Abstract
Memory B cells provide rapid protection to previously encountered antigens; however, how these cells develop from germinal center B cells is not well understood. A previously described in vitro culture system using human tonsillar germinal center B cells was used to study the transcriptional changes that occur during differentiation of human memory B cells. Kinetic studies monitoring the expression levels of several known late B cell transcription factors revealed that BCL-6 is not expressed in memory B cells generated in vitro, and gene expression profiling studies confirmed that BCL-6 is not expressed in these memory B cells. Furthermore, ectopic expression of BCL-6 in human B cell cultures resulted in formation of fewer memory B cells. In addition, the expression profile of in vitro memory B cells showed a unique pattern that includes expression of genes encoding multiple costimulatory molecules and cytokine receptors, antiapoptotic proteins, T cell chemokines, and transcription factors. These studies establish new molecular criteria for defining the memory B cell stage in human B cells.
When B cells respond to a T cell–dependent protein antigen, extrafollicular plasma cells expressing low-affinity B cell receptors (BCRs) form early in the primary response. However, 10–12 d after immunization, after B cell maturation in germinal centers (GCs), two kinds of differentiated effectors expressing high affinity Igs are formed (1). These are non-Ig–secreting memory B cells and Ig-secreting plasma cells. Memory B cells differ from plasma cells in that they do not secrete Ig, have a low basal level of proliferation, and have the ability to differentiate rapidly into Ig-secreting cells after secondary antigenic challenge (2). Most plasma cells are short lived, although a subset receives survival signals in the bone marrow and continues secreting Ig for extended periods (3). Transcription factors that are critical for the maturation of B cells in GCs and for formation and function of plasma cells have been identified. In addition, gene expression profiles GC B cells and plasma cells have been determined by several groups (4–7). Significantly less is known about regulation and gene expression patterns in memory B cells.
Plasma cell differentiation is under the control of several transcription factors that coordinately drive terminal differentiation and Ig secretion. These transcription factors include Blimp-1, IRF-4, and XBP-1 (for review see reference 8). The targets of these factors and the complex patterns of gene expression in plasma cells have been well studied. When plasma cell transcription factors are induced, GC or activated B cell factors such as Bcl-6, Pax5, Bach2, MiTF (1, 9), and IRF-8 (10) are repressed. Indeed, the plasma cell transcription factors and the GC transcription factors appear to establish mutually exclusive regulatory programs enforced by gene repression. For example, in GC B cells Bcl-6 represses Prdm1 (encoding Blimp-1) (11), BSAP may repress Xbp-1 (12), and MiTF represses IRF4 (13), whereas in plasma cells, Blimp-1 represses Bcl-6 and Pax5 (14).
In contrast, much less is known about gene expression patterns in memory B cells, and transcription factors required for their differentiation have not been identified. It is known that memory B cells have increased expression of CD80 and CD86 (15), allowing efficient costimulation of T helper cells and elevated expression of antiapoptotic genes BCL-2 and BCL-xL, aiding long-term survival (4, 16). More recently, it was demonstrated that Stat5-dependent induction of BCL-6 in human memory B cells resulted in self-renewal properties (17). There is also evidence for different subsets of memory B cells (for review see reference 18).
Mice lacking Bcl-6 demonstrate that Bcl-6 is required for the function of GC B cells (19, 20). However, Bcl-6 may also have a role in memory B cells. It has been suggested that BCL-6 functions in memory B cells by providing replicative potential and repressing terminal differentiation to plasma cells (21). Indeed, two studies have demonstrated that ectopic expression of BCL-6 in B cells maintains cell survival over a long period of time (17, 22). Scheeren et al. demonstrated in human B cells that phosphorylated Stat5 directly induces BCL-6 expression and that ectopic expression of STAT5 or BCL-6 in peripheral B cells increased their self-renewal capacity (17). However, in Bcl-6 −/− mice, functional memory B cells develop and are capable of responding to secondary antigenic stimulation, providing evidence that Bcl-6 is not strictly required for memory B cell differentiation or function (23). Additionally, several studies have shown that Bcl-6 RNA and protein are expressed primarily in GC B cells (10, 24–26). Thus, a role for Bcl-6 in memory B cells is still unclear.
We are interested in understanding gene expression patterns in memory B cells, the role of Bcl-6 in memory B cells, and the changes in transcriptional regulators that allow GC B cells to differentiate into plasma and memory B cells. Because it is difficult to address these questions in vivo, we have taken advantage of an in vitro culture system originally described by Choi and colleagues (27–29). In this system, centroblasts purified from human tonsils are cultured with CD40L, cytokines, and a follicular dendritic cell–like cell line called HK (30), thus mimicking a GC environment. The addition of IL-4 into the culture drives memory B cell differentiation and the addition of IL-10 drives plasma cell differentiation (27, 28). Various stages of cells in the cultures have been defined previously and can be distinguished by flow cytometry: centroblasts are CD20hiCD38hi, memory B cells are CD20+CD38−, and plasma cells are CD20−CD38+ (29, 31). IL-4 drives the in vitro differentiation of human tonsillar GC B cells to memory B cells, characterized by surface proteins found on human memory B cells that are IgD−CD20+CD38−CD44+CD86+ (15, 27). Memory B cells expressing these surface proteins in vivo are quiescent, completely isotype switched, have hypermutations of the heavy chain locus, and do not secrete Ig (15, 32, 33). Additionally, purified cells with these phenotypes produce specific IgG antibodies when challenged with tetanus toxoid (15, 29). Although the CD38−CD20+ cells in this culture have many characteristics of in vivo memory B cells, their ability to provide memory in vivo cannot be demonstrated. Therefore, we refer to them as in vitro memory B cells.
Using this culture system, we monitored changes in the expression of transcription factors known to be important in GC B cells and plasma cells. These studies showed that none of the GC or plasma cell transcription factors studied, including BCL-6, were induced or highly expressed during formation of memory B cells in vitro. Furthermore, ectopic expression of BCL-6 in cultured GC B cells blocked the majority of the cells from differentiating into memory B cells in response to IL-4, providing evidence that BCL-6 repression is necessary for in vitro memory B cell differentiation. Microarray studies confirmed that BCL-6 was not expressed in in vitro memory B cells. However, a unique pattern of gene expression was identified for in vitro memory B cells, including genes encoding multiple costimulatory molecules and cytokine receptors, antiapoptotic proteins, T cell chemokines, and transcription factors.
RESULTS
Changes in transcription factor mRNAs during differentiation of memory B cells and plasma cells in culture
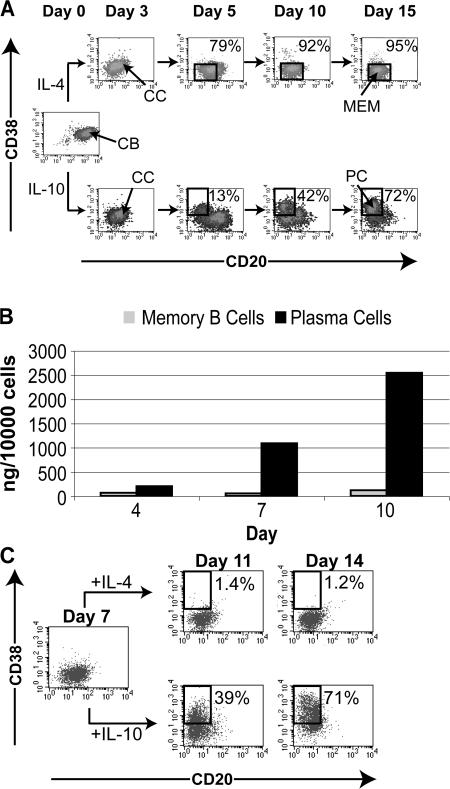
To explore changes in expression of transcription factors during the differentiation of memory B cells and plasma cells, we took advantage of a previously described in vitro culture system for human tonsillar B cells (27–29). This system allowed us to compare the kinetics and magnitude of changes in gene expression during post-GC differentiation of plasma and memory B cells in vitro. As previously reported, purified human tonsillar centroblasts cultured on HK cells (30) with CD40L, IL-2 and IL-4, or IL-10 develop into memory B cells or plasma cells, respectively (28) (Fig. 1 A). Using antibodies to CD20 and CD38, we detected over 90% CD20+CD38− memory B cells in cultures of centroblasts with IL-4 and at least 70% CD20−CD38+ plasma cells in cultures with IL-10 after 10–15 d of culture. For simplicity, stimulation of centroblasts with CD40L, IL-2, and IL-4 on HK cells will be referred to as the memory B cell culture and stimulation with CD40L, IL-2, and IL-10 on HK cells will be referred to as the plasma cell culture. Based on previous studies, CD20hiCD38hi, CD20+CD38+, CD20+CD38−, and CD20−CD38+ will be referred to as centroblasts, centrocytes, in vitro memory B cells, and plasma cells, respectively. Consistent with previous observations, only plasma cell cultures contain substantial levels of IgG in the supernatant (34). Compared with memory B cell cultures, plasma cell cultures contain 25-fold more IgG secreted into the culture supernatant by the end of 10 d (Fig. 1 B).
Figure 1.
GC B cell cultures. (A) Flow cytometric analyses of representative plasma and memory B cell cultures. Purified CD20hiCD38hi centroblasts (CB) were cultured with CD40L and IL-2, with IL-4 or IL-10 on HK cells to develop CD20+CD38− memory B cells (MEM) or CD20−CD38+ plasma cells (PC), respectively. Cells were harvested on the days indicated and stained with fluorescent-tagged antibodies to CD20 and CD38. The percentages indicate the proportion of plasma and memory B cells. Centrocytes (CC) are CD20+CD38+ and are present in both cultures. (B) Plasma cells but not memory B cells secrete high levels of IgG. Supernatant of plasma and memory B cell cultures were harvested on the days indicated. ELISA was performed to detect IgG. Cells were counted on the days supernatant was collected, and the concentration of IgG per ml was normalized to the number of cells. (C) On day 7 of a memory B cell culture, cells were divided for culture with CD40L, IL-2, and IL-4 (memory B cell culture) or washed and cultured with CD40L, IL-2, and IL-10 (plasma cell culture). Unswitched and switched cells were analyzed with antibodies to CD20 and CD38 on day 11 and day 14. This is representative of two independent experiments.
In vitro memory B cells can quickly differentiate into plasma cells upon reculturing with IL-10 instead of IL-4. The rapid induction of plasma cells from in vitro memory B cells occurs within 2–3 d compared with 5–10 d required for the differentiation of centroblasts into plasma cells (Fig. 1 C) (28). This is consistent with the characteristics of in vivo memory B cells, which undergo rapid plasmacytic differentiation upon challenge (35, 36).
Changes in steady-state mRNAs encoding key transcription factors during the differentiation of centroblasts to in vitro memory B and plasma cells were monitored by quantitative RT-PCR, normalized to cyclophilin A, and compared with levels in centroblasts at day 0 of the culture (Fig. 2). In the plasma cell cultures, there is a gradual increase in mRNA for BLIMP-1, XBP-1, and IRF-4 (Fig. 2), consistent with previous reports (37–40). The mRNA levels of BLIMP-1, XBP-1, and IRF-4 increase by 15-, 30-, and 30-fold, respectively by the end of the plasma cell culture compared with centroblasts. Concomitantly, mRNA encoding two well-known GC transcription factors, BCL-6 (19) and PAX-5 (41), drop ∼33- and 5-fold, respectively, in plasma cells compared with centroblasts. The mRNA for BCL-6 and PAX-5 fall to a low level before BLIMP-1 and XBP-1 mRNA levels increase significantly. Additionally, mRNA for BACH2, a GC transcription factor known to have a role in somatic hypermutation and class switching (42), decreases in the plasma cell culture. Although the length of time necessary for plasma cell and memory B cell differentiation varied among cultures, the trends in the increase and decrease of the transcript levels of transcription factors were the same in different cultures. These observations are consistent with the transcriptional network described for murine plasma cells (8).
Figure 2.
Expression kinetics of steady-state mRNA encoding transcription factors during memory B cell and plasma cell differentiation in vitro. Cells from plasma and memory B cell cultures were harvested on the days indicated. Total cells were harvested and RNA was prepared. Quantitative RT-PCR for BCL-6, Pax5, Bach2, BLIMP-1, XBP-1, and IRF-4 was performed. All values were normalized to cyclophilin A and are shown relative to the value at day 0. Each point is the mean of triplicates in qRT-PCR for each experiment. These profiles are representative of three separate experiments.
In the memory B cell cultures the mRNA levels of all three plasma cell factors, BLIMP-1, XBP-1, and IRF-4, are lower than in the plasma cell cultures. BLIMP-1 transcripts oscillate between 2–3-fold below the level in centroblasts. XBP-1 and IRF-4 mRNA levels increase modestly and peak on day 4 to 10-fold above the level found in centroblasts. After day 4 the mRNA for XBP-1 and IRF-4 decreases gradually to be 5–7-fold above that in centroblasts. However the modest increase in XBP-1 and IRF-4 transcript levels in in vitro memory B cells compared with centroblasts suggests that they might play a role in memory B cell function. This supports a recent study showing that switched memory B cells deficient in IRF-4 do not survive well, suggesting a role for IRF-4 in maintaining memory B cell pool (43).
Interestingly, PAX5, BCL-6, and BACH2 mRNAs decrease rapidly during differentiation of memory B cells in vitro; the kinetics and magnitude of the decrease are similar to that observed during plasma cell differentiation. mRNAs for PAX5, BCL-6, and BACH2 drop 3-, 30-, and 5-fold, respectively, below the level expressed in centroblasts. Thus, none of the transcription factors known to be important for GC B cells or plasma cells were highly expressed in memory B cells in this culture system.
Ectopic expression of BCL-6 represses memory B cell differentiation in vitro
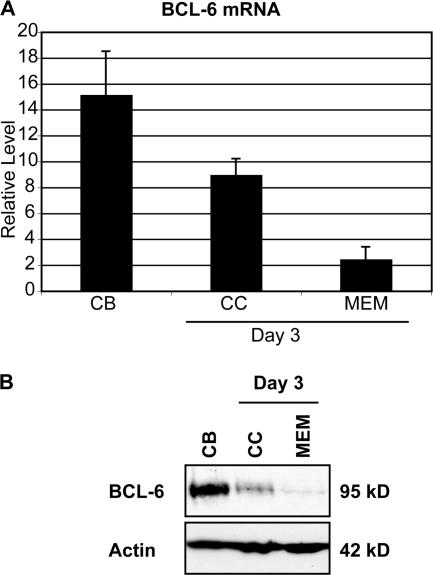
The functional role of BCL-6 in memory B cells has been controversial (17, 21, 44). Our data show that BCL-6 transcripts fall significantly in memory B cell cultures; however, because BCL-6 can be regulated posttranscriptionally (45), we also quantitated BCL-6 protein levels in our cultures. Centroblasts were cultured for 3 d with or without IL-4, generating centrocytes or memory B cells, respectively, as described previously (27). Consistent with steady-state mRNA levels (Fig. 3 A), BCL-6 protein was also reduced in both centrocytes and memory B cells generated in vitro (Fig. 3 B). The substantial decrease in BCL-6 protein at day 3 in the in vitro memory B cells suggests that BCL-6 is unlikely to play a role in memory B cell differentiation.
Figure 3.
Expression of Bcl-6 mRNA and protein is low in centrocyte and memory cultures. (A) Purified centroblasts at Day 0 (centroblasts [CB]) or cultured for 3 d with CD40L, IL-2 (centrocytes [CC]), or CD40L, IL-2, IL-4 (memory B cells [MEM]) were harvested and qRT-PCR was performed for BCL-6 and cyclophilin A. Representative of at least five independent experiments. Error bars correspond to the standard deviation. (B) Whole cell extracts of 100,000 cells from each culture described (see Materials and methods) were used for immunoblotting with antibodies to BCL-6 and actin. Representative of two independent experiments.
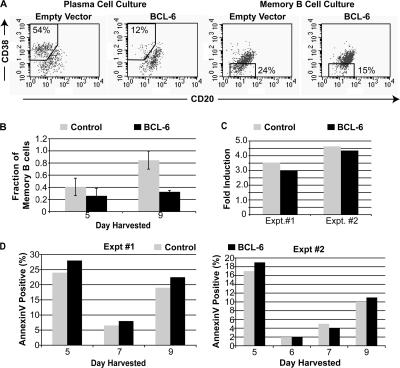
Because our data showed that BCL-6 levels decrease in in vitro memory B cells, we wished to determine if reduction of BCL-6 is required for memory B cell differentiation. To do this we generated lentiviruses expressing genes encoding eGFP alone or a fusion protein of BCL-6 and eGFP. The functionality of the BCL-6–eGFP fusion protein was confirmed in Wil-2 cells by observing a decrease of CD69 mRNA after infection (unpublished data); CD69 is a known target of BCL-6 transcriptional repression (46). On day 2 of IL-4 and IL-10 cultures, the cells were infected with the control lentivirus expressing eGFP (Fig. 4, B–D, Control) or the lentivirus expressing BCL-6–eGFP (Fig. 4, B–D, BCL-6). 3 d after infection, cells were harvested and analyzed by flow cytometry, using GFP to identify infected cells. As expected (11), expression of BCL-6 blocked the formation of plasma cells (Fig. 4 A, left), further confirming the biological activity of the BCL-6 fusion protein. Interestingly, ectopic expression of BCL-6 also blocked in vitro memory B cell formation (Fig. 4 A, right). When cells were analyzed 7 d after infection, the block was more dramatic (Fig. 4 B). 9 d after infection, an average of 85% of the cells were memory B cells in cultures infected with the control virus, whereas only an average of 32% of the cells were memory B cells in cultures infected with the BCL-6 virus.
Figure 4.
Ectopic expression of BCL-6 represses memory B cell differentiation in vitro. Plasma and memory B cell cultures were infected with lentivirus encoding GFP alone (Empty Vector, Control) or a fusion protein of GFP and BCL-6 (BCL-6) on day 2 and harvested for analyses on day 5. (A) Cells were first gated for GFP expression and analyzed with antibodies to CD20 and CD38. Percentages represent proportion of cell types (memory B cells and plasma cells) in each gated population. Data are representative of three experiments. (B) Graphical representation of the fraction of GFP+CD20+CD38− memory B cells in infected IL-4 B cell cultures 3 and 7 d after infection (5 and 9 total days in culture). Data shown are the mean of four experiments for day 5 and 3 experiments for day 9. Error bars correspond to the standard deviation. (C) BCL-6–infected memory B cell cultures are similarly proliferative compared with memory B cells infected with the control virus. The fold induction in proliferation was calculated by dividing the number of GFP+ cells 7 d after infection by the number 3 d after infection from in vitro memory B cells infected with the control or BCL-6 virus. Shown are two independent experiments. (D) Memory B cell cultures with forced expression of BCL-6 and control cells have similar amounts of cell death. Infected memory B cell cultures were harvested on day 5, 6 (for Expt #2), 7, and 9. Cells were analyzed for apoptosis with annexin V and 7-AAD staining of GFP+ cells. Graphical representation of annexinV+7AAD−GFP+ cells 3, 4, 5, and 7 d after infection. Shown are two independent experiments.
Inhibition of in vitro memory B cell differentiation in this experiment could have been caused by cell death or a developmental block. To investigate further, we counted live GFP+ cells 3 and 7 d after infection (Fig. 4 C). Cultures infected with the BCL-6 virus had a slightly lower number of infected cells 3 d after infection. This is most likely because of differences in infection efficiency of different virus stocks. However, 7 d after infection, BCL-6–infected cells increased in cell number, similar to cells infected with the control virus. Results from two separate experiments show that cells in both cultures were similarly proliferative (Fig. 4 C). We also stained infected cells with annexin V and 7-amino actinomycin D (7-AAD) to monitor apoptosis (Fig. 4 D). Five out of seven times analyzed, in two experiments, BCL-6–infected cells had small increases in numbers of annexin V+ cells; two of out seven times the BCL-6 cultures had equal or less apoptotic cells (Fig. 4 D). The small difference are insufficient to explain the ∼50% reduction in memory B cells in BCL-6 cultures versus control cultures on day 9. These data are not consistent with the possibility that apoptosis of memory B cells is solely responsible for their absence in the BCL-6 cultures but provide strong support for the idea that Bcl-6 blocks memory B cell differentiation. This is consistent with previous reports (17, 22, 47, 48) showing that Bcl-6 in fact provides survival signals to GC B cells.
Global gene expression patterns in tonsillar B cell cultures
Because none of the GC or plasma cell transcription factors we studied were expressed in the in vitro memory B cell cultures, we explored global gene expression patterns in these cultures using cDNA microarrays (5). RNA was prepared from sort-purified CD38−CD20+ memory B cells and CD38+CD20− plasma cells generated in vitro and compared with CD38+CD20+ centrocytes. The relative abundance of transcripts in plasma cells and in vitro memory B cells was compared with centrocytes, and mRNAs with >1.5-fold differential expression (P < 0.05) are listed. Complete datasets are provided in Tables S1–S5 (available at http://www.jem.org/cgi/content/full/jem.20062104/DC1).
Plasma cell and centrocyte gene expression patterns
mRNAs encoding transcription factors PAX5, BACH2, STAT6, ID3, SPIB, ZNFN1A1 (Ikaros), EBF, and POU2F2 (OCT2) decreased in plasma cells (Table S1). All of these transcription factors except for BACH2 are known to be down-regulated by Blimp-1 (14, 49). mRNAs for BLIMP-1, XBP-1, and IRF4 increased in plasma cells. In addition, the mRNAs for numerous XBP-1 targets including ARP, PPIB, ribophorin I, FKBP19, signal sequence receptor gamma, protein kinase inhibitor protein p58 and pA3 also increased in plasma cells (50). Ig heavy and light chain and J chain mRNAs were also elevated. Thus, the gene expression patterns observed are consistent with previous studies of ex vivo plasma cells in human and mice (4, 6, 7) and of B cells with forced expression of Blimp-1 to drive plasmacytic differentiation (14, 49). All the expression profiles show that during plasma cell differentiation B cell and proliferation gene expression programs are extinguished, whereas genes that mediate efficient immunoglobulin secretion are induced (Tables S1 and S2).
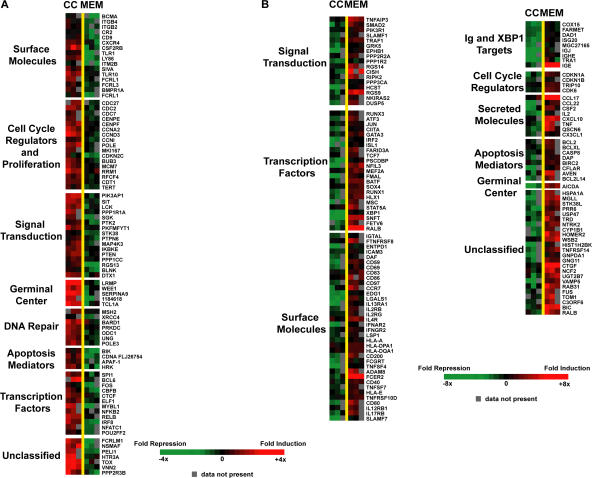
Transcripts that decreased in both plasma cells and in vitro memory B cells indicate genes specifically expressed in GC B cells. Among such mRNAs were MME (CD10), LRMP (JAW1), Serpina9 (centerin), MKI67 (Ki67), wee1 kinase, BCL-6, POU2F2, SPI1, and cell cycle genes (Tables S1 and S3 and Fig. 5 A), which have previously been identified as GC transcripts (5). Overall, the gene expression profiles for plasma cells and GC B cells in culture provide further evidence that the developmental changes in this system closely mimic those that occur in vivo and further suggest many commonalities between human and murine B cells.
Figure 5.
DNA microarray analysis comparing memory B cells and GC B cells cultured in vitro. Each column represents data from an independent experiment. Each row represents a gene significantly elevated (red) or decreased (green). A color bar shows the magnitude of gene expression changes as a ratio of expression in memory B cells versus GC B cells. Values shown are log2. Genes were organized into broad functional categories and were limited to genes with at least 1.5-fold difference between memory B cells and GC B cells. Unclassified genes were limited to genes with at least fourfold difference between memory B cells and GC B cells. (A) Genes decreased in memory B cells compared with GC B cells. (B) Genes elevated in memory B cells to GC B cells. The lists of other unclassified genes with less than 4-fold difference but above 1.5-fold are available in Tables S3 and S4.
Gene expression patterns in in vitro memory B cells
Consistent with previous findings, transcript levels of antiapoptotic genes BCL-2 and BCL-xL (16) and costimulatory molecules CD80 and CD86 (15) were elevated in in vitro memory B cells compared with GC B cells (Fig. 5 B). Additionally, mRNAs for BIRC2 (c-IAP1) and CFLAR (c-Flip), which inhibit caspase activation and function (51), were elevated. However, mRNAs for some proapoptotic genes, such as BCL-G (BCL2L14) and caspase 8, were also elevated (Fig. 5 B). Caspase 8 has recently been shown to play a role in the activation of NF-κB upon antigen receptor signaling and, thus, may not be playing a role in the apoptotic pathway in these cells (52). Interestingly, expression of both proapoptotic and antiapoptotic genes in ex vivo memory B cells has been observed in a previous study (4).
Memory B cells undergo activation and proliferation more rapidly than naive B cells (36). Consistent with this phenotype, mRNAs were elevated for proteins involved in antigen presentation (MHC class II and CIITA), T cell costimulation (CD80, CD86, OX40L, CD70, and CD40), chemokines to attract T cells (CXCL10, CCL17, CCL22, CX3C, and CCL22), and for proteins involved in JNK signaling (c-jun, ATF-3, and B-ATF) and certain phosphatases. A striking increase in mRNAs encoding cytokine receptors (IL-2RB, IL-4RA, IL-12RB1, IL-13RA1, and IL-17R) was observed (Fig. 5 B), consistent with a heightened response to the environment.
Two comparable gene expression profiling studies are available from ex vivo human memory B cells to provide comparison with our results for in vitro memory B cell gene expression. Klein et al. (4) analyzed an ex vivo purified CD27+CD38loCD10−CD3−CD14− population from human tonsillar B cells that they considered memory B cells. Alizadeh et al. analyzed an ex vivo purified CD20+CD27+ population from human peripheral blood that they considered memory B cells and CD38+CD77− centrocytes from the tonsils (53). This complete dataset is presented in Table S5. In agreement with our data, IL-2RB (4) and IL-13RA1 mRNA levels were found to be elevated in ex vivo–purified human memory B cells compared with centrocytes (Table S5). Stimulation via IL-4 (54), IL-2 (54), IL-12 (55), and IL-13 (56) receptors on B cells is known to provide proliferation signals, but the role IL-17 in B cells is currently not clear.
Interestingly, a group of transcription factor mRNAs (Stat5, TCF7, and MEF2A) known to play a role in self-renewal were elevated in in vitro memory B cells (Fig. 5 B). Stat5 has been shown by Scheeren et al. to provide self-renewal properties for human memory B cells (17). In a recent study to identify a shared transcriptional program among self-renewal cells, hematopoietic stem cells, memory B and T cells, Mef2a and members of the TCF family were shown to be elevated in memory B cells and hematopoietic stem cells (57). Although neither Mef2a nor TCF7 has been shown to provide self-renewal properties in memory B cells, this has been shown in neurons (58, 59) and stem cells (60). Thus, there is potential importance for the role of these transcription factors in self-renewal of memory B cells as well.
Unexpectedly, activation-induced cytidine deaminase (AID) mRNA was significantly increased in in vitro memory B cells compared with centrocytes. AID is required for somatic hypermutation that occurs in GC B cells and is known to be highly expressed in centroblasts (61). However, affinity maturation continues in a primary response after the GC response has apparently waned (62, 63). Although it has been assumed that ongoing affinity maturation is the result of ongoing selection of memory cells, our observation of high levels of AID in in vitro memory B cells suggests the intriguing possibility that diversification and selection may continue in memory B cells.
Some mRNAs showed opposite expression patterns in plasma cells and in vitro memory B cells (Fig. 6); those specifically elevated in the memory cultures may indicate genes required for memory B cells. Specific elevation of mRNAs encoding costimulatory molecules (CD40, CD80, and MHCII) and cytokine receptors in in vitro memory B cells is consistent with the rapid reactivation of memory B cells (Fig. 6 A). Stat5 mRNA was also increased in memory B cells and decreased in plasma cells generated in vitro, consistent with the role of Stat5 for self-renewal (17). However, BCL-6 mRNA was not higher in memory B cells compared with plasma cells generated in vitro, consistent with our previous data (Figs. 2 and 4) and microarray of ex vivo memory B cells (64). Other mRNAs specifically elevated in in vitro memory B cells include RUNX3, CCL17, TNFAIP3 (A20), NCOR2 (SMRT), and TRIP10 (CIP4). Elevated transcript levels of TNFAIP3 have also been seen in ex vivo memory B cells (Table S5). TNFAIP3 regulates NF-κB–dependent gene activation and apoptosis and may be important for memory B cell survival (65). Interestingly, transcripts encoding TNFRSF17 (B cell maturation antigen [BCMA]), a receptor for B cell factors BAFF and APRIL are increased in plasma cells but decreased in in vitro memory B cells (Fig. 6 B). Both c-fos and NFAT also decreased in in vitro memory B cells, and although not significantly elevated in our in vitro–generated plasma cells, have been shown to be expressed in plasma cells. Thus, a decrease of these factors may be unique to memory B cells (6, 66). Another molecule known to be critical for plasma cells is the cyclin-dependent kinase inhibitor CDKN2C (p18-INK6) (67), which is increased in plasma cells and decreased in memory B cells generated in culture (Fig. 6 B). These data provide important new insights into gene expression patterns in human memory B cells and establish new molecular criteria for defining memory B cell differentiation.
Figure 6.
Genes with opposite expression patterns in plasma cells and memory B cells generated in vitro. (A) Bar graphs showing the expression level of genes elevated in memory B cells compared with GC B cells but decreased in plasma cells compared with GC B cells. (B) Bar graphs showing the expression level of genes elevated in plasma cells compared with GC B cells but decreased in memory B cells. Log2 values are shown.
DISCUSSION
Multiple studies have contributed to a general consensus about gene expression patterns in GC B cells and plasma cells, showing induction in plasma cells of genes encoding proteins involved in Ig production and secretion and repression of genes encoding proteins necessary for B cell function and cell division (7, 14, 50). Much less, however, is known about gene expression in memory B cells. We have studied specific transcription factors and global gene expression profiles in in vitro cultures of human B cells in which both plasma cells and memory B cells can be generated from GC B cells (27–29). Our studies reveal the kinetics with which key GC and plasma cell transcription factors change during plasma cell and memory B cell differentiation in vitro. Gene profiling studies in this system confirm previously reported centrocyte and plasma cell gene expression programs and reveal gene expression patterns in human memory B cells. These include increased transcripts encoding cytokine receptors, costimulatory molecules and transcription factors including c-jun, and decreased expression of centrocyte transcription factors including BCL-6. Not only is BCL-6 mRNA extremely low in in vitro memory B cells, we show that ectopic expression of BCL-6 leads to formation of fewer memory B cells. This supports the idea that BCL-6 repression is necessary for memory B cell differentiation and plasma cell differentiation.
In vitro–generated memory B cells and the role of IL-4
The hallmarks of memory B cells include self-renewal, longevity, and rapid response upon secondary antigen encounter. Increased expression of known memory B cell genes CD80, CD86, BCL-2, and BCL-xL in our IL-4 cultures supports the notion that CD20+CD38− cells in this culture are similar to memory B cells formed in vivo.
The memory B cells that develop in our in vitro system are similar to those formed in vivo with respect to surface phenotype and the ability to generate Ig-secreting plasma cells rapidly (Fig. 1 C) (28). Although IL-4 drives differentiation of memory B cells in vitro, the role of IL-4 in the differentiation of memory B cells in vivo is unclear. No defects in primary and secondary B cell responses were reported in IL-4–deficient mice (68); however, IL-13, a closely related cytokine that shares similar signaling pathways, could be compensating for the loss of IL-4 (69). It is evident from our microarray analysis of in vitro memory B cells that IL-4– dependent genes are induced and it is unclear which may also be characteristic of in vivo memory B cells. IL-4–induced genes are likely to be important for memory B cells in vivo because IL-13RA, a known target of IL-4 (70), is up-regulated both in memory B cells derived from our in vitro culture in the presence of IL-4 and in ex vivo memory B cells in the absence of IL-4 (Fig. S1). Additionally, the decreased expression of many GC B cell and plasma cell genes in the memory B cell culture indicates differentiative changes and changes in IL-4–dependent genes (Fig. 5).
Changes in transcription factors during memory B cell and plasma cell differentiation in vitro
In vitro differentiation of GC B cells provided an opportunity to understand the dynamic changes in specific transcription factors that occur during memory B and plasma cell formation. The induction of mRNA encoding late B cell regulators BLIMP-1, XBP-1, and IRF-4 confirmed our current understanding of the transcriptional network in plasma cells. During memory B cell differentiation we found that BCL-6 and PAX5 mRNAs decreased to low levels, suggesting that these transcription factors do not play a role in in vitro memory B cells. Interestingly, decreased expression of BCL-6 and PAX5 mRNA in in vitro memory B cells was not sufficient to induce BLIMP-1 and XBP-1, although both are reported to repress BLIMP-1 and XBP-1 (46, 71). Two nonmutually exclusive possibilities could explain these observations: (a) other repressors may control BLIMP-1 and XBP-1 transcription in in vitro memory B cells or (b) in addition to removal of repressors, induction of activators for BLIMP-1 and/or XBP-1 transcription may be required for plasma cell differentiation. For example, IL-21 is known to be a strong inducer of Blimp-1 mRNA and plasma cell differentiation (72). The latter explanation would suggest that for memory B cells to differentiate quickly into plasma cells upon antigen rechallenge, only one signal, induction of Blimp-1 activator(s), would be required to initiate plasmacytic differentiation, because the repressors Bcl-6 and PAX5 would already be absent.
Gene expression profile in in vitro memory B cells and plasma cells
Compared with GC B cells, the in vitro–generated memory B cells showed elevated transcripts of genes encoding costimulatory molecules for T cells (CD40, CD80, CD86, MHCII), chemokines to attract T cells (CCL17, CX3C, CCL12), survival molecules (BCL-2, BCL-xL, BIRC2, and CFLAR), and self-renewal molecules (Stat5, Mef2a, TCF7). Expression of these genes is consistent with the phenotype of memory B cells. Interestingly, a comparison of our data with two other memory B cell gene expression profiles shows 12 similarly regulated genes between the study performed by Klein et al. (4) and our study and 18 similarly regulated genes between our study and the study by Alizadeh et al. (reference 53 and Table S5). Three genes were similarly increased in all three studies: c-jun, HSP70, and HLA-E. A complete list of genes similar among all three studies is shown in Fig. S1. The limited similarity among these studies is most likely because of differences in the source and identification of the memory B cells, which could represent memory B cells at different stages during their differentiation. It will be interesting to perform a time course gene expression profiling study using memory B cells from this in vitro culture system to determine the gene expression profile during memory B cell differentiation and to see if there will be more similarities to the other ex vivo gene-profiling studies.
Several interesting transcripts increased in plasma cells but decreased in memory B cells generated in vitro, including BCMA and p18-INK6. BCMA provides survival signals for long-lived plasma cells (73). The induction of BCMA transcripts in plasma cells suggests that all/many post-GC plasma cells have the capability of becoming long-lived plasma cells if they find a niche in the bone marrow. Decreased BCMA mRNA in memory B cells suggests that memory B cells survive through activation of another receptor for BAFF and APRIL, probably BAFF-R (74) and/or another mechanism. p18-INK6 is a known regulator of cell cycle arrest in plasma cells (67). Decreased p18 expression in in vitro memory B cells suggests that p27kip, which is increased in both in vitro memory B cells and plasma cells, may be more important for dampening cell cycle in memory B cells, whereas in plasma cells p18 is critical.
Transcripts encoding c-fos and NFATc1 decreased only in in vitro memory B cells. Interestingly, both genes play a role in plasma cells (6, 66). Stimulation of splenic B cells from c-fos transgenic mice with CD40L and IL-4 resulted in the induction of Blimp-1 transcripts and differentiation into Ig-secreting cells (66), whereas similar treatment of wild-type B cells resulted in no Blimp-1 induction and minimal formation of Ig-secreting cells.
BCL-6 blocks memory B cell differentiation in vitro
BCL-6 protein is most highly expressed in GC B cells and is known to functionally inhibit plasma cell differentiation. One way it does this is by inhibiting Blimp-1 transcription (19, 46). This is consistent with our finding that ectopic expression of BCL-6 in plasma cell cultures blocks plasma cell differentiation (Fig. 4). It was hypothesized that BCL-6 might be required for memory B cells because BCL-6 can block terminal differentiation and provide self-renewal properties (21). Our data show the contrary. BCL-6 mRNA in in vitro memory B cells is as low as it is plasma cells and BCL-6 protein is barely detectable (Fig. 6). Additionally, the in vitro memory B cell gene expression profile shows an increase in CDKN1B (p27), a target of BCL-6 (46). Thus, a function for BCL-6 in memory B cells is unlikely. Indeed, our data show that ectopic expression of BCL-6 in memory B cell cultures blocks the majority of the GC B cells from differentiating into memory B cells. Our data, in combination with earlier studies showing Bcl-6 expression primarily in GC B cells (4, 10, 24, 25, 75–77) and a memory response in Bcl-6−/− mice (23), argue strongly against a requirement for Bcl-6 in memory B cell differentiation. The inability of ectopic Bcl-6 to fully block memory B cell differentiation in vitro may be caused by developmental heterogeneity of isolated tonsillar B cells that have spent varying amounts of time in GCs undergoing affinity maturation and class switch recombination. Alternatively, there may be a subset of cells (18) which can become memory cells in the presence of Bcl-6.
In contrast to our results, a recent study reported that BCL-6 provides self-renewal properties to human memory B cells (17). The discrepancies between the Scheeren study and our study are most likely because of differences in memory B cell subpopulations being studied. To demonstrate the self-renewal property of BCL-6, Scheeren et al. ectopically expressed BCL-6 in peripheral blood B cells cultured with cells expressing CD40L, IL-2, and IL-4 for 70 d. At day 70 the cells expressed surface molecules characteristic of GC B cells in our in vitro culture (CD20+CD38+) (31), although the authors identified these cells as memory B cells. Further studies using the same source of memory B cells and the same memory cell isolation will be necessary to resolve these differences and definitively establish the role of BCL-6 in memory B cell development.
In conclusion, our studies have revealed dynamic changes in transcription factors during terminal B cell differentiation and have identified new gene expression programs in in vitro memory B cells. We show that BCL-6 is normally expressed at very low levels in in vitro memory B cells and that forced expression of BCL-6 blocks memory B cell differentiation in culture. Our data provide validation for using this in vitro culture system for further analysis of B cell differentiation.
MATERIALS AND METHODS
Purification of GC B cells.
Tonsillar B cells were prepared and purified as described previously (34). Tonsils were obtained from tonsillectomies performed at the Children's Hospital of Columbia University. Tissue collection was approved by the Institutional Review Board of Columbia University Medical Center. Tonsils were shredded in RPMI plus 10% FBS (complete media). Media containing cells were passed through 70-μm filters. Tonsillar cells were spun over Histopaque 1077 (Sigma-Aldrich) to obtain mononuclear cells. Pelleted cells were subjected to sheep red blood cells (Colorado Serum Company) rosetting to deplete T cells. Aminoethylisothiouronium bromide (AET)–treated (Sigma-Aldridch) sheep red blood cell were incubated with tonsillar B cells on ice for 1 h. Resuspended AET-treated cells were spun over Histopaque 1077. The resulting nonrosetted cells in the interphase contained >95% CD19+ cells. Centroblasts were isolated by magnetic cell separation (Miltenyi Biotec), and purification was performed according to manufacturer's protocol. CD19+ cells were incubated with antibodies to IgD (1.5 μg/30 × 106 cells) and CD44 (1.5 μg/30 × 106 cells) (Southern Biotechnology Associates, Inc.), washed, and then incubated with rat anti–mouse IgG2a/IgG2b microbeads (Miltenyi Biotec) in MACS buffer for 15 min on ice. The negatively selected cells were collected.
GC B cell culture.
HK cells were a kind gift from Dr. Yong Sung Choi (Alton Ochsner Medical Foundation, New Orleans, LA). Cytokines IL-2, IL-4, and IL-10 were obtained from Peprotech. Concentrations used for the cultures were 25 ng/ml for IL-2, 50 ng/ml for IL-4, and 100 ng/ml for IL-10. Soluble human trimeric CD40L was obtained from Amgen and used at a concentration of 400 ng/ml. In brief, 3,000 HK cells/well/ml were plated in 24-well plates the day before culture of tonsillar B cells. On day 0, 600,000 centroblasts were cocultured with HK cells per ml of ISCOV media with relevant cytokines depending on culture, and cells were recultured every 3–5 d with new HK cells and cytokines. Subsequent reculturing of cells was plated at 300,000 cells/ml/well.
The following antibodies were used for flow cytometry: FITC-CD19 (Becton Dickinson), PE-CD19 (Southern Biotechnology Associates, Inc.), FITC-CD44 (Becton Dickinson), PE-CY7-CD20 (eBioscience), PE-CD20 (Becton Dickinson), and CD38 (Becton Dickinson). All antibodies were used at 1:100 except for PE-CY7-CD20, which was used at 1:5.
Lentiviral infections.
FUW-hBCL6 was constructed by inserting hBCL-6 into SalI–BamHI site of pEGFP-C1. The construct was digested and blunted at the NheI and BamHI sites to generate a fragment containing eGFP–BCL-6. Blunted eGFP–BCL-6 fragment was cloned into blunted BamHI–EcoRI sites of FUGW (a gift from Dr. Jeremy Luban, Columbia University).
293T cells (at 75–80% confluency) were cotransfected with 10 μg of pHD8.9, a lentiviral packaging construct (provided by Dr. Jeremy Luban, Columbia University), 5 μg of vesicular stomatitis virus G protein expression vector (provided by Dr. S.P. Goff, Columbia University), and 10 μg of either FUW–BCL-6 or FUGW using calcium phosphate precipitation. Lentiviral supernatants were collected 48 h after transfection concentrated and titered. GC B cells were infected at a multiplicity of infection of 1–2.
Quantitative PCR.
RNAs were isolated using TRIzol Reagent (Life Technologies), and cDNAs were prepared using SuperScript III reverse Trasncriptase (Invitrogen), according to the manufacturer's protocol. Quantitative RT-PCRs were performed on Abi7700 (Applied Biosystems). Concentrations for stock reagents are as follows: PCR buffer, 200 μM dNTP, 0.4× SYBR Green (Sigma-Aldrich), 150 nM 6-carboxy-x-rhodamine, ROX (Sigma-Aldrich), 1% DMSO, and 1.25 U Taq polymerase in 25 μl. Conditions and primer concentrations are listed in Table S6. The amplification programs are as follows: 95°C for 5 min; 95°C for 20 s, 59°C for 1 min, and 80–87°C (depending on primer) for 20 s (data collected) for 40 cycles. The melting curves are as follows: 95°C for 20 s, 59°C for 15 s, and up to 95°C for 20 s with a 19-min ramping time.
Apoptosis staining of cells with annexin V and 7-AAD.
Cells were infected on day 2 and harvested on day 5, washed twice with cold PBS. Cells were resuspended in annexin V buffer (Becton Dickinson) and incubated with annexin (Becton Dickinson) at 5 μl/100 μl buffer/105 cells and 5 μl VIA-PROBE (7AAD; Becton Dickinson) for 15 min at room temperature.
Protein expression analysis.
Detection of BCL-6 (Santa Cruz Biotechnology) and actin (Sigma-Aldrich) by Western blotting were performed using whole cell lysates of 100,000 cultured cells. BCL-6 antibody was used at 1:500 and actin antibody was used at 1:1,000.
Microarray procedures.
Fluorescent cDNA probes were prepared from an experimental mRNA sample (Cy5 labeled) and a control mRNA sample (Cy3 labeled) isolated from a pool of lymphoma cell lines. The use of a common control cDNA probe allows the relative expression of each gene to be compared across all samples. Microarray analysis was performed as described in Shaffer et al. (46) using 25–50 μg of polyA+ RNA (FastTrack; Invitrogen) for each cellular population. Arrays for each culture systems were repeated three times with independent pools.
Microarray analysis.
All listed genes are statistically significant (P < 0.05) as determined by three microarray replicates for each population of cells. Genes are grouped into functional categories with at least 1.5-fold difference between two cellular populations, and uncategorized genes have at least a fourfold difference between two memory B cells and GC B cells. The raw data is available at GEO (http://www.ncbi.nlm.nih.gov/geo) under series accession no. GSE7128.
Online supplemental material.
DNA microarray analysis comparing plasma or memory B cells to centrocytes. Values from three independent arrays for each cell population were averaged and then the averages for two populations were compared. Genes were limited to genes with at least 1.5-fold difference between two populations of cells with a p-value < 0.05. Log2 values are shown for each gene. The genes are arranged in order of the most to least difference in level between two cellular populations. Figure S1 shows similar genes elevated in memory B cells compared with GC B cells among three independent studies. A Venn diagram showing genes elevated in memory B cells comparing two studies and all three studies. Group A is genes from the current study, group B is data from Alizadeh et al. (53), and group C represents the study performed as described in reference 4. Table S1 shows genes decreased in plasma cells as compared with centrocytes. Table S2 shows genes elevated in centrocytes compared with plasma cells. Table S3 shows genes decreased in in vitro memory B cells compared with centrocytes. Table S4 shows genes elevated in centrocytes compared with in vitro memory B cells. Table S5 shows genes elevated in ex vivo CD20+CD27+ memory B cells compared with ex vivo CD38+CD77− centrocytes. Table S6 shows sequences and concentration of primers used for qRT-PCR.
Supplemental Material
Acknowledgments
We thank members of the Calame Laboratory for helpful discussions, Kristie Gordon for her help with flow cytometry, and Amgen (Thousand Oaks, CA) for providing trimeric CD40L.
A.L. Shaffer and L.M. Staudt are supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. T.C. Kuo and K. Calame are supported by RO1AI50659, and RO1AI43576 to K. Calame.
The authors have no conflicting financial interests.
Abbreviations used: 7-AAD, 7-amino actinomycin D; AID, activation-induced cytidine deaminase; BCMA, B cell maturation antigen; BCR, B cell receptor; GC, germinal center.
References
- 1.Lin, K.I., C. Tunyaplin, and K. Calame. 2003. Transcriptional regulatory cascades controlling plasma cell differentiation. Immunol. Rev. 194:19–28. [DOI] [PubMed] [Google Scholar]
- 2.McHeyzer-Williams, L.J., and M.G. McHeyzer-Williams. 2005. Antigen-specific memory B cell development. Annu. Rev. Immunol. 23:487–513. [DOI] [PubMed] [Google Scholar]
- 3.Manz, R.A., A.E. Hauser, F. Hiepe, and A. Radbruch. 2005. Maintenance of serum antibody levels. Annu. Rev. Immunol. 23:367–386. [DOI] [PubMed] [Google Scholar]
- 4.Klein, U., Y. Tu, G.A. Stolovitzky, J.L. Keller, J. Haddad Jr., V. Miljkovic, G. Cattoretti, A. Califano, and R. Dalla-Favera. 2003. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. USA. 100:2639–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaffer, A.L., A. Rosenwald, E.M. Hurt, J.M. Giltnane, L.T. Lam, O.K. Pickeral, and L.M. Staudt. 2001. Signatures of the immune response. Immunity. 15:375–385. [DOI] [PubMed] [Google Scholar]
- 6.Underhill, G.H., D. George, E.G. Bremer, and G.S. Kansas. 2003. Gene expression profiling reveals a highly specialized genetic program of plasma cells. Blood. 101:4013–4021. [DOI] [PubMed] [Google Scholar]
- 7.Tarte, K., F. Zhan, J. De Vos, B. Klein, and J. Shaughnessy Jr. 2003. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 102:592–600. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro-Shelef, M., and K. Calame. 2005. Regulation of plasma-cell development. Nat. Rev. Immunol. 5:230–242. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, K., M. Shapiro-Shelef, C. Tunyaplin, and K. Calame. 2005. Regulatory events in early and late B-cell differentiation. Mol. Immunol. 42:749–761. [DOI] [PubMed] [Google Scholar]
- 10.Lee, C.H., M. Melchers, H. Wang, T.A. Torrey, R. Slota, C.F. Qi, J.Y. Kim, P. Lugar, H.J. Kong, L. Farrington, et al. 2006. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J. Exp. Med. 203:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tunyaplin, C., A.L. Shaffer, C.D. Angelin-Duclos, X. Yu, L.M. Staudt, and K.L. Calame. 2004. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J. Immunol. 173:1158–1165. [DOI] [PubMed] [Google Scholar]
- 12.Reimold, A.M., P.D. Ponath, Y.S. Li, R.R. Hardy, C.S. David, J.L. Strominger, and L.H. Glimcher. 1996. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J. Exp. Med. 183:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, L., A.J. Gerth, and S.L. Peng. 2004. Active inhibition of plasma cell development in resting B cells by microphthalmia-associated transcription factor. J. Exp. Med. 200:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaffer, A.L., K.I. Lin, T.C. Kuo, X. Yu, E.M. Hurt, A. Rosenwald, J.M. Giltnane, L. Yang, H. Zhao, K. Calame, and L.M. Staudt. 2002. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 17:51–62. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y.J., C. Barthelemy, O. de Bouteiller, C. Arpin, I. Durand, and J. Banchereau. 1995. Memory B cells from human tonsils colonize mucosal epithelium and directly present antigen to T cells by rapid up-regulation of B7-1 and B7-2. Immunity. 2:239–248. [DOI] [PubMed] [Google Scholar]
- 16.Bovia, F., A.C. Nabili-Tehrani, C. Werner-Favre, M. Barnet, V. Kindler, and R.H. Zubler. 1998. Quiescent memory B cells in human peripheral blood co-express bcl-2 and bcl-x(L) anti-apoptotic proteins at high levels. Eur. J. Immunol. 28:4418–4423. [DOI] [PubMed] [Google Scholar]
- 17.Scheeren, F.A., M. Naspetti, S. Diehl, R. Schotte, M. Nagasawa, E. Wijnands, R. Gimeno, F.A. Vyth-Dreese, B. Blom, and H. Spits. 2005. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat. Immunol. 6:303–313. [DOI] [PubMed] [Google Scholar]
- 18.Tarlinton, D. 2006. B-cell memory: are subsets necessary? Nat. Rev. Immunol. 6:785–790. [DOI] [PubMed] [Google Scholar]
- 19.Dent, A.L., A.L. Shaffer, X. Yu, D. Allman, and L.M. Staudt. 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 276:589–592. [DOI] [PubMed] [Google Scholar]
- 20.Ye, B.H., G. Cattoretti, Q. Shen, J. Zhang, N. Hawe, R. de Waard, C. Leung, M. Nouri-Shirazi, A. Orazi, R.S. Chaganti, et al. 1997. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 16:161–170. [DOI] [PubMed] [Google Scholar]
- 21.Fearon, D.T., P. Manders, and S.D. Wagner. 2001. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science. 293:248–250. [DOI] [PubMed] [Google Scholar]
- 22.Shvarts, A., T.R. Brummelkamp, F. Scheeren, E. Koh, G.Q. Daley, H. Spits, and R. Bernards. 2002. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 16:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyama, H., S. Okada, M. Hatano, Y. Takahashi, N. Takeda, H. Ichii, T. Takemori, Y. Kuroda, and T. Tokuhisa. 2002. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity. 17:329–339. [DOI] [PubMed] [Google Scholar]
- 24.Cattoretti, G., C.C. Chang, K. Cechova, J. Zhang, B.H. Ye, B. Falini, D.C. Louie, K. Offit, R.S. Chaganti, and R. Dalla-Favera. 1995. BCL-6 protein is expressed in germinal-center B cells. Blood. 86:45–53. [PubMed] [Google Scholar]
- 25.Allman, D., A. Jain, A. Dent, R.R. Maile, T. Selvaggi, M.R. Kehry, and L.M. Staudt. 1996. BCL-6 expression during B-cell activation. Blood. 87:5257–5268. [PubMed] [Google Scholar]
- 26.Cattoretti, G., M. Buttner, R. Shaknovich, E. Kremmer, B. Alobeid, and G. Niedobitek. 2006. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood. 107:3967–3975. [DOI] [PubMed] [Google Scholar]
- 27.Choe, J., H.S. Kim, R.J. Armitage, and Y.S. Choi. 1997. The functional role of B cell antigen receptor stimulation and IL-4 in the generation of human memory B cells from germinal center B cells. J. Immunol. 159:3757–3766. [PubMed] [Google Scholar]
- 28.Choe, J., and Y.S. Choi. 1998. IL-10 interrupts memory B cell expansion in the germinal center by inducing differentiation into plasma cells. Eur. J. Immunol. 28:508–515. [DOI] [PubMed] [Google Scholar]
- 29.Arpin, C., J. Dechanet, C. Van Kooten, P. Merville, G. Grouard, F. Briere, J. Banchereau, and Y.J. Liu. 1995. Generation of memory B cells and plasma cells in vitro. Science. 268:720–722. [DOI] [PubMed] [Google Scholar]
- 30.Kim, H.S., X. Zhang, E. Klyushnenkova, and Y.S. Choi. 1995. Stimulation of germinal center B lymphocyte proliferation by an FDC-like cell line, HK. J. Immunol. 155:1101–1109. [PubMed] [Google Scholar]
- 31.Choi, Y.S. 1997. Differentiation and apoptosis of human germinal center B-lymphocytes. Immunol. Res. 16:161–174. [DOI] [PubMed] [Google Scholar]
- 32.Pascual, V., Y.J. Liu, A. Magalski, O. de Bouteiller, J. Banchereau, and J.D. Capra. 1994. Analysis of somatic mutation in five B cell subsets of human tonsil. J. Exp. Med. 180:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagresle, C., C. Bella, and T. Defrance. 1993. Phenotypic and functional heterogeneity of the IgD- B cell compartment: identification of two major tonsillar B cell subsets. Int. Immunol. 5:1259–1268. [DOI] [PubMed] [Google Scholar]
- 34.Choe, J., H.S. Kim, X. Zhang, R.J. Armitage, and Y.S. Choi. 1996. Cellular and molecular factors that regulate the differentiation and apoptosis of germinal center B cells. Anti-Ig down-regulates Fas expression of CD40 ligand-stimulated germinal center B cells and inhibits Fas-mediated apoptosis. J. Immunol. 157:1006–1016. [PubMed] [Google Scholar]
- 35.Tangye, S.G., D.T. Avery, and P.D. Hodgkin. 2003. A division-linked mechanism for the rapid generation of Ig-secreting cells from human memory B cells. J. Immunol. 170:261–269. [DOI] [PubMed] [Google Scholar]
- 36.Tangye, S.G., D.T. Avery, E.K. Deenick, and P.D. Hodgkin. 2003. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J. Immunol. 170:686–694. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro-Shelef, M., K.I. Lin, L.J. McHeyzer-Williams, J. Liao, M.G. McHeyzer-Williams, and K. Calame. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 19:607–620. [DOI] [PubMed] [Google Scholar]
- 38.Reimold, A.M., N.N. Iwakoshi, J. Manis, P. Vallabhajosyula, E. Szomolanyi-Tsuda, E.M. Gravallese, D. Friend, M.J. Grusby, F. Alt, and L.H. Glimcher. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 412:300–307. [DOI] [PubMed] [Google Scholar]
- 39.Mittrucker, H.W., T. Matsuyama, A. Grossman, T.M. Kundig, J. Potter, A. Shahinian, A. Wakeham, B. Patterson, P.S. Ohashi, and T.W. Mak. 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 275:540–543. [DOI] [PubMed] [Google Scholar]
- 40.Sciammas, R., A.L. Shaffer, J.H. Schatz, H. Zhao, L.M. Staudt, and H. Singh. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236. [DOI] [PubMed] [Google Scholar]
- 41.Horcher, M., A. Souabni, and M. Busslinger. 2001. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 14:779–790. [DOI] [PubMed] [Google Scholar]
- 42.Muto, A., S. Tashiro, O. Nakajima, H. Hoshino, S. Takahashi, E. Sakoda, D. Ikebe, M. Yamamoto, and K. Igarashi. 2004. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 429:566–571. [DOI] [PubMed] [Google Scholar]
- 43.Klein, U., S. Casola, G. Cattoretti, Q. Shen, M. Lia, T. Mo, T. Ludwig, K. Rajewsky, and R. Dalla-Favera. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782. [DOI] [PubMed] [Google Scholar]
- 44.Niu, H. 2002. The proto-oncogene BCL-6 in normal and malignant B cell development. Hematol. Oncol. 20:155–166. [DOI] [PubMed] [Google Scholar]
- 45.Niu, H., B.H. Ye, and R. Dalla-Favera. 1998. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 12:1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaffer, A.L., X. Yu, Y. He, J. Boldrick, E.P. Chan, and L.M. Staudt. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 13:199–212. [DOI] [PubMed] [Google Scholar]
- 47.Kusam, S., F.H. Vasanwala, and A.L. Dent. 2004. Transcriptional repressor BCL-6 immortalizes germinal center-like B cells in the absence of p53 function. Oncogene. 23:839–844. [DOI] [PubMed] [Google Scholar]
- 48.Logarajah, S., P. Hunter, M. Kraman, D. Steele, S. Lakhani, L. Bobrow, A. Venkitaraman, and S. Wagner. 2003. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene. 22:5572–5578. [DOI] [PubMed] [Google Scholar]
- 49.Sciammas, R., and M.M. Davis. 2004. Modular nature of Blimp-1 in the regulation of gene expression during B cell maturation. J. Immunol. 172:5427–5440. [DOI] [PubMed] [Google Scholar]
- 50.Shaffer, A.L., M. Shapiro-Shelef, N.N. Iwakoshi, A.H. Lee, S.B. Qian, H. Zhao, X. Yu, L. Yang, B.K. Tan, A. Rosenwald, et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. [DOI] [PubMed] [Google Scholar]
- 51.Marsden, V.S., and A. Strasser. 2003. Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annu. Rev. Immunol. 21:71–105. [DOI] [PubMed] [Google Scholar]
- 52.Su, H., N. Bidere, L. Zheng, A. Cubre, K. Sakai, J. Dale, L. Salmena, R. Hakem, S. Straus, and M. Lenardo. 2005. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science. 307:1465–1468. [DOI] [PubMed] [Google Scholar]
- 53.Alizadeh, A.A., M.B. Eisen, R.E. Davis, C. Ma, I.S. Lossos, A. Rosenwald, J.C. Boldrick, H. Sabet, T. Tran, X. Yu, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403:503–511. [DOI] [PubMed] [Google Scholar]
- 54.Pound, J.D., and J. Gordon. 1997. Maintenance of human germinal center B cells in vitro. Blood. 89:919–928. [PubMed] [Google Scholar]
- 55.Li, L., D. Young, S.F. Wolf, and Y.S. Choi. 1996. Interleukin-12 stimulates B cell growth by inducing IFN-gamma. Cell. Immunol. 168:133–140. [DOI] [PubMed] [Google Scholar]
- 56.Banchereau, J., F. Briere, Y.J. Liu, and F. Rousset. 1994. Molecular control of B lymphocyte growth and differentiation. Stem Cells. 12:278–288. [DOI] [PubMed] [Google Scholar]
- 57.Luckey, C.J., D. Bhattacharya, A.W. Goldrath, I.L. Weissman, C. Benoist, and D. Mathis. 2006. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 103:3304–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao, Z., A. Bonni, F. Xia, M. Nadal-Vicens, and M.E. Greenberg. 1999. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 286:785–790. [DOI] [PubMed] [Google Scholar]
- 59.Gaudilliere, B., Y. Shi, and A. Bonni. 2002. RNA interference reveals a requirement for myocyte enhancer factor 2A in activity-dependent neuronal survival. J. Biol. Chem. 277:46442–46446. [DOI] [PubMed] [Google Scholar]
- 60.Reya, T., and H. Clevers. 2005. Wnt signalling in stem cells and cancer. Nature. 434:843–850. [DOI] [PubMed] [Google Scholar]
- 61.Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi, Y., P.R. Dutta, D.M. Cerasoli, and G. Kelsoe. 1998. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. J. Exp. Med. 187:885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furukawa, K., A. Akasako-Furukawa, H. Shirai, H. Nakamura, and T. Azuma. 1999. Junctional amino acids determine the maturation pathway of an antibody. Immunity. 11:329–338. [DOI] [PubMed] [Google Scholar]
- 64.Phan, R.T., M. Saito, K. Basso, H. Niu, and R. Dalla-Favera. 2005. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat. Immunol. 6:1054–1060. [DOI] [PubMed] [Google Scholar]
- 65.Beyaert, R., K. Heyninck, and S. Van Huffel. 2000. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem. Pharmacol. 60:1143–1151. [DOI] [PubMed] [Google Scholar]
- 66.Ohkubo, Y., M. Arima, E. Arguni, S. Okada, K. Yamashita, S. Asari, S. Obata, A. Sakamoto, M. Hatano, J. O-Wang, M. Ebara, H. Saisho, and T. Tokuhisa. 2005. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J. Immunol. 174:7703–7710. [DOI] [PubMed] [Google Scholar]
- 67.Tourigny, M.R., J. Ursini-Siegel, H. Lee, K.M. Toellner, A.F. Cunningham, D.S. Franklin, S. Ely, M. Chen, X.F. Qin, Y. Xiong, et al. 2002. CDK inhibitor p18(INK4c) is required for the generation of functional plasma cells. Immunity. 17:179–189. [DOI] [PubMed] [Google Scholar]
- 68.Kuhn, R., K. Rajewsky, and W. Muller. 1991. Generation and analysis of interleukin-4 deficient mice. Science. 254:707–710. [DOI] [PubMed] [Google Scholar]
- 69.Jiang, H., M.B. Harris, and P. Rothman. 2000. IL-4/IL-13 signaling beyond JAK/STAT. J. Allergy Clin. Immunol. 105:1063–1070. [DOI] [PubMed] [Google Scholar]
- 70.Hajoui, O., R. Janani, M. Tulic, P. Joubert, T. Ronis, Q. Hamid, H. Zheng, and B.D. Mazer. 2004. Synthesis of IL-13 by human B lymphocytes: regulation and role in IgE production. J. Allergy Clin. Immunol. 114:657–663. [DOI] [PubMed] [Google Scholar]
- 71.Nera, K.P., P. Kohonen, E. Narvi, A. Peippo, L. Mustonen, P. Terho, K. Koskela, J.M. Buerstedde, and O. Lassila. 2006. Loss of Pax5 promotes plasma cell differentiation. Immunity. 24:283–293. [DOI] [PubMed] [Google Scholar]
- 72.Ozaki, K., R. Spolski, R. Ettinger, H.P. Kim, G. Wang, C.F. Qi, P. Hwu, D.J. Shaffer, S. Akilesh, D.C. Roopenian, et al. 2004. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 173:5361–5371. [DOI] [PubMed] [Google Scholar]
- 73.O'Connor, B.P., V.S. Raman, L.D. Erickson, W.J. Cook, L.K. Weaver, C. Ahonen, L.L. Lin, G.T. Mantchev, R.J. Bram, and R.J. Noelle. 2004. BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med. 199:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ng, L.G., A.P. Sutherland, R. Newton, F. Qian, T.G. Cachero, M.L. Scott, J.S. Thompson, J. Wheway, T. Chtanova, J. Groom, et al. 2004. B cell-activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. J. Immunol. 173:807–817. [DOI] [PubMed] [Google Scholar]
- 75.Flenghi, L., B.H. Ye, M. Fizzotti, B. Bigerna, G. Cattoretti, S. Venturi, R. Pacini, S. Pileri, F. Lo Coco, E. Pescarmona, et al. 1995. A specific monoclonal antibody (PG-B6) detects expression of the BCL-6 protein in germinal center B cells. Am. J. Pathol. 147:405–411. [PMC free article] [PubMed] [Google Scholar]
- 76.Onizuka, T., M. Moriyama, T. Yamochi, T. Kuroda, A. Kazama, N. Kanazawa, K. Sato, T. Kato, H. Ota, and S. Mori. 1995. BCL-6 gene product, a 92- to 98-kD nuclear phosphoprotein, is highly expressed in germinal center B cells and their neoplastic counterparts. Blood. 86:28–37. [PubMed] [Google Scholar]
- 77.Cattoretti, G., R. Shaknovich, P.M. Smith, H.M. Jack, V.V. Murty, and B. Alobeid. 2006. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J. Immunol. 177:6930–6939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.