistent with our observation that CSR is mostly completed after 72 h of in vitro stimulation.
Figure 2.
APE1 is constitutively expressed and APE2 expression is induced in the nuclei of switching B cells. 10 μg of nuclear and cytoplasmic protein extracts from unstimulated ex vivo splenic B cells (lysed immediately after B cell enrichment; t = 0) and from splenic B cells stimulated with LPS plus IL-4 or anti–δ-dextran for 24, 48, or 72 h were electrophoresed in each lane and immunoblotted with rabbit–anti-APE1 (A) and rabbit–anti-APE2 (B). Whole-cell extracts of LPS plus anti–δ-dextran–stimulated splenic B cells from WT, ape1+/−, ape2Y/−, and DBL mice (48 h) were immunoblotted with the indicated antibodies (C). TATA box–binding protein 1 (TBP1) and GAPDH were used as steady-state/loading controls for nuclear and cytoplasmic/whole-cell extracts, respectively.
Fig. 2 C demonstrates that APE1 protein levels are reduced in stimulated splenic B cells from ape1+/− and DBL mice but not in ape2Y/− B cells. We found no difference in APE2 protein levels between WT and ape1+/− B cells. Moreover, expression of AID and UNG is not affected by APE deficiency.
Class switching is reduced in ape1+/−, ape2Y/−, and DBL B cells
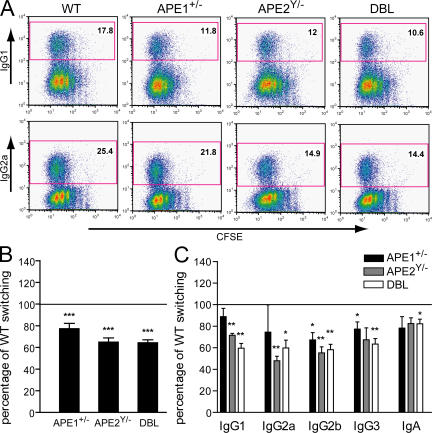
To determine the function of APE1 and APE2 in CSR, CFSE-stained splenic B cells from ape1+/−, ape2Y/−, and DBL mice were cultured for 3 d with LPS and specific combinations of cytokines that induce CSR to different isotypes, and analyzed by flow cytometry for surface Ig expression and cell division. In each experiment, WT littermates from ape1+/− and ape2+/− crossbreedings were used as controls. An example of a FACS analysis showing switching to IgG1 and IgG2a in the different genotypes is shown in Fig. 3 A. Overall, CSR is reduced in ape1+/− B cells to a mean of 77 ± 4.9% (SEM) of WT (Fig. 3 B). Ape2Y/− B cells show a reduction to 65 ± 3.9% of WT, comparable to the reduction observed in DBL mice (64 ± 2.7%). The reduction in CSR is highly significant for all three genotypes (P < 0.001). Fig. 3 C shows that switching to each of the isotypes tested is reduced in the APE-deficient B cells. The reduction in switching reaches significance for the IgG2b and IgG3 isotypes in ape1+/− B cells and for the IgG1, IgG2a, and IgG2b isotypes in ape2Y/− B cells, whereas all isotypes are significantly reduced in DBL B cells. These results indicate that both APE1 and APE2 are involved in CSR.
Figure 3.
CSR is reduced in splenic B cells from mice with reduced levels of APEs. CFSE-stained splenic B cells were cultured for 3 d with LPS plus IL-4 (IgG1 switching); LPS plus IFN-γ (IgG2a); LPS plus dextran sulfate (IgG2b); LPS plus anti–δ-dextran (IgG3); LPS plus IL-4 plus IL-5 plus anti–δ-dextran plus TGF-β (IgA); and BlyS. Ig class switching was assessed by flow cytometry using Ig isotype–specific antibodies. At least four independent experiments were performed for each mouse and all isotypes, except for IgG2a (n = 3). For each experiment, splenic B cells from a WT littermate were used as a control. (A) Examples of FACS analyses for IgG1 and IgG2a switching in CFSE-stained cells. The percentage of switched cells is indicated in each plot. (B) The mean percentages (+SEM) of overall switching (all isotypes pooled) relative to WT are shown. WT switching was set at 100% (horizontal line). ***, P < 0.001, as determined by the one-sample t test. (C) The mean percentages (+SEM) of switching relative to WT for the different isotypes are shown. WT switching was set at 100% (horizontal line). *, P < 0.05; and **, P < 0.01, as determined by the one-sample t test.
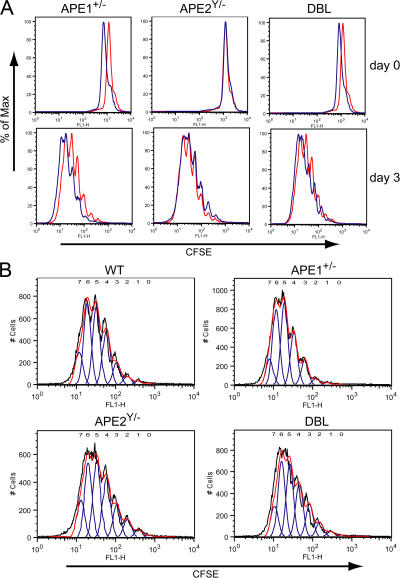
To ascertain whether the reduction in CSR was caused by perturbed proliferation or cell-cycle progression, CFSE was used to track cell division and to determine CSR after equal numbers of cell divisions. Fig. 4 shows that splenic B cells from ape1+/−, ape2Y/−, and DBL mice divided equally well compared with WT cells in response to LPS plus IL-4 treatment, therefore excluding a proliferation defect as the cause of the decreased CSR. In addition, Fig. 3 A presents an example of results obtained when we analyzed isotype expression in CFSE-stained cells, further illustrating that CSR is not reduced because of an effect on cell division by APE deficiency. Staining of DNA content by propidium iodide revealed a moderate increase in G2/M phase in stimulated ape2Y/− and DBL splenic B cells (not depicted), consistent with a previous report on mitogen-stimulated ape2Y/− thymocytes and splenocytes (20), but this has no apparent effect on proliferation as assessed by CFSE staining (Fig. 4) and [3H]thymidine incorporation (not depicted). Perhaps APE2-deficient B cells compensate for the G2/M phase delay by a shortened G1 and S phase, resulting in unaltered overall proliferation.
Figure 4.
Splenic B cells from ape1+/−, ape2Y/−, and DBL mice proliferate normally upon CSR induction. Splenic B cells were stained with CFSE and cultured for 3 d with LPS plus IL-4 and analyzed by FACS. (A) Overlay CFSE histograms of ex vivo B cells immediately after CFSE staining and after 3 d of stimulation are shown. CFSE fluorescence is represented by red profiles for WT splenic B cells, and by blue profiles for ape1+/−, ape2Y/−, and DBL splenic B cells. (B) Computational integration of the level of CFSE fluorescence in B cells stimulated for IgG1 switching. The numbers of cell divisions are given above the CFSE peaks.
Reduced CSR in B cells treated with the APE inhibitor 7-nitro-1H-indole-2-carboxylic acid (CRT0044876)
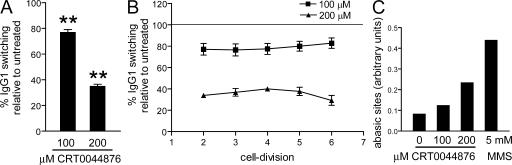
To further confirm that APEs are important for CSR, splenic B cell cultures were treated with the small molecule inhibitor CRT0044876, which was previously shown to selectively inhibit APE1 (27). Whether this compound also inhibits APE2 has not been demonstrated, but active-site comparisons of APE1 and APE2 indicate a high degree of similarity in their abasic-site binding pockets, which are the targets of this inhibitor (15). CFSE-stained splenic B cells from WT mice were cultured for 3 d with LPS plus IL-4 and various doses of CRT0044876. Overall, IgG1 class switching in cells treated with 100 and 200 μM CRT0044876 was 77 and 35%, respectively, of that in untreated cells (Fig. 5 A). Although some apoptosis and a decrease in the proliferation of CRT0044876-treated B cells was observed (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20071289/DC1), the effect of APE inhibition on CSR was not caused by toxicity or differences in cell proliferation, as IgG1 switching was reduced when comparing treated cells that had divided the same number of times as untreated cells (Fig. 5 B). However, we could not test the effect of higher doses of the inhibitor on CSR because of increased apoptosis. Also, we were unable to further reduce CSR by treating APE-deficient cells with this inhibitor. We verified that treatment with the APE inhibitor results in the accumulation of abasic sites by measuring the binding of a biotinylated aldehyde-reactive probe (ARP) to DNA from switching cells in an ELISA-based assay (Fig. 5 C).
Figure 5.
The APE small-molecule inhibitor CRT0044876 inhibits CSR in a dose-dependent manner. CFSE-stained WT splenic B cells were cultured for 3 d with LPS plus IL4 (IgG1 switching). CRT0044876 or DMSO was added at the start of the culture and again after 24 h. IgG1 switching per cell division was determined by flow cytometry. (A) Mean (+SEM) overall IgG1 switching in B cells treated with CRT0044876 is shown as a percentage of DMSO-treated B cells. **, P < 0.01, as determined by the one-sample t test. (B) Mean percentage (±SEM) of IgG1+ cells relative to DMSO-treated B cells, set at 100% (horizontal line), is shown for each cell division. The data are the mean of six independent experiments. (C) Abasic sites were measured in living cells using the ARP assay (see Materials and methods). Cells treated with 5 mM methyl methanesulfonate (MMS) before ARP incubation were used as a positive control. The data are representative of two independent experiments.
S–S junctions are similar in WT and APE-deficient B cells
To determine if APE1 or APE2 deficiency altered the microhomologies at the S–S junctions, we cloned Sμ–Sγ3 junctions from B cells induced to switch to IgG3 from DBL and WT cells. No significant differences were detected in the lengths of junctional microhomology (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20071289/DC1), suggesting that APE1 and APE2 are not involved in the processing of DNA ends during the formation of S–S junctions.
DSBs in Sμ are reduced in ape1+/−, ape2Y/−, and DBL B cells
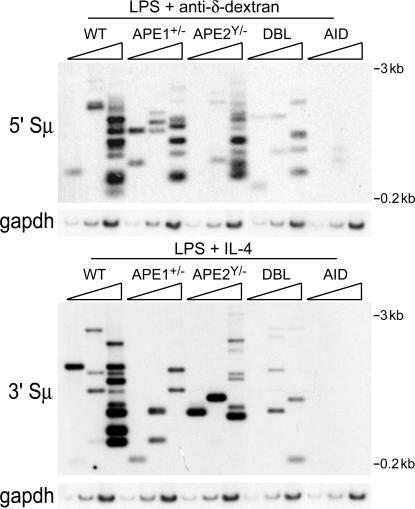
To determine whether the DSBs in S regions observed during CSR are dependent on APE1 and APE2, splenic B cells from WT, ape1+/−, ape2Y/−, and DBL mice were stimulated with LPS plus IL-4 or LPS plus anti–δ-dextran for 2 d, genomic DNA was isolated, and DSBs in the Sμ region were detected by LM-PCR using primers specific for the 5′ or 3′ ends of Sμ. Using this assay, we have previously demonstrated that blunt DSBs in Sμ are AID- and UNG-dependent in switching B cells (7). Fewer blunt Sμ breaks are detected in switching B cells from ape1+/− and ape2Y/− mice than from WT mice, although the reduction varies between experiments (Fig. 6). However, we consistently detect very few breaks in B cells from DBL mice. Moreover, treatment of DNA with T4 polymerase to fill in or excise SS overhangs from staggered DSBs before linker ligation increased the number of detected breaks, but they were still reduced in B cells from ape1+/−, ape2Y/−, and DBL mice relative to WT B cells (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20071289/DC1). These results suggest that APE1 and APE2 act in CSR by cutting at abasic sites rather than functioning as exonucleases converting staggered DSBs into blunt breaks.
Figure 6.
B cells from mice with reduced levels of APEs have fewer DSBs in Sμ upon CSR induction. LM-PCR was performed on threefold dilutions of DNA isolated from splenic B cells stimulated to undergo CSR for 2 d. Sμ-specific primers were used in conjunction with a linker-specific primer. PCR products were blotted and hybridized to an internal Sμ-specific probe. Input of genomic DNA was normalized by performing PCR for gapdh on the same threefold DNA dilutions. (top) A representative experiment for LPS plus anti–δ-dextran–stimulated B cells. LM-PCR was performed for the 5′ portion of Sμ; the 5′ Sμ primer allows detection of blunt DSBs downstream of this primer. (bottom) A representative experiment for LPS plus IL-4–stimulated B cells. In this experiment, LM-PCR was performed for the 3′ portion of Sμ, detecting blunt breaks upstream of the 3′ Sμ primer. The sizes of the fragments detected on the blots range from 3 to 0.2 kb and, thus, occur over the entire Sμ segment.
Sites of DSBs in Sμ in ape1+/−, ape2Y/−, and DBL B cells
To provide further evidence that APE1 and APE2 function as APEs during CSR, the nt's at which the breaks occur were determined by cloning and sequencing LM-PCR products. Deamination of dC residues by AID followed by UNG activity and APE incision would create breaks at the cytosine (or its complementary G), predominantly in the AID hotspot motif WRC/GYW. In agreement with our previous report (7), Sμ breaks in WT B cells occur preferentially at the G:C bp (82.6%) and at the G:C bp within AID hotspots (48%), a highly significant difference relative to the Sμ sequence itself (consisting of 56.8% G:C bp and 23.2% AID hotspots; Table I). In contrast, we found that only 69.4% of Sμ breaks occur at the G:C bp and 30.6% at AID hotspots in B cells from DBL mice, which is not significantly different than the frequency of these elements in the Sμ sequence. The Sμ breaks in ape1+/− B cells showed a significant preference for the G:C bp but not for AID hotspots. Breaks in ape2Y/− B cells, however, do not display a G:C bp preference and occur at AID hotspots only with borderline significance (P = 0.047; Table I). Likewise, the staggered Sμ breaks in DBL B cells show a reduced preference for the G:C bp and AID hotspots relative to WT cells (Table S1, available at http://www.jem.org/cgi/content/full/jem.20071289/DC1). Collectively these data suggest that APE1 and APE2 are functioning during CSR to incise abasic sites created by AID–UNG activity, and in the DBL cells, some of the few DSBs created are probably AID–independent.
Table I.
Sμ DSBs in APE-deficient B cells show reduced targeting to the G:C bp and AID hotspots
| nt at break | WT | ape1+/− | ape2Y/− | DBL | Sequencea |
|---|---|---|---|---|---|
| G | 73.3%b (55) | 79% (45) | 64.3% (18) | 67.4% (33) | 40.7% |
| C | 9.3% (7) | 1.8% (1) | 10.7% (3) | 2% (1) | 16.1% |
| A | 8% (6) | 8.8% (5) | 21.4% (6) | 14.3% (7) | 21.4% |
| T | 9.3% (7) | 10.5% (6) | 3.6% (1) | 16.3% (8) | 21.8% |
| Total | 75 breaks | 57 breaks | 28 breaks | 49 breaks | 2,000 nt's |
| G + C | 82.6% | 80.7% | 75% | 69.4% | 56.8% |
| p-valuec | 0.001 | 0.007 | NS | NS | |
| Hotspots | 48% | 40.4% | 46.4% | 30.6% | 23.2% |
| p-valuec | 0.002 | NS | 0.047 | NS |
Frequency of nt's in the analyzed genomic sequence.
Percentage of the DSBs at each nt reading the top-strand sequence.
Fisher's exact t test showing the significance of difference from the Sμ sequence.
Mutations in unrecombined Sμ in APE-deficient cells
In switching B cells from APE-deficient mice, one would expect the number of abasic sites in Sμ to be increased compared with WT B cells, which might result in increased mutations at the G:C bp after replication. However, the frequency of mutations in the 5′ portion of unrecombined (germline) Sμ segments is not altered in DBL B cells (Table II). Interestingly, the proportion of mutations at A:T is decreased in DBL cells (P = 0.012). Such a G:C bias would be consistent with the hypothesis that mutations at A:T are introduced during repair DNA synthesis initiated at SSBs, which are reduced in the DBL B cells.
Table II.
Mutation frequency and spectrum in germline Sμ segments from splenic B cells cultured with LPS plus anti–δ-dextran and LPS plus IL-4
| WT | DBL | p-valuea | |
|---|---|---|---|
| Total mutations | 24.6 (51)b | 19 (124) | NS |
| Mutations at A:T | 70.6% (36)c | 49.2% (61) | 0.012 |
| Mutations at G:C | 29.4% (15) | 50.8% (63) | |
| nt's analyzed | 20,718 | 65,344 |
Fisher's exact t test showing the significance of difference between WT and DBL mutations.
Mutation frequency ×10−4 (number).
Percentage of total mutations (number).
As abasic sites are lethal, they must be repaired, and yet the DBL B cells show unperturbed viability and proliferation. The remaining APE1 in the DBL cells could be responsible for correct repair of some of these abasic sites. We conclude that the DNA breaks in Sμ occurring during CSR can originate from both APE1 and APE2 activity. However, the finding that the level of CSR in DBL cells is not further reduced compared with ape2Y/− cells suggests that APE2 activity might be more important than APE1 for DSB formation in Sμ.
DISCUSSION
In this study, we provide the first evidence that APEs act in CSR. Moreover, we report the unexpected finding that APE2 plays a significant part in CSR. It has been shown previously that both AID and UNG are essential components of the CSR machinery, initiating DSBs in S regions (3, 6–8, 28). Our findings that (a) both blunt and staggered Sμ breaks are reduced in B cells from ape1+/−, ape2Y/−, and DBL mice; (b) unlike DSBs in WT cells, the remaining DSBs in the DBL mice do not specifically occur at the G:C bp in AID-targeting hotspots; and (c) the lack of significant alterations in S–S junctional microhomology indicate that APE1 and APE2 do not function as exonucleases in DSB end processing during CSR, but rather act as endonucleases upstream of the generation of DSBs in S regions.
The finding that APE2 plays an important role in CSR is surprising, as it has been reported that APE2 has low endonuclease activity despite its highly conserved nuclease domain (15, 21). However, it must be noted that in these studies DNA incision activity was determined using partially purified human APE2 produced in E. coli (15, 21). In a more recent study, it was shown that purified human APE2 produced in S. cerevisiae possessed considerable endonuclease activity at physiological salt conditions (14). It is conceivable that cellular cofactors increase the endonuclease activity of APE2.
APE1 is a ubiquitous protein and is crucial during early embryogenesis and development, whereas APE2 is not required. The phenotype of APE2-deficient mice appears to be restricted to the lymphoid compartment, suggesting a special role for this protein in lymphoid cells (20). In agreement, we have shown in this study that APE2 is induced in B cells upon CSR stimulation, whereas APE1 appears to be constitutively expressed in B cell nuclei. These data suggest that APE2 might fulfill a specialized function related to B cell activation, resulting in the generation of DNA DSBs needed for recombination, whereas APE1 is required for the maintenance of global genome stability, which is APE2 independent.
By the use of in vitro cleavage assays with purified proteins, it has been suggested that Mre11–Rad50 cleaves DNA at abasic sites after AID and UNG action (29). However, whether the Mre11–Rad50 complex is actually involved in creating DNA breaks in S regions in vivo is unknown. The MRN complex, consisting of Mre11, Rad50, and Nbs1, is essential for the repair of DSBs and, therefore, is likely to be important for recombination between S regions during CSR (30–32). Cleavage of apurinic/apyrimidinic sites by Mre11–Rad50 would result in 3′-phospho-α,β-unsaturated aldehydes, which must be removed before DNA polymerases can extend from the 3′ end. Both APE1 and APE2 are capable of removing such 3′ groups either through their lyase or exonuclease activity (14, 33); thus, in principle it is possible that Mre11–Rad50, APE1, and APE2 act in the same pathway during CSR.
Our observation that CSR is not ablated in B cells from DBL mice or in CRT0044876-treated B cells can be explained in several ways. First, the remaining APE1 allele in DBL mice might be responsible for sufficient APE1 activity, resulting in reduced but sufficient DSBs for recombination. It has been recently shown that even a single DSB in Sμ and a downstream S region allows CSR (34). The notion that S region breaks are not limiting during CSR provides an explanation for the relatively mild CSR defect observed in our study (∼35% reduction in DBL B cells), whereas the number of DSBs are reduced to a much greater extent in B cells from DBL mice. Conversely, it was recently found that in DNA polymerase β–deficient B cells, a two- to threefold increase in DSBs results in only a modest CSR increase (35).
Elimination of all APE1 protein or function leads to rapid cell death (36), hampering assessment of the contribution of the remaining APE1 to DSB formation in CSR (12). Thus, complete inhibition of APE function was probably not achieved by the CRT0044876 dosage used in this study, but a further dose increase resulted in decreased proliferation and considerable cell death, preventing analysis of CSR.
Second, persistent abasic sites, as would be expected in these B cells, might have an intrinsic fragility in vivo, which when closely spaced might result in DSB formation. It has been shown that abasic DNA is highly unstable in vitro and undergoes spontaneous strand cleavage 3′ to the abasic site, especially under alkaline conditions (37, 38).
Third, abasic sites might be cleaved by topoisomerase IIα during DNA replication (39). However, we have found that AID-dependent S region DSBs are made and resolved in the G1 phase of the cell cycle, arguing against a role for replication in the generation of DSBs (40).
Fourth, transcription is arrested when it encounters an abasic site, which might recruit transcription-coupled nt excision repair (NER) proteins capable of cleaving the DNA backbone (41). However, we have found that the gene for xeroderma pigmentosum A, which encodes an essential component of the NER pathway, is dispensable for CSR (40). It is possible that this pathway might be used when APEs are reduced or absent, but whether this is physiologically relevant in WT cells is questionable (42). Furthermore, SSBs introduced by NER would not specifically occur at the G:C bp (43), whereas the DSBs and staggered DSBs detected in Sμ regions in WT B cells occur specifically at the G:C bp (7, 44).
Fifth, the recently described endonuclease activity of MutLα (Mlh1-Pms2 heterodimer) might be responsible for some SSBs, although the possibility that this activity leads to DSBs is unlikely, as the endonuclease activity is targeted only to previously nicked DNA strands, and the sites of SSBs introduced by MutLα are not likely to be restricted to the G:C bp (45). Moreover, we found that B cells from pms2−/− mice did not show a greater reduction in CSR when treated with CRT0044876 than B cells from WT mice (unpublished data).
Finally, a novel APE termed PALF/APLF/XIP1 (PNK- and APTX-like FHA protein), which was recently identified in human cells (46–48), might contribute to the remaining abasic-site cleavage activity in CRT0044876-treated or DBL B cells. It is unknown, however, if this protein is expressed in B cells.
APE deficiency did not affect the frequency of mutations in the 5′ region of the unrecombined (germline) Sμ segment but did result in a decreased proportion of mutations at A:T bp's. It was possible that accumulation of abasic sites in the Sμ region followed by replication would result in an increased mutation frequency, especially at G:C bp's. However, the decrease in SSBs at abasic sites in APE-deficient B cells might also reduce Sμ mutations, as there are fewer entry sites for exonuclease1 and subsequent repair by error-prone polymerase.
In conclusion, our studies are consistent with a model for CSR in which abasic sites generated by AID and UNG activity are converted to SSBs in an APE1- and APE2- dependent fashion. Further studies are required to determine the relative contribution of APE1 and APE2, and to establish if APE2 has special functions that favor its participation in CSR.
MATERIALS AND METHODS
Mice.
All mouse strains were backcrossed to C75BL/6 for at least four generations. ape1+/− mice (25) were obtained from E. Friedberg (University of Texas Southwestern Medical Center, Dallas, TX). ape2Y/− mice were previously described (20). These mice were crossed to generate DBL mice. WT littermates of the DBL mice were used as controls for all experiments reported in this study. AID-deficient mice were obtained from T. Honjo (Kyoto University, Kyoto, Japan). Mice were housed in the Institutional Animal Care and Use Committee–approved specific pathogen-free facility at the University of Massachusetts Medical School. The mice were bred and used according to the guidelines from the University of Massachusetts Animal Care and Use Committee.
Splenic B cell isolation and culture.
Single-cell suspensions were prepared from the spleens of 2–5-mo-old mice by mechanical dispersion, and RBCs were lysed in Gey's solution. B cells were enriched by guinea pig complement lysis of T cells using a cocktail of anti–T cell antibodies, as previously described (49). For CSR analysis, cells stained with CFSE (Invitrogen) were cultured at 105 cells per milliliter in 24-well plates and activated for switching to the different Ig isotypes. Cultures contained 50 μg/ml LPS (Sigma-Aldrich) and 100 ng/ml human BLyS (Human Genome Sciences). For IgG1 switching, 800 U/ml IL-4 was added; for IgG2a switching, 10 U/ml IFN-γ was added; for IgG2b switching, 30 μg/ml dextran sulfate (GE Healthcare) was added; for IgG3 switching, 0.3 ng/ml anti–δ-dextran (a gift from C. Snapper, Uniformed Services University of the Health Sciences, Bethesda, MD) was added; and for IgA switching, 2 ng/ml TGF-β (R&D Systems), 800 U/ml IL-4, 1.5 ng/ml IL-5 (BD Biosciences), and 0.3 ng/ml anti–δ-dextran were added. For LM-PCR analysis, cells were cultured at 2 × 105 cell per milliliter in sixwell plates and activated for 2 d, as previously described (7). A 1-M stock solution of the selective APE1 inhibitor CRT0044876 (Maybridge Trevillett) was made in DMSO, and working stocks of 100 and 200 mM (1,000×) were prepared freshly in DMSO for each experiment. CRT0044876 was added at the start of the culture and again after 24 h of culture. An equal amount of DMSO was added to untreated control cultures.
Flow cytometry.
For FACS analysis, cells were washed twice with PBS, 1% FCS, and 0.2% NaN3, and were incubated for 30 min on ice with PE–goat F(ab')2 anti–mouse IgG1, IgG2b, and IgG3 and PE–goat anti–mouse IgA (SouthernBiotech). For CFSE labeling, cells were washed in HBSS (Invitrogen) and resuspended at 40 × 106 cells per milliliter. An equal volume of 2 μM CFSE was added, and cells were incubated at 37°C for 15 min, quenched in 100% FCS, and washed twice with medium containing 10% FCS. For splenic B cell subset analysis, cells were stained with anti-B220 allophycocyanin (RA3-6B2; Invitrogen), anti-CD23 PE (2G8; SouthernBiotech), anti-CD21 FITC (7G6; BD Biosciences), and biotinylated anti-CD24 (Invitrogen), followed by streptavidin-PerCP (BD Biosciences). CFSE fluorescence and antibody staining were acquired on a flow cytometer (LSR; BD Biosciences) and analyzed using the FlowJo software package (Tree Star Inc.).
Western blotting.
Cytoplasmic and nuclear extracts were prepared by resuspending cells in hypotonic buffer (10 mM Hepes [pH 8], 1 mM EDTA, 10 mM KCl, 0.1 mM EGTA, 1 mM DTT, 2 μM pepstatin, and complete protease inhibitor cocktail [Roche]). After a 15-min incubation on ice, cells were lysed by the addition of NP-40 to a final concentration of 0.625%. Supernatants were taken as cytoplasmic extracts. Nuclei were washed once with hypotonic buffer and resuspended in hypertonic buffer (20 mM Hepes [pH 8], 1 mM EDTA, 400 mM NaCl, 1 mM EGTA, 1 mM DTT, 2 μM pepstatin, and complete protease inhibitor cocktail). For whole-cell extracts, cells were lysed in RIPA buffer (150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 1% NP-40, 50 mM Tris-Cl, and complete protease and phosphatase inhibitor cocktail) and subjected to three freeze–thaw cycles. The protein content of cytoplasmic, nuclear, and whole-cell extracts was determined using the Bradford assay (Bio-Rad Laboratories). Equal amounts of proteins were electrophoresed on 8% SDS-polyacrylamide gels or 4–20% gradient SDS-polyacrylamide gels (Thermo Fisher Scientific) and blotted onto polyvinylidene fluoride membranes (Immobilon-P; Millipore). Immunoblotting was done using rabbit–-anti-APE1, rabbit–anti-APE2 (AnaSpec Inc.), rabbit–anti-GAPDH (Santa Cruz Biotechnology, Inc.), and mouse–anti–TATA box–binding protein 1 (Abcam), followed by goat–anti–rabbit horseradish peroxidase (HRP) or donkey–anti–mouse HRP (Santa Cruz Biotechnology, Inc.), and enhanced chemiluminescent substrate (Thermo Fisher Scientific).
ARP assay for quantification of abasic sites in DNA.
Abasic sites in living cells were measured as previously described (50). B cells stimulated with LPS and IL-4 for 2 d were resuspended in PBS plus 5 mM glucose at 106 cells per milliliter and incubated with 1 mM of biotin-containing ARP N'-aminooxy-methylcarbonyl hydrazino-D-biotin (Invitrogen) for 60 min at 37°C. Cells incubated with 5 mM methyl methanesulfonate (Sigma-Aldrich) were used as a positive control. Cells were washed twice with ice-cold PBS, and DNA was isolated using DNeasy columns (QIAGEN). For each sample, 700 ng DNA was resuspended in 200 μl of 10-mM Tris (pH 9), 15 μl of 5-M NaCl, and 30 μl of a 1:120 dilution of avidin-conjugated HRP (Thermo Fisher Scientific) was added, and the mixture was incubated for 1 h at room temperature. DNA–HRP complexes were precipitated by the addition of 65 μl of 1-mM DNA precipitation agent (DAPER; Sigma-Aldrich). Precipitated DNA–HRP was washed twice with wash buffer (1% BSA, 0.17 M NaCl, 20 mM Tris [pH 8], 0.25% Tween 20), resuspended in 500 μl of ice-cold 50-mM sodium-citrate (pH 5.3), and sonicated 10 times for 2 s at an output of 3 W. HRP activity was measured by use of a substrate kit (TMB ImmunoPure; Thermo Fisher Scientific).
Genomic DNA preparation and linker LM-PCR.
After culture for 2 d, viable cells were isolated by flotation on Ficoll-Hypaque gradients (P = 1.09) or Lympholyte (Cedar Lane), cells were imbedded in low-melt agarose plugs, and DNA was isolated as previously described (7). For linker ligation, 50 μl of 1× ligase buffer was added to the plugs, which were then heated to 62°C to melt the agarose. 20 μl DNA (∼10,000 cell equivalents) was added to 2 μl T4 DNA ligase (2 Weiss units; MBI Fermentas), 10 μl of DS annealed linker in 1× ligase buffer, 3 μl of 10× ligase buffer, and 30 μl dH2O and incubated overnight at 18°C. Linker was prepared by annealing 5 nmol each of LMPCR.1 (5′-GCGGTGACCCGGGAGATCTGAATTC-3′) and LMPCR.2 (5′-GAATTCAGATC-3′) in 300 μl of 1× ligase buffer, which results in a DS oligo with a 14-nt SS overhang that can only ligate unidirectionally. Ligated DNA samples were heated at 70°C for 10 min, diluted 5× in dH2O, and assayed for gapdh DNA by PCR to adjust DNA input before LM-PCR. The primers 5′ Sμ (5′-GCAGAAAATTTAGATAAAATGGATACCTCAGTGG-3′; Integrated DNA Technologies) or 3′ Sμ (5′-GCTCATCCCGAACCATCTCAACCAGG-3′) were used in conjunction with the linker primer (LMPCR.1) to amplify DNA breaks. Threefold dilutions of input DNA (0.5, 1.5, and 4.5 μl) were amplified by HotStar Taq (QIAGEN) using a touchdown PCR program. PCR products were electrophoresed on 1.25% agarose gels and vacuum blotted (VacuGene XL; GE Healthcare) onto nylon membranes (GeneScreen Plus; PerkinElmer). Blots were hybridized with an Sμ-specific oligonucleotide probe (μ probe 5′, AGGGACCCAGGCTAAGAAGGCAAT for 5′ Sμ LM-PCR; or μ probe 3′, CCTGTCCTGCTTGGCTTCCCTCTG for 3′ Sμ LM-PCR) end labeled with γ-[32P]ATP at 37°C overnight and washed at 55°C with 2× SSC/0.1% SDS. LM-PCR products were cloned into the vector pCR4-TOPO (Invitrogen) and sequenced by Macrogen using T3 and T7 primers. Cloned breaks in Sμ were aligned with germline Sμ sequenced from C57BL/6 chromosome 12 (available from GenBank/EMBL/DDBJ under accesion no. AC073553), with numbering starting at nt 136,645. This is the 5′ Sμ primer binding site and is ∼800 nt's upstream of the beginning of the tandem repeats.
PCR amplification of germline Sμ segments.
Sμ PCR was performed as previously described (51). Genomic DNA was isolated from B cells stimulated for 4 d with either LPS plus IL-4 or LPS plus anti–δ-dextran. The primers used for germline Sμ PCR were 5μ3 (5′-AATGGATACCTCAGTGGTTTTTAATGGTGGGTTA-3′) and 3μ2 (5′-AGAGGCCTAGATCCTGGCTTCTCAAGTAG-3′). The 3-kb PCR products were excised from agarose gels and cloned into pCR4-TOPO and sequenced by Macrogen using T7 primers.
Online supplemental material.
Fig. S1 shows the overlay CFSE fluorescence histograms of ex vivo B cells immediately after CFSE staining and after 3 d of culturing with LPS, BLyS, and IL-4 and different doses of CRT0044876. Fig. S2 shows the Sμ–Sγ3 junction microhomology in splenic B cells from WT and DBL mice cultured for 4 d with LPS, BLyS, and anti–δ-dextran. Fig. S3 shows a Southern blot to compare the blunt and staggered Sμ DSBs detected by LM-PCR after T4 DNA–polymerase polishing. Table S1 shows the sites of staggered DSBs in WT and APE-deficient cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071289/DC1.
Supplemental Material
Acknowledgments
We thank Anna Ucher and Sameer Taneja for excellent technical support. We thank Dr. Michael Volkert for helpful discussions.
This work was supported by National Institutes of Health grants RO1 AI065639 (to C.E. Schrader), and RO1 AI23283 and RO1 AI63026 (to J. Stavnezer). J.E.J. Guikema is supported by a postdoctoral fellowship from the Cancer Research Institute.
The authors have no conflicting financial interests.
Abbreviations used: AID, activation-induced cytidine deaminase; APE, apurinic/apyrimidinic endonuclease; ARP, aldehyde-reactive probe; CSR, class switch recombination; DBL, APE double deficient; DS, double stranded; DSB, DS break; HRP, horseradish peroxidase; LM-PCR, ligation-mediated PCR; NER, nt excision repair; S, switch; SS, single stranded; SSB, SS break; UNG, uracil DNA glycosylase.
References
- 1.Manis, J.P., M. Tian, and F.W. Alt. 2002. Mechanism and control of class-switch recombination. Trends Immunol. 23:31–39. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 3.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 4.Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri, J., and F.W. Alt. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541–552. [DOI] [PubMed] [Google Scholar]
- 6.Rada, C., G.T. Williams, H. Nilsen, D.E. Barnes, T. Lindahl, and M.S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 7.Schrader, C.E., E.K. Linehan, S.N. Mochegova, R.T. Woodland, and J. Stavnezer. 2005. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J. Exp. Med. 202:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai, K., G. Slupphaug, W.I. Lee, P. Revy, S. Nonoyama, N. Catalan, L. Yel, M. Forveille, B. Kavli, H.E. Krokan, et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 4:1023–1028. [DOI] [PubMed] [Google Scholar]
- 9.Strauss, P.R., W.A. Beard, T.A. Patterson, and S.H. Wilson. 1997. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem. 272:1302–1307. [DOI] [PubMed] [Google Scholar]
- 10.Strauss, P.R., and N.E. O'Regan. Abasic site repair in higher eukaryotes. In DNA Damage and Repair, vol. III. J.A. Nickoloff and M.F. Hoekstra, editors. 2001. Humana Press, Totowa, NJ. 43–86.
- 11.Christmann, M., M.T. Tomicic, W.P. Roos, and B. Kaina. 2003. Mechanisms of human DNA repair: an update. Toxicology. 193:3–34. [DOI] [PubMed] [Google Scholar]
- 12.Fung, H., and B. Demple. 2005. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 17:463–470. [DOI] [PubMed] [Google Scholar]
- 13.Xanthoudakis, S., R.J. Smeyne, J.D. Wallace, and T. Curran. 1996. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. USA. 93:8919–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkovics, P., V. Szukacsov, I. Unk, and L. Haracska. 2006. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 34:2508–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadi, M.Z., K. Ginalski, L.H. Nguyen, and D.M. Wilson III. 2002. Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J. Mol. Biol. 316:853–866. [DOI] [PubMed] [Google Scholar]
- 16.Hanson, S., E. Kim, and W. Deppert. 2005. Redox factor 1 (Ref-1) enhances specific DNA binding of p53 by promoting p53 tetramerization. Oncogene. 24:1641–1647. [DOI] [PubMed] [Google Scholar]
- 17.Hirota, K., M. Matsui, S. Iwata, A. Nishiyama, K. Mori, and J. Yo. 1997. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc. Natl. Acad. Sci. USA. 94:3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayaraman, L., K.G. Murthy, C. Zhu, T. Curran, S. Xanthoudakis, and C. Prives. 1997. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 11:558–570. [DOI] [PubMed] [Google Scholar]
- 19.Xanthoudakis, S., G. Miao, F. Wang, Y.C. Pan, and T. Curran. 1992. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 11:3323–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ide, Y., D. Tsuchimoto, Y. Tominaga, M. Nakashima, T. Watanabe, K. Sakumi, M. Ohno, and Y. Nakabeppu. 2004. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood. 104:4097–4103. [DOI] [PubMed] [Google Scholar]
- 21.Hadi, M.Z., and D.M. Wilson III. 2000. Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ. Mol. Mutagen. 36:312–324. [PubMed] [Google Scholar]
- 22.Akbari, M., M. Otterlei, J. Pena-Diaz, and H.E. Krokan. 2007. Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress. Neuroscience. 145:1201–1212. [DOI] [PubMed] [Google Scholar]
- 23.Tsuchimoto, D., Y. Sakai, K. Sakumi, K. Nishioka, M. Sasaki, T. Fujiwara, and Y. Nakabeppu. 2001. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 29:2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raffoul, J.J., D.C. Cabelof, J. Nakamura, L.B. Meira, E.C. Friedberg, and A.R. Heydari. 2004. Apurinic/apyrimidinic endonuclease (APE/REF-1) haploinsufficient mice display tissue-specific differences in DNA polymerase beta-dependent base excision repair. J. Biol. Chem. 279:18425–18433. [DOI] [PubMed] [Google Scholar]
- 25.Meira, L.B., S. Devaraj, G.E. Kisby, D.K. Burns, R.L. Daniel, R.E. Hammer, S. Grundy, I. Jialal, and E.C. Friedberg. 2001. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 61:5552–5557. [PubMed] [Google Scholar]
- 26.Izumi, T., W.D. Henner, and S. Mitra. 1996. Negative regulation of the major human AP-endonuclease, a multifunctional protein. Biochemistry. 35:14679–14683. [DOI] [PubMed] [Google Scholar]
- 27.Madhusudan, S., F. Smart, P. Shrimpton, J.L. Parsons, L. Gardiner, S. Houlbrook, D.C. Talbot, T. Hammonds, P.A. Freemont, M.J. Sternberg, et al. 2005. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 33:4711–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rada, C., J.M. Di Noia, and M.S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171. [DOI] [PubMed] [Google Scholar]
- 29.Larson, E.D., W.J. Cummings, D.W. Bednarski, and N. Maizels. 2005. MRE11/RAD50 cleaves DNA in the AID/UNG-dependent pathway of immunoglobulin gene diversification. Mol. Cell. 20:367–375. [DOI] [PubMed] [Google Scholar]
- 30.Kracker, S., Y. Bergmann, I. Demuth, P.O. Frappart, G. Hildebrand, R. Christine, Z.Q. Wang, K. Sperling, M. Digweed, and A. Radbruch. 2005. Nibrin functions in Ig class-switch recombination. Proc. Natl. Acad. Sci. USA. 102:1584–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahdesmaki, A., A.M. Taylor, K.H. Chrzanowska, and Q. Pan-Hammarstrom. 2004. Delineation of the role of the Mre11 complex in class switch recombination. J. Biol. Chem. 279:16479–16487. [DOI] [PubMed] [Google Scholar]
- 32.Reina-San-Martin, B., M.C. Nussenzweig, A. Nussenzweig, and S. Difilippantonio. 2005. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc. Natl. Acad. Sci. USA. 102:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915–948. [DOI] [PubMed] [Google Scholar]
- 34.Zarrin, A.A., C. Del Vecchio, E. Tseng, M. Gleason, P. Zarin, M. Tian, and F.W. Alt. 2007. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 315:377–381. [DOI] [PubMed] [Google Scholar]
- 35.Wu, X., and J. Stavnezer. 2007. DNA polymerase β is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J. Exp. Med. 204:1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izumi, T., D.B. Brown, C.V. Naidu, K.K. Bhakat, M.A. Macinnes, H. Saito, D.J. Chen, and S. Mitra. 2005. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. USA. 102:5739–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kupfer, P.A., and C.J. Leumann. 2007. The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res. 35:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindahl, T., and A. Andersson. 1972. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 11:3618–3623. [DOI] [PubMed] [Google Scholar]
- 39.Wilstermann, A.M., and N. Osheroff. 2001. Base excision repair intermediates as topoisomerase II poisons. J. Biol. Chem. 276:46290–46296. [DOI] [PubMed] [Google Scholar]
- 40.Schrader, C.E., J.E. Guikema, E.K. Linehan, E. Selsing, and J. Stavnezer. 2007. Activation-induced cytidine deaminase–dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J. Immunol. 179:6064–6071. [DOI] [PubMed] [Google Scholar]
- 41.Tornaletti, S., L.S. Maeda, and P.C. Hanawalt. 2006. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem. Res. Toxicol. 19:1215–1220. [DOI] [PubMed] [Google Scholar]
- 42.Spivak, G., and P.C. Hanawalt. 2006. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst.). 5:13–22. [DOI] [PubMed] [Google Scholar]
- 43.Hoeijmakers, J.H. 2001. Genome maintenance mechanisms for preventing cancer. Nature. 411:366–374. [DOI] [PubMed] [Google Scholar]
- 44.Stavnezer, J., and C.E. Schrader. 2006. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 22:23–28. [DOI] [PubMed] [Google Scholar]
- 45.Kadyrov, F.A., L. Dzantiev, N. Constantin, and P. Modrich. 2006. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 126:297–308. [DOI] [PubMed] [Google Scholar]
- 46.Kanno, S., H. Kuzuoka, S. Sasao, Z. Hong, L. Lan, S. Nakajima, and A. Yasui. 2007. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 26:2094–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bekker-Jensen, S., K. Fugger, J.R. Danielsen, I. Gromova, M. Sehested, J. Celis, J. Bartek, J. Lukas, and N. Mailand. 2007. Human Xip1 (C2ORF13) is a novel regulator of cellular responses to DNA strand breaks. J. Biol. Chem. 282:19638–19643. [DOI] [PubMed] [Google Scholar]
- 48.Iles, N., S. Rulten, S.F. El Khamisy, and K.W. Caldecott. 2007. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 27:3793–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrader, C.E., W. Edelmann, R. Kucherlapati, and J. Stavnezer. 1999. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 190:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atamna, H., I. Cheung, and B.N. Ames. 2000. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc. Natl. Acad. Sci. USA. 97:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrader, C.E., S.P. Bradley, J. Vardo, S.N. Mochegova, E. Flanagan, and J. Stavnezer. 2003. Mutations occur in the Ig Smu region but rarely in Sgamma regions prior to class switch recombination. EMBO J. 22:5893–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.