Abstract
The long pentraxin (PTX) 3 is produced by macrophages and myeloid dendritic cells in response to Toll-like receptor agonists and represents a nonredundant component of humoral innate immunity against selected pathogens. We report that, unexpectedly, PTX3 is stored in specific granules and undergoes release in response to microbial recognition and inflammatory signals. Released PTX3 can partially localize in neutrophil extracellular traps formed by extruded DNA. Eosinophils and basophils do not contain preformed PTX3. PTX3-deficient neutrophils have defective microbial recognition and phagocytosis, and PTX3 is nonredundant for neutrophil-mediated resistance against Aspergillus fumigatus. Thus, neutrophils serve as a reservoir, ready for rapid release, of the long PTX3, a key component of humoral innate immunity with opsonic activity.
Innate immunity is the first line of defense against pathogens and plays a key role in the initiation, activation, and orientation of adaptive immunity. Innate immunity receptors, also called pattern recognition receptors (PRRs), recognize a few highly conserved structures, called pathogen-associated molecular patterns, expressed by microorganisms (1). PRRs are either cell associated (expressed intracellularly or on the cell surface) or present in body fluids. There are two functional classes of cell-associated PRRs: endocytic PRRs (i.e., scavenger receptors and mannose receptors) involved in microorganism binding and uptake; and signaling PRRs (members of the Toll-like receptor [TLR], nucleotide-binding oligomerization domain, and helicase families) involved in cell activation upon contact with pathogens (2). The humoral arm of the innate immunity includes soluble PRRs, such as collectins, ficolins, complement components, and pentraxins (PTXs) (3).
Members of the PTX superfamily are usually characterized by a pentameric structure and are highly conserved during evolution (3–6). This family is subdivided into two subclasses that depend on the length and structure of the molecules. The classical short PTXs C-reactive protein (CRP) and serum amyloid P component (SAP) are acute-phase proteins in humans and mice, respectively (7, 8), that are produced in the liver in response to inflammatory mediators, most prominently IL-6. CRP and SAP bind, in a calcium-dependent manner, different ligands and are involved in innate resistance to microbes and scavenging of cellular debris and extracellular matrix components (4, 7). Long PTXs are characterized by an unrelated N-terminal domain coupled to a PTX-like C-terminal domain (3, 6, 9). The prototypic long PTX3 is rapidly produced and released by diverse cell types, in particular by mononuclear phagocytes, DCs, and endothelial and epithelial cells in response to primary inflammatory signals (e.g., TLR engagement, TNFα, and IL-1β) (10–14). With high affinity, PTX3 binds the complement component C1q, the extracellular matrix TNF-inducible protein 6, and selected microorganisms (e.g., Aspergillus fumigatus, Pseudomonas aeruginosa, Salmonella typhimurium, Paracoccidioides brasiliensis, and zymosan) (15–18).
Recent studies in ptx3-deficient (PTX3−/−) mice have shown that this molecule performs complex nonredundant functions in vivo, ranging from the assembly of a hyaluronic acid–rich extracellular matrix to female fertility to innate immunity against diverse microorganisms (15, 16, 19, 20). PTX3 also binds apoptotic cells (21) and may contribute to editing recognition of apoptotic cells versus infectious nonself (22). In addition, there is evidence for a regulatory role of PTX3 in noninfectious inflammatory reactions (Latini, R., personal communication) (23). Outer membrane protein A (OmpA) is a conserved constituent of the outer membrane of Enterobacteriaceae. A search of moieties recognized by PTX3 has discovered that it binds OmpA from Klebsiella pneumoniae and that it represents a nonredundant humoral amplification loop of the innate immunity response to this microbial moiety (24).
Among innate cells, neutrophils play an important role because of their ability to be rapidly recruited in tissues during infections and to produce mediators that kill or inhibit microbial growth (25–27). As PTX3 is important in infectious and inflammatory responses (3, 28), we evaluated whether neutrophils produce PTX3. In previous studies, PTX3 was found to be expressed in HL60 cells (11) and bone marrow myelocytes (29) as mRNA and, by proteomic analysis, in neutrophil granules (30). In this paper, we report that PTX3 is stored in a ready-made form in neutrophils but not in eosinophils and basophils. PTX3 is localized in specific granules and is secreted in response to recognition of microbial moieties and inflammatory signals. PTX3 can localize in neutrophil extracellular traps (NETs) (31), and PTX3-deficient neutrophils have defective phagocytic activity. In addition, injection of wild-type neutrophils restores protective immunity against A. fumigatus in PTX3−/− mice. Thus, neutrophils serve as a reservoir, ready for rapid release, of a key component of humoral innate immunity and complement its subsequent delayed neosynthesis by macrophages and DCs.
RESULTS
Storage of preformed PTX3 in resting neutrophils
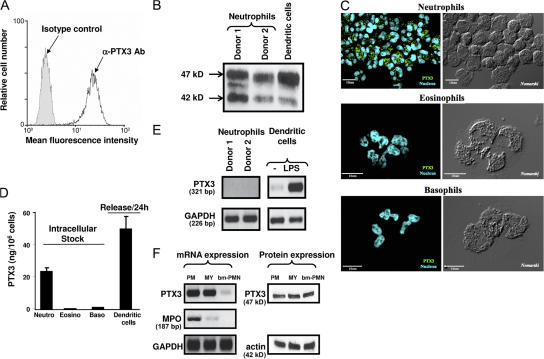
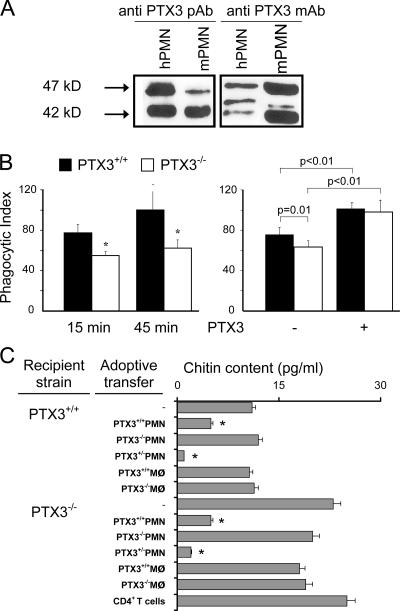
FACS analysis revealed constitutive expression of PTX3 in freshly isolated human neutrophils, as assessed by intracellular labeling (mean fluorescence intensity = 89 ± 34 and 4 ± 1.5, using anti-PTX3 and control mAbs, respectively; mean ± SD; n = 8; Fig. 1 A). No expression of PTX3 was observed on the surface of human neutrophils (not depicted). Recent studies reported that nonspecific binding of polyclonal Ig within neutrophils may give false positive results, as observed using antigranzyme antibody (32–34). Following the methodologies described in these studies, we observed similar levels of intracellular PTX3 using 20 and 200 μg/ml of human IgG for saturation, showing that the detection of intracellular PTX3 in neutrophils is not related to nonspecific binding of the anti-PTX3 mAb (not depicted). To confirm this observation, PTX3 expression was analyzed by Western blotting. Human neutrophils and DCs express two or three immunoreactive forms of PTX3, depending on the donor (with one major band at 47 kD and two minor bands at 44 and 42 kD; Fig. 1 B). The presence of intracellular PTX3 in neutrophils was also confirmed by confocal microscopy (Fig. 1 C). We assessed whether other circulating elements containing granules store PTX3. PTX3 could not be detected in eosinophils and basophils (Fig. 1 C), nor in large granular lymphocytes (NK cells; not depicted).
Figure 1.
PTX3 is constitutively expressed in human neutrophils. (A) FACS analysis of PTX3 expression in permeabilized human neutrophils isolated from peripheral blood. (B) Western blot analysis of PTX3 expression in neutrophils and LPS-stimulated DCs. (C) Analysis of PTX3 expression in freshly isolated neutrophils, eosinophils and basophils by confocal microscopy. Fluorescence (left) and differential interference contrasts (right, Nomarski technique) are shown. Bars, 10 μm. (D) Analysis of PTX3 content in freshly isolated neutrophils, eosinophils, and basophils, as well as release by LPS-stimulated DCs, determined by ELISA (mean ± SD). (E) Analysis of PTX3 mRNA expression in freshly isolated human neutrophils by RT-PCR. Results obtained in 2 out of 10 subjects tested are presented. LPS-stimulated DCs are used as a positive control. RNA integrity and cDNA synthesis were verified by amplifying GAPDH cDNA. (F) Analysis of PTX3 mRNA and protein in neutrophil precursors. Promyelocytes (PM), myelocytes/metamyelocytes (MY), and bone marrow–segmented neutrophils (bm-PMN) were analyzed for PTX3 and MPO mRNA expression (left). Expression of PTX3 was evaluated by Western blotting using the anti-PTX3 mAb 16B5 in the three populations of neutrophil precursors (right), and total protein loading was evaluated by analyzing actin expression.
PTX3 was detected in LPS-activated DCs (Fig. 1 B) (10, 35). By ELISA, resting human neutrophils contain 24.9 ± 3.8 ng PTX3 per 106 cells (n = 4) corresponding to ∼0.55 pmol (Fig. 1 D), calculated based on the protomer molecular mass. Over a period of 24 h, DCs release ∼50.2 ± 8.1 ng PTX3 per 106 cells (n = 5; Fig. 1 D) (10). These results show that neutrophils contain considerable amounts of preformed PTX3. Finally, unlike PTX3, the short PTXs CRP and SAP could not be localized in neutrophils (not depicted).
In agreement with a previous study (11), PTX3 mRNA is not expressed in freshly isolated human neutrophils, as assessed by RT-PCR analysis (Fig. 1 E). As a positive control, PTX3 mRNA was evident in LPS-stimulated DCs (Fig. 1 E), as previously reported (10). We thus evaluated whether neutrophil precursors express PTX3 mRNA. Promyelocytes, myelocytes/metamyelocytes, and bone marrow–segmented neutrophils were isolated by density centrifugation on a Percoll gradient, followed by magnetic cell sorting (29, 36). RT-PCR analysis revealed that PTX3 mRNA is expressed in promyelocytes and myelocytes/metamyelocytes but not or at low levels in bone marrow–segmented neutrophils (Fig. 1 F, left), a result in accordance with the absence of PTX3 mRNA expression in mature peripheral neutrophils. As a positive control, myeloperoxidase (MPO) mRNA was mainly detected in promyelocytes and, at a lower level, in myelocytes but not in bone marrow–segmented neutrophils, as previously reported (29, 36), confirming the purity of the isolated cell populations. Western blot analysis showed that PTX3 is expressed in the three populations of neutrophil precursors (Fig. 1 F, right). In agreement with previous observations (11), the human promyelocytic cell line HL60 constitutively expresses PTX3 mRNA (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20061301/DC1). HL60 cells express mRNA encoding MPO, a marker of primary granules, but not mRNA encoding lactoferrin and matrix metalloproteinase 9 (MMP-9; Fig. S1), as previously reported (37, 38). Moreover, HL60 cells spontaneously produce PTX3 protein (Fig. S1).
Localization of PTX3 in neutrophil-specific granules
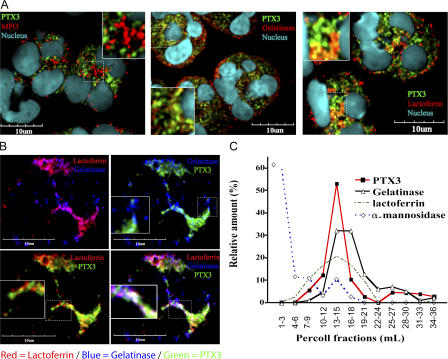
Confocal microscopy and subcellular fractionation of neutrophil-derived nitrogen cavitates were used to specifically localize PTX3 within freshly isolated neutrophils. As shown in Fig. 2 A, confocal studies pointed to complete colocalization of PTX3 with lactoferrin, a constituent of specific granules, and, at least in part, with gelatinase (tertiary granules; Fig. 2 A). In contrast, no colocalization with MPO, a marker of azurophilic granules, was observed (Fig. 2 A). Confocal microscopy was combined with quantitative analysis to measure the percentage of PTX3 colocalization, as assessed by Pearson's coefficient of correlation. Higher degrees of colocalization were measured in different experiments (n = 5) in lactoferrin+ and gelatinase+ vesicles, which display coefficients of 72.22 ± 7.32% (r = 0.78) and 27.8 ± 8.73% (r = 0.45), respectively. Little or no colocalization of PTX3 was found with MPO+ granules (5.90 ± 2.21%; r = 0.08).
Figure 2.
Localization of PTX3 within neutrophil granules. (A) Localization of PTX3 in freshly isolated neutrophils by confocal microscopy. Cells were fixed and stained for human PTX3 and MPO (left), PTX3 and gelatinase (middle), and PTX3 and lactoferrin (right; see Materials and methods). DNA labeling is also shown (Hoechst 33258). Insets show enlargements of the indicated areas. Bars, 10 μm. (B) Localization of PTX3 in neutrophil-specific granules. Cells were stained for PTX3, gelatinase, and lactoferrin (see Materials and methods). A representative cell is shown. Specific granules were identified as lactoferrin+ and lactoferrin+/gelatinase+; tertiary granules were identified as gelatinase+. Insets show enlargements of the indicated areas. Bars, 10 μm. (C) Analysis of PTX3 content in neutrophil subcellular fractions. PTX3, gelatinase, lactoferrin, and α-mannosidase content in each fraction were determined by ELISA or using a functional assay for α-mannosidase. Results are expressed as a percentage of the relative amount in the collected fractions.
Among the MPO-negative granules, ∼16% contain only lactoferrin (specific) and 24% contain only gelatinase (tertiary), whereas 60% contain both markers (specific) (39). Fig. 2 B shows lactoferrin+ granules, gelatinase+ granules, or double- stained granules in a representative cell. PTX3 was found to colocalize with lactoferrin+ granules and double-stained granules (specific), but no colocalization was found with tertiary granules that were only gelatinase+.
In an effort to confirm these observations, subcellular fractionation of neutrophil-derived nitrogen cavitates was performed. PTX3 was detected in fractions that contain lactoferrin and part of the gelatinase distribution but not in fractions containing α-mannosidase (azurophilic granules; Fig. 2 C) or albumin (secretory vesicles/light membranes; not depicted) (40). Collectively, these results demonstrate the selective association of PTX3 with specific (lactoferrin+ and lactoferrin/gelatinase+) granules.
PTX3 release by stimulated neutrophils
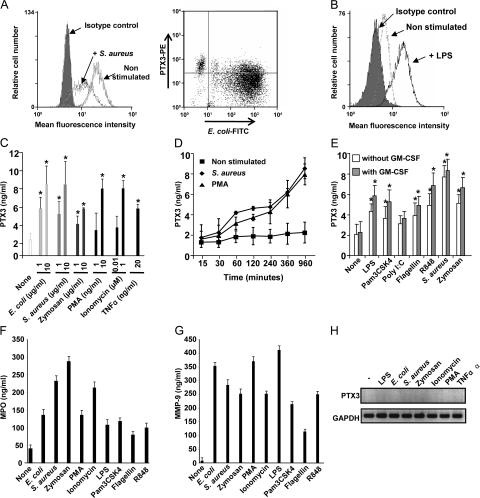
We next evaluated whether neutrophils release PTX3 upon stimulation. FACS analysis showed that the relative intracellular level of PTX3 decreased upon 2 h of stimulation with 10 μg/ml Staphylococcus aureus and Escherichia coli (Fig. 3 A). In contrast, the intracellular level of PTX3 increased in DCs stimulated for 8 h with 80 ng/ml LPS compared with nonstimulated cells (Fig. 3 B). The levels of PTX3 were also quantified by ELISA in the cell culture supernatants. E. coli, S. aureus, and, to a lesser extent, zymosan increased the release of PTX3 by neutrophils in a dose-dependent manner, with a maximal effect observed with the highest concentration used (10 μg/ml; Fig. 3 C). A similar increase of PTX3 was observed using 1 or 10 ng/ml PMA, 0.01 or 1 μM ionomycin, and 20 ng/ml of the proinflammatory cytokine TNFα (Fig. 3 C). 20 ng/ml IL-1β was inactive (not depicted). Propidium iodide staining and lactate dehydrogenase release assay excluded that PTX3 secretion resulted from cell death (not depicted). Latex beads failed to trigger PTX3 release (not depicted). The release of PTX3 is associated with a decrease in the level of intracellular PTX3 in neutrophils, as assessed by Western blot analysis (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20061301/DC1). PMA and S. aureus induced a time-dependent release of PTX3 by neutrophils, significant at 1 h and maximal at the latest time analyzed (16 h; Fig. 3 D). It is noteworthy that TNF and PMA induce selective release of specific granules (41, 42). Among the different activators tested, microorganisms appear to be the most potent inducers of PTX3 release by human neutrophils. We therefore evaluated the ability of TLR agonists to induce PTX3 release. LPS, R848, Pam3CSK4, and, to a lesser extent, flagellin (but not poly (I:C)) induced the release of PTX3 by neutrophils (Fig. 3 E). In agreement with previous studies (43–45), we observed that a stimulation with TLR agonists delays neutrophil apoptosis, confirming that the release of PTX3 was not a consequence of cell death (not depicted). Previous experiments reported that GM-CSF increases the sensitivity of neutrophils to TLR agonists (46). Neutrophils were thus primed with 50 ng/ml GM-CSF for 2 h before stimulation with zymosan, S. aureus, and TLR agonists. The levels of PTX3 released upon stimulation with TLR agonists or microorganisms were weakly but significantly increased in GM-CSF–primed neutrophils (Fig. 3 E). In addition, microorganisms and TLR agonists induced the release of MPO (Fig. 3 F) and MMP-9 (Fig. 3 G). Independent of the stimulus used, PTX3 mRNA was not induced in stimulated neutrophils, as assessed by RT-PCR analysis after 8 h (Fig. 3 H) or 16 h (not depicted) of activation. These data show that microorganisms and TLR agonists trigger substantial release of PTX3 by human neutrophils.
Figure 3.
Secretion of PTX3 by activated neutrophils. (A and B) FACS analysis of intracellular PTX3 expression in neutrophils stimulated for 2 h with 10 μg/ml S. aureus or 10 μg/ml FITC-labeled E. coli (A) or in DCs stimulated or not for 8 h with 80 ng/ml LPS (B). Representative results from one to five experiments are shown. (C–E) Analysis of PTX3 release upon neutrophil stimulation. (C) 2 × 106 cells/ml of neutrophils were activated for 16 h with 1 or 10 μg/ml E. coli, S. aureus, or zymosan; 1 or 10 ng/ml PMA; 0.01 or 1 μM ionomycin; or 20 ng/ml TNFα. (D) Time-dependent release of PTX3 in neutrophils (nonstimulated or stimulated with 10 μg/ml S. aureus or 10 ng/ml PMA) is shown. Supernatants were collected at the indicated time points. (E) Induction of PTX3 release by TLR agonists in neutrophils pretreated or not with 50 ng/ml GM-CSF. Supernatants were collected at 16 h. PTX3 was quantified in the supernatants by ELISA. Results are expressed as ng/ml (mean ± SD; n = 6). MPO (F) and MMP-9 (G) were quantified in the supernatants of neutrophils stimulated for 16 h with the indicated stimuli; results are expressed in ng/ml, showing the mean of two representative experiments. (H) PTX3 mRNA expression analyzed by RT-PCR in neutrophils untreated or stimulated for 8 h with 100 ng/ml LPS, 10 μg/ml E. coli, 10 μg/ml S. aureus, 10 μg/ml zymosan, 20 ng/ml TNFα, 10 ng/ml PMA, or 1 μM iononmycin. RNA integrity and cDNA synthesis were verified by amplifying GAPDH cDNA. *, P < 0.01 for untreated versus GM-CSF–treated cells.
Association of PTX3 with NETs
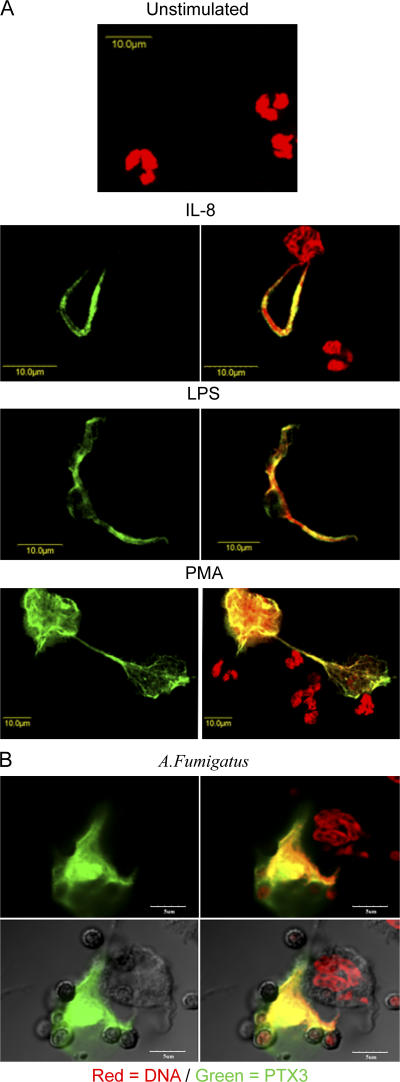
NETs are extracellular structures formed by the extrusion of DNA from viable neutrophils upon stimulation, and they act as focal points to focus antimicrobial effector molecules (31). We analyzed whether PTX3 colocalizes with NETs upon stimulation. As shown in Fig. 4, after 40 min of stimulation with IL-8, LPS, or PMA (Fig. 4 A) or the conidia of A. fumigatus (Fig. 4 B), PTX3 was found associated with NETs. PTX3 binds conidia from A. fumigatus and plays a nonredundant role in resistance against this fungus (15). Interestingly, both PTX3 and conidia were found associated to NETs (Fig. 4 B). Thus, PTX3 is rapidly released by neutrophils (Fig. 4 A and Fig. 3 D) and localizes in NETs.
Figure 4.
PTX3 is localized in NETs. (A) Neutrophils were exposed to 100 ng/ml IL-8, 100 ng/ml LPS, or 2.5 ng/ml PMA for 40 min. (left) PTX3 staining. (right) PTX3 and DNA staining. Bars, 10 μm. (B) Neutrophils exposed to conidia from A. fumigatus; a differential interference contrast (Nomarski technique) is shown in the bottom panels. In both A and B, PTX3 immunostaining was done on nonpermeabilized neutrophils. Bars, 5μm.
Characterization of neutrophil PTX3
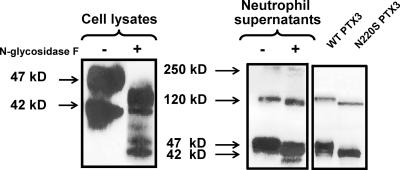
As mentioned above, Western blot analysis revealed two or three immunoreactive PTX3 isoforms in human neutrophils, depending on the donor tested (Fig. 1 B and Fig. 5, left). Three immunoreactive bands were also evident in the supernatants of activated neutrophils (Fig. 5, right), with molecular masses ranging from 47 to 250 kD. A previous study reported that PTX3 is glycosylated at the Asn 220 residue (18). A recent characterization of the PTX3 glycosidic moiety revealed three antennary structures and a role in the interaction with C1q (47). We thus analyzed the level of PTX3 glycosylation in neutrophils. Treatment of neutrophil cell lysates and culture supernatants with N-glycosidase F resulted in a decrease of the apparent molecular mass of PTX3 (Fig. 5). As expected on the basis of PTX3 synthesis during neutrophil differentiation but not in mature PMN, tunicamycin did not modify the Western blot profile in resting and activated neutrophils. In contrast, it prevented the glycosylation of newly synthesized PTX3 in LPS-activated DCs, resulting in a decrease of PTX3 release in the supernatant (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20061301), as previously reported (48). Moreover, an Asn→Ser substitution at position 220 was introduced in PTX3 (N220S mutant). Recombinant wild-type and N220S PTX3 were detected in the supernatants of Chinese hamster ovary (CHO) cells in a multimeric form (Fig. 5, right). Collectively, these data show that neutrophils contain a mature glycosylated form of PTX3 that associates in the extracellular milieu to form multimers independently of glycosylation.
Figure 5.
Biochemical analysis of PTX3 in neutrophils. Cell lysates from nonstimulated cells (left) and supernatants from S. aureus–stimulated neutrophils (right) were either untreated (−) or treated with N-glycosidase F (+). Supernatants from CHO cells transfected with wild-type or N220S mutant PTX3 were collected after a 24-h culture (right). PTX3 was analyzed by Western blotting using rabbit polyclonal anti-PTX3 and revealed by peroxidase-labeled anti–rabbit IgG antibody and ECL.
In vitro and in vivo relevance of PTX3 expression in neutrophils
We evaluated whether neutrophil-derived PTX3 is functional in recognizing ligands of self or microbial origin and may play a role in microbial recognition and destruction. First, we found that PTX3 purified by immunoaffinity from human PMN lysate bound to immobilized C1q and OmpA from K. pneumoniae; similarly, neutrophil-released PTX3, obtained by concentration of PMN supernatant, bound to A. fumigatus conidia, as did the recombinant protein (Fig. S4, A and B, available at http://www.jem.org/cgi/content/full/jem.20061301/DC1) (15, 18, 24).
Second, we studied neutrophil expression of PTX3 in the mouse. Mature mouse neutrophils constitutively express PTX3, as assessed by Western blotting using anti-PTX3 pAbs or mAbs (Fig. 6 A). Bone marrow–derived (two experiments; not depicted) and peritoneal (five experiments performed in two different laboratories; Fig. 6 B) neutrophils from PTX3−/− mice showed a significantly lower phagocytosis of conidia from A. fumigatus, with a 31 ± 7% reduction compared with PTX3-competent cells (Fig. 6 B, left). 1.1 μM of exogenous PTX3 caused a significant increase in the phagocytosis of conidia by PTX3-competent and incompetent neutrophils (phagocytic index = 102 and 76% for PTX3+/+ cells with and without PTX3, respectively; phagocytic index = 99 and 64% for PTX3−/− cells with and without PTX3, respectively). In the presence of exogenous PTX3, no difference was detected in the phagocytic activity of PTX3-competent and incompetent neutrophils (Fig. 6 B, right). Similarly, the conidicidal activity of PTX3−/− PMN was severely impaired compared with that of PTX3+/+ PMN (10 and 45%, respectively) and was increased 30% by the addition of recombinant PTX3 (not depicted).
Figure 6.
Functional role of neutrophil-derived PTX3. (A) Analysis of PTX3 expression in mouse bone marrow–segmented neutrophils from C57BL/6 mice by Western blotting. Results are representative of three independent experiments. A pAb or mAb (16B5) was used. (B, left) Phagocytosis of A. fumigatus conidia by peritoneal neutrophils from PTX3−/− or PTX3+/+ mice after 15 or 45 min. One representative experiment out of five performed is shown. *, P < 0.01. (B, right) Effect of 50 μg/ml of exogenous PTX3 in the phagocytosis of conidia by PTX3-competent and incompetent neutrophils (45 min of incubation). One representative experiment out of five performed is shown. (C) Role of PTX3 in neutrophil-mediated resistance against A. fumigatus. 108 A. fumigatus conidia per 20 μl were given intranasally to PTX3-deficient or -competent mice pretreated with cyclophosphamide. 3 h later, mice were given 106 neutrophils, macrophages, or CD4+ T cells i.v. from PTX3+/+, PTX3+/−, or PTX3−/− mice. Chitin content was measured as a correlation of fungal growth (mean ± SEM; n = 3). *, P < 0.05 using the Student's t test.
In an effort to assess the actual in vivo relevance of neutrophil-associated PTX3, cyclophosphamide-treated PTX3−/− and PTX3+/+ mice were infected with Aspergillus conidia and given PTX3−/−, PTX3+/+, or PTX3+/− PMN, macrophages, or purified CD4+T cells 3 h later. Mice were monitored for fungal growth 2 d later by quantifying the chitin content in the lung (49). Results showed that injection of PTX3+/+ and PTX3+/− neutrophils, but not PTX3−/− cells, in PTX3−/− mice decreased fungal growth (Fig. 6 C). No protective effect was observed using macrophages or CD4+ T cells. As a control, PTX3+/+ or PTX3+/− neutrophils also prevented fungal growth in PTX3+/+ mice, but the effect was less pronounced than in highly susceptible PTX3−/− mice (Fig. 6 C). No such effect was observed with PTX3−/− neutrophils or macrophages (Fig. 6 C).
DISCUSSION
The prototypic long PTX3 has long been known to be produced by diverse cell types on demand, i.e., in a gene expression–dependent fashion in response to extracellular signals (e.g., LPS, IL-1β, TNFα, and TLR agonists) (3). The finding that PTX3 is stored in neutrophil granules is therefore unexpected. PTX3 is not stored in MPO+ granules (primary or azurophilic). By confocal analysis among the MPO− granules, PTX3 was found to localize in lactoferrin+ and in lactoferrin+/gelatinase+ (specific) but not in gelatinase+ (tertiary) granules. Storage of PTX3 in neutrophil granules is selective, inasmuch as short PTXs are absent and other granulated circulating elements (eosinophils, basophils, and NK cells) do not contain preformed PTX3. In addition to the diversity generated during granulopoiesis, granules are secreted in a targeted manner, with a timing hierarchy in granule exocytosis (50, 51). PTX3 is localized in granules that are rapidly mobilized and secreted upon stimulation, in agreement with its early detection in the supernatants of stimulated neutrophils. Expression of PTX3 transcripts is confined to immature myeloid elements, and mature neutrophils only act as a reservoir of preformed PTX3. Thus, neutrophils represent a reservoir of this PRR and release it in response to microbial or inflammatory signals.
The release of PTX3 by neutrophils is induced by microorganisms and TLR agonists and, to a lower extent, by proinflammatory cytokines. Latex beads do not induce secretion of PTX3, suggesting that phagocytosis is not sufficient and that TLR recruitment is required to trigger its release by neutrophils. This dichotomy of PTX3-inducing mediators between neutrophils and other cell types is of physiological significance. Indeed, neutrophils are the first cells recruited into tissues in response to microorganism entry. Neutrophils are thus highly sensitive to microbes and microbe-derived components, suggesting that PTX3, rapidly released by infiltrating neutrophils, interacts with microorganisms and facilitates their internalization by neutrophils themselves, as well as resident and/or recruited professional APCs (19). Microorganisms and TLR agonists, the main inducers of PTX3 release, also induced the release of MPO and MMP-9, which are stored mainly in azurophilic and tertiary granules, respectively. These results suggest that microorganisms and TLR agonists induce the release of the molecules stored within the three types of granules.
Upon exposure to microbial or inflammatory signals, viable neutrophils extrude nuclear components that form an extracellular DNA fibrillary network. NETs trap microbes and retain neutrophil antimicrobial molecules (31). Therefore, NETs serve as a focal point to focus the action of antimicrobial molecules. In this paper, we report that part of exocytosed PTX3 can localize in NETs. Therefore, in addition to antimicrobial molecules, NETs can concentrate and focus the action of PTX3, a functional ancestor of antibodies.
The PTX3 protein is expressed in circulating PMN in the absence of transcripts, whereas NF-κB–driven PTX3 production (52) is induced in a variety of cells by microbial sensing and inflammatory cytokines (3). PTX3 mRNA was detected in promyelocytes and myelocytes/metamyelocytes but not in bone marrow–segmented neutrophils. In agreement with this result and a previous report (11), we observed that the promyelocytic cell line HL60 expresses PTX3 mRNA. A previous study reported that PTX3 is expressed only at the myelocyte stage (29), and on this basis localization of PTX3 in specific granules was postulated, a hypothesis confirmed by our results. Collectively, we can hypothesize that, in accordance with the targeting-by-timing hypothesis (the protein content into distinct granules is determined by the time of their biosynthesis), PTX3 mRNA is expressed at a late stage of promyelocyte differentiation and in myelocytes/metamyelocytes and that most PTX3 is synthesized at the myelocytes/metamyelocytes stage, a result in agreement with its preferential localization in secondary granules.
PTXs usually form multimers with a discoid arrangement of five subunits (3). PTX3 assembles as a decamer and can be produced as 10–20 subunit multimer proteins (18). PTX3 in neutrophil granules is mainly in the monomer form, and multimeric forms are detected in the supernatants of activated neutrophils. The formation of PTX3 multimers is not dependent on glycosylation (47). However, glycosylation appears crucial for the release of neo-synthetized PTX3, as observed in DCs (this study) and fibroblasts (48). These results show that human neutrophils contain a mature glycosylated form of PTX3 that assembles as multimers in the extracellular milieu.
Myeloid, but not plasmacytoid, DCs and macrophages are major producers of PTX3 (10). Over a period of 24 h, DCs release ∼50 ng of PTX3 per 106 cells (10). We report that neutrophils contain 24.9 ± 3.8 ng of this PRR per 106 cells (n = 5). Upon stimulation, they release ∼25% of stored PTX3, with a part of it remaining cell associated, presumably with NETs. Given the abundance of neutrophils in the circulation and in the early phases of inflammatory reactions in tissues, these cells represent a major source of PTX3 covering a temporal window preceding gene expression–dependent production. Under conditions of tissue damage (e.g., myocardial infarction) or infection (e.g., sepsis), PTX3 levels increase rapidly. For instance, in acute myocardial infarction with ST elevation, PTX3 reaches a peak in 6–8 h, compared with 36–48 h for CRP (53). Under these conditions, high PTX3 is an independent marker associated with death (54). The results reported in this paper shed new light on PTX3 elevations in pathological conditions and on their pathophysiological implications. It is likely that rapid release of stored PTX3 by activated neutrophils plays a role in the early phases of its elevation in pathology, preceding gene expression–dependent production.
Finally, we evaluated the in vivo relevance of PTX3 expression in neutrophils. PTX3-deficient mice are highly sensitive to A. fumigatus infection (15). Effector mechanisms of innate immunity are crucial to prevent aspergillosis (55), and among innate cells, neutrophils are essential in the initiation of the acute inflammatory response. Moreover, susceptibility to fungal infection can be associated, among other parameters, to neutropenia (56), and impairment of neutrophil antifungal responses results in an increase of fungal burden (57). These data underline the essential role played by neutrophils in controlling A. fumigatus infection. We report that PTX3 expressed by neutrophils is essential to control fungal growth in vitro and in vivo. Innate and adaptive immunity are both essential for the development of a protective antifungal immune response. Generation of a Th1-oriented A. fumigatus–specific immune response is associated with protection (58, 59). Injection of PTX3 in PTX3−/− mice favors the generation of a protective Th1 anti-Aspergillus immune response (15). Neutrophil-derived PTX3, in addition to DC-derived PTX3, may be involved in the orientation of the immune response toward a protective Th1 cell phenotype. Neutrophils, an innate cell type without professional antigen-presenting functions, may participate in the orientation of specific antimicrobial immune responses via the release of this preformed soluble PRR.
PTX3 is a long PTX conserved in evolution, with functional properties (e.g., complement activation and opsonization) that qualify it as a functional ancestor of antibodies. This fluid-phase PRR binds diverse microbial agents, activates the classic pathway of complement, and facilitates ingestion by innate immunity cells (3, 17, 24, 28), including neutrophils (Fig. 6 B). Gene-modified mice unequivocally indicate that PTX3 represents a nonredundant humoral amplification loop of the innate immune response to diverse microbial agents (3, 15, 19, 24). Neutrophils store a variety of constituents in their granules, including adhesion receptors (e.g., CD11b and CD18), proteolytic enzymes (e.g., cathepsin G), effectors and regulators of matrix degradation (e.g., gelatinase), and antimicrobial molecules (25–27). The results reported in this study broaden the repertoire of effector molecules stored and released by neutrophils to include a humoral PRR with functional properties of a predecessor of antibodies. PTX3-deficient neutrophils have defective recognition, phagocytosis, and killing of conidia, completely restored by PTX3, and this molecule is nonredundant for protection against A. fumigatus by neutrophils. Thus, PTX3 stored in neutrophil granules amplifies microbial recognition by neutrophils themselves as well as presumably by neighboring innate immunity cells.
MATERIALS AND METHODS
Leukocyte purification.
Monocytes were isolated and differentiated into DCs by a 5-d culture with 20 ng/ml IL-4 and 20 ng/ml GM-CSF (R&D Systems) (60). CD14−CD86− immature DCs were used. After Ficoll-Paque centrifugation, neutrophils were separated from erythrocytes by 3% dextran (GE Healthcare) density gradient sedimentation. Purity, determined by FACS analysis on forward scatter/side scatter parameters, was routinely >98%. Spontaneous activation of purified neutrophils was evaluated by analyzing CD11b and l-selectin expression by FACS; only l-selectin+CD11blow neutrophils were used. Neutrophils were also isolated from whole blood collected from healthy donors using a two-step buoyant density centrifugation on a Ficoll gradient (61).
Bone marrow cells (obtained from healthy donors) at different myeloid differentiation stages were isolated as previously described (36). In brief, bone marrow cells were separated by density sedimentation on a discontinuous Percoll gradient (GE Healthcare) of 1.065 g/ml and 1.08 g/ml. Three bands of cells, numbered in order of decreasing density, were harvested: band 1 primarily contained segmented neutrophils, band 2 primarily contained metamyelocytes and myelocytes, and band 3 contained promyelocytes (36). The three populations were subjected to immunomagnetic depletion of nongranulocytic cells, as previously described (29) using MACS (Miltenyi Biotec). The purity of cell populations was assessed by microscopy and cell surface phenotype (29). Eosinophils and basophils were enriched by depletion of magnetically labeled cells (MACS).
Blood and bone marrow samples were obtained with written informed consent in accordance with the Angers University Hospital ethical committee requirements.
Cell act ivation.
2 × 106 neutrophil cells/ml in RPMI 1640 (2% FCS) were either nonstimulated or stimulated with 1–10 μg/ml E. coli, S. aureus, or zymosan (all obtained from Invitrogen); 20 ng/ml IL-1β or TNFα (R&D Systems); 1–10 ng/ml PMA; 0.01–1 μM ionomycine; 100 ng/ml LPS (from E. coli serotype O55:B5; all purchased from Sigma-Aldrich); 500 ng/ml Pam3CSK4 (a TLR2 agonist); 5 μg/ml R848 (a TLR7/8 agonist), 2 μg/ml flagellin (a TLR5 agonist; all obtained from InvivoGen); and 10 μg/ml poly (I:C) (a TLR3 agonist; Sigma-Aldrich). 2 × 106 DCs/ml were stimulated for 8, 16, or 24 h with the stimuli indicated in the figures. PTX3 levels in supernatants were quantified by ELISA.
NET formation was induced as previously described (31). In brief, neutrophils (400 μl of cells at 2 × 106 cells/ml and 8 × 105 cells/well) were seeded on rounded glass coverslips treated with 1% poly-l-lysine (Sigma-Aldrich), allowed to settle, and treated for 40 min with 100 ng/ml IL-8 (PeproTech), 2.5 ng/ml PMA, 100 ng/ml LPS, and A. fumigatus conidia at a 1:5 ratio.
Cell lysates.
106 cells were lysed in 10 mM Tris-HCl, pH 7.6, 5 mM EDTA, 1% Triton X-100, or Nonidet P40 (Sigma-Aldrich) plus protease inhibitors (Roche Diagnostics). Lysates were centrifuged at 14,000 rpm for 15 min at 4°C to remove cellular debris. For N-glycosidase F treatment, SDS (0.2% final, vol/vol) was added to cell lysates or cell culture supernatants before heating at 100°C for 5 min. Lysates were treated with 5 U/ml N-glycosidase F (Roche Diagnostics) for 16 h at 37°C before analysis by Western blotting.
Site-directed mutagenesis.
The complete PTX3 cDNA was cloned into the pCDNA3.1+ vector (Invitrogen). The N220S substitution was introduced using a kit (QuickChange XL; Stratagene). CHO cells were transfected using Lipofectamine (Invitrogen).
Subcellular fractionation.
Subcellular fractionation of neutrophils was performed as previously described (62). In brief, fresh neutrophils were disrupted by nitrogen cavitation (Parr Instruments Co.), and the resulting cavitates were centrifuged to eliminate pellet nuclei and the remaining intact cells (63). The postnuclear supernatant containing cytosol, granules, and light membranes was immediately separated by centrifugation on a three-layer Percoll density gradient. 3-ml gradient fractions were collected after a sonication, frozen, and thawed for two times and analyzed for PTX3 content and for localization of subcellular organelles by marker assays (62). An α-mannosidase functional assay was used for azurophil granules (64); gelatinase, lactoferrin, and albumin ELISA were used to identify, respectively, gelatinase+ (tertiary) granules, lactoferrin+ (specific) granules, and secretory vesicles/light membranes (40).
FACS analysis.
PTX3 expression was analyzed on fixed (2% paraformaldehyde) and permeabilized (0.1% saponin; Sigma-Aldrich) cells with anti-PTX3 mAb (MNB4; Qbiogene) in PBS containing 0.01% saponin. Bound antibodies were revealed by PE-labeled anti–rat IgG mAb (BD Biosciences). Isotype control mAb was obtained from BD Biosciences. Fluorescence was analyzed using a cytofluorometer (FACScan; BD Biosciences), and results are expressed in mean fluorescence intensity values.
ELISA and Western blotting.
PTX3 (53), MPO (sensitivity = 0.4 ng/ml; HyCult Biotechnology), and total gelatinase (MMP-9, sensitivity = 30 pg/ml; R&D Systems) were quantified by ELISA. For Western blotting, proteins (corresponding to 0.4 × 106 cells or 5 μl of supernatants) were electrophoretically separated on a 10% polyacrylamide gel in reducing conditions and transferred to a membrane (Immobilon; Millipore). After saturation, membranes were incubated for 16 h at 4°C with 3 μg/ml anti-PTX3 pAbs or mAbs (16B5) and with 1 μg/ml peroxydase-labeled anti–rabbit IgG antibody or peroxydase-labeled anti–rat IgG antibody (Biosource International). Protein loading was verified with an anti-actin pAb (Sigma-Aldrich) revealed by the peroxydase-labeled anti–rabbit IgG antibody. Bound antibodies were detected using the ECL system (GE Healthcare).
Confocal microscopy.
Cytospins were fixed with 4% paraformaldehyde and permeabilized for 5 min with 0.2% Triton X-100 (Sigma-Aldrich) in PBS, pH 7.4, before incubation for 1 h at 4°C with 10% normal goat serum (Sigma-Aldrich) and then for 2 h at 4°C with 0.5 μg/ml biotin-conjugated rat anti–human PTX3 mAb (1 μg/ml; MNB4), or with IgG2a control mAb. Slides were incubated with streptavidin–Alexa Fluor 488 conjugate, followed by 1 μg/ml Hoechst 33258 or by 5 μg/ml propidium iodide and RNase (Invitrogen). The following reagents were also used: rabbit anti-MPO pAb (Invitrogen), mouse antilactoferrin mAb (clone NI25; Calbiochem), a rabbit antigelatinase pAb (Chemicon), and Alexa Fluor 647–conjugated goat anti–rabbit and anti–mouse IgG secondary antibodies. In each step, cells were washed with 0.2% BSA/0.05% Tween 20 in PBS, pH 7.4.
To stain NETs, neutrophils were fixed, blocked, and washed as described in the previous paragraph, without permeabilization. Cells were incubated with 1 μg/ml of biotin-conjugated PTX3 affinity-purified rabbit IgG, followed by streptavidin–Alexa Fluor 647 conjugate (Invitrogen). For DNA detection, Syto 13 (Invitrogen) plus RNase (Sigma-Aldrich) was used. Sections were mounted with a reagent (FluorSave; Calbiochem) and analyzed with a laser scanning confocal microscope (FluoView FV1000; Olympus). Images (1,024 × 1,024 pixels) were acquired with an oil immersion objective (100× 1.4 NA Plan-Apochromat; Olympus). Differential interference contrast (Nomarski technique) was also used. Overlay images were assembled, and ImarisColoc (version 4.2; Bitplane AG) software was used for quantitative colocalization and statistical analysis.
Analysis of PTX3 and MPO mRNA expression by RT-PCR.
Total RNA from DCs and peripheral blood neutrophils was extracted using TRIzol reagent (Life Technologies). Total RNA from neutrophil precursors and HL60 (American Type Culture Collection) was purified using the RNeasy mini kit (QIAGEN). cDNA was synthesized from 1 μg of total RNA using an oligo-dT primer and reverse transcriptase (GE Healthcare). PCR amplification was performed with an amount of cDNA corresponding to 25 ng of the starting total RNA using specific oligonucleotides (PTX3, 5′-GTGCAGGGCTGGGCTGCCCG-3′ and 5′-GCCGCACAGGTGGGTCCACC-3′; MPO, 5′-CCTTCATGTTCCGCCTGGACAATCG-3′ and 5′-CGGATCTCATCCACTGCAATTTGG-3′). RNA integrity was assessed by GAPDH cDNA amplification. The PCR products were analyzed on a 1% agarose gel by electrophoresis and visualized with ethidium bromide.
Mouse neutrophils and phagocytosis assay.
Neutrophils from C57BL/6 mice (Charles River Laboratories) were isolated from bone marrow by Percoll step gradient (52, 65, and 75% Percoll). An enriched neutrophil population was recovered at the 65–75% interface. Purity was analyzed by morphology, Giemsa staining, and FACS using anti-Ly6G (clone 1A8; BD Biosciences), and antineutrophil mAb (clone 7/4; Serotec) was routinely >96%. Cells were also isolated from wild-type and PTX3−/− total bone marrow (15, 65) or from the peritoneal cavity 4 h after a 1.5-ml 3% thioglycollate (Difco) injection and plated at 5 × 106 cells/ml in a final volume of 0.5 ml RPMI 1640 with human serum in a 24-well plate. A. fumigatus conidia were added at 2.5 × 107 cells/well. After 15 or 45 min of incubation at 37°C, phagocytosis was blocked by using NaF (final concentration = 0.2 M). In some of the experiments, 50 μg/ml of recombinant PTX3 was added. Cytospins were stained with Diff Quick (Dade, Biomap). At least 200 neutrophils per sample were counted under oil immersion microscopy (100× objective). Results are expressed as a phagocytic index: (percentage of neutrophils containing at least one conidia) × (mean number of conidia per positive cell).
Infection.
Mice were administered 150 mg/kg cyclophosphamide i.p. 2 d before the intranasal infection with 108 A. fumigatus conidia per 20 μl (15). Mice received 106 cells per 500 μl of peritoneal neutrophils, splenic macrophages, or CD4+ T cells i.v. 3 h after the infection. Quantification of fungal growth in the lungs was done by the chitin assay (49), and results are expressed as micrograms of glucosamine per pair of lungs. Peritoneal neutrophils were obtained 18 h after the i.p. injection of 1 ml of endotoxin-free 10% thioglycolate solution (Difco). Endotoxin was depleted from all solutions with Detoxi-Gel (Pierce Chemical Co.). Purity was >98%, as determined by Cytospin and FACS analysis (GR-1+ and CD11b+). Macrophages were obtained by 2 h of plastic adherence of spleen cells at 37C°. CD4+ T cells were purified from spleens of mice using anti-CD4 magnetic MicroBeads (Miltenyi Biotec).
Procedures involving animals and their care conformed to institutional guidelines in compliance with national (4D.L. N.116, G.U., suppl. 40, 18-2-1992) and international (EEC Council Directive 86/609, OJ L 358,1,12-12-1987; National Institutes of Health Guide for the Care and Use of Laboratory Animals) law and policies. All efforts were made to minimize the number of animals used and their suffering.
Statistical analysis.
Statistical analysis was performed using the Student's t test.
Online supplemental material.
Supplemental materials and methods describes the binding assay and the analysis of PTX3, MPO, lactoferrin, and MMP-9 mRNA expression in HL60 cells and human bone marrow. Fig. S1 shows the constitutive expression of PTX3 (mRNA and protein) in the human promyelocytic cell line HL60. Fig. S2 shows the relative content of PTX3, as assessed by Western blotting, in neutrophils stimulated for 16 h with the indicated stimuli. Fig. S3 shows that the release of PTX3 by neutrophils is not affected by tunicamycin, in contrast to PTX3 produced by activated DCs. Fig. S4 shows PMN-released PTX3 binding to (A) immobilized C1q and OmpA and to (B) A. fumigatus conidia. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20061301/DC1.
Supplemental Material
Acknowledgments
We sincerely acknowledge Dr. Odile Blanchet and Ms. Irène Dobo for giving human bone marrow cells.
S. Jaillon is supported by a grant from the Conseil Général du Maine et Loire. This study is supported by the Institut National de la Santé et de la Recherche Médicale (Avenir program), the Ministère de la Recherche (ACI program), Cancéropôle Grand-Ouest, the Sixth Research Framework Programme of the European Union (projects MUGEN LSHB-CT-2005-005203 and MUVAPRED), the Ministero dell'Istruzione, Università e della Ricerca (project FIRB), Telethon (grant GGP05095), the Fondazione CARIPLO (project Nobel), and the Italian Association for Cancer Research.
G. Peri, A. Doni, C. Garlanda, and A. Mantovani are inventors in patent applications concerning PTX3. The authors have no other conflicting financial interests.
Abbreviations used: CHO, Chinese hamster ovary; CRP, C-reactive protein; MMP-9, matrix metalloproteinase 9; MPO, myeloperoxidase; NET, neutrophil extracellular trap; OmpA, outer membrane protein A; PRR, pattern recognition receptor; PTX, pentraxin; SAP, serum amyloid P component; TLR, Toll-like receptor.
S. Jaillon, G. Peri, P. Jeannin, and A. Mantovani contributed equally to this work.
References
- 1.Janeway, C.A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197–216. [DOI] [PubMed] [Google Scholar]
- 2.Gordon, S. 2002. Pattern recognition receptors: doubling up for the innate immune response. Cell. 111:927–930. [DOI] [PubMed] [Google Scholar]
- 3.Garlanda, C., B. Bottazzi, A. Bastone, and A. Mantovani. 2005. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu. Rev. Immunol. 23:337–366. [DOI] [PubMed] [Google Scholar]
- 4.Szalai, A.J., A. Agrawal, T.J. Greenhough, and J.E. Volanakis. 1999. C-reactive protein: structural biology and host defense function. Clin. Chem. Lab. Med. 37:265–270. [DOI] [PubMed] [Google Scholar]
- 5.Hirschfield, G.M., and M.B. Pepys. 2003. C-reactive protein and cardiovascular disease: new insights from an old molecule. QJM. 96:793–807. [DOI] [PubMed] [Google Scholar]
- 6.Bottazzi, B., A. Bastone, A. Doni, C. Garlanda, S. Valentino, L. Deban, V. Maina, A. Cotena, F. Moalli, L. Vago, et al. 2006. The long pentraxin PTX3 as a link among innate immunity, inflammation, and female fertility. J. Leukoc. Biol. 79:909–912. [DOI] [PubMed] [Google Scholar]
- 7.Pepys, M.B., and M.L. Baltz. 1983. Acute phase proteins with special reference to C-reactive protein and related proteins (pentaxis) and serum amyloid A protein. Adv. Immunol. 34:141–212. [DOI] [PubMed] [Google Scholar]
- 8.Baumann, H., and J. Gauldie. 1994. The acute phase response. Immunol. Today. 15:74–80. [DOI] [PubMed] [Google Scholar]
- 9.Goodman, A.R., T. Cardozo, R. Abagyan, A. Altmeyer, H.G. Wisniewski, and J. Vilcek. 1996. Long pentraxins: an emerging group of proteins with diverse functions. Cytokine Growth Factor Rev. 7:191–202. [DOI] [PubMed] [Google Scholar]
- 10.Doni, A., G. Peri, M. Chieppa, P. Allavena, F. Pasqualini, L. Vago, L. Romani, C. Garlanda, and A. Mantovani. 2003. Production of the soluble pattern recognition receptor PTX3 by myeloid, but not plasmacytoid, dendritic cells. Eur. J. Immunol. 33:2886–2893. [DOI] [PubMed] [Google Scholar]
- 11.Alles, V.V., B. Bottazzi, G. Peri, J. Golay, M. Introna, and A. Mantovani. 1994. Inducible expression of PTX3, a new member of the pentraxin family, in human mononuclear phagocytes. Blood. 84:3483–3493. [PubMed] [Google Scholar]
- 12.Breviario, F., E.M. d'Aniello, J. Golay, G. Peri, B. Bottazzi, A. Bairoch, S. Saccone, R. Marzella, V. Predazzi, M. Rocchi, et al. 1992. Interleukin-1-inducible genes in endothelial cells. Cloning of a new gene related to C-reactive protein and serum amyloid P component. J. Biol. Chem. 267:22190–22197. [PubMed] [Google Scholar]
- 13.Han, B., M. Mura, C.F. Andrade, D. Okutani, M. Lodyga, C.C. dos Santos, S. Keshavjee, M. Matthay, and M. Liu. 2005. TNFalpha- induced long pentraxin PTX3 expression in human lung epithelial cells via JNK. J. Immunol. 175:8303–8311. [DOI] [PubMed] [Google Scholar]
- 14.Vouret-Craviari, V., C. Matteucci, G. Peri, G. Poli, M. Introna, and A. Mantovani. 1997. Expression of a long pentraxin, PTX3, by monocytes exposed to the mycobacterial cell wall component lipoarabinomannan. Infect. Immun. 65:1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garlanda, C., E. Hirsch, S. Bozza, A. Salustri, M. De Acetis, R. Nota, A. Maccagno, F. Riva, B. Bottazzi, G. Peri, et al. 2002. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. 420:182–186. [DOI] [PubMed] [Google Scholar]
- 16.Salustri, A., C. Garlanda, E. Hirsch, M. De Acetis, A. Maccagno, B. Bottazzi, A. Doni, A. Bastone, G. Mantovani, P. Beck Peccoz, et al. 2004. PTX3 plays a key role in the organization of the cumulus oophorus extracellular matrix and in in vivo fertilization. Development. 131:1577–1586. [DOI] [PubMed] [Google Scholar]
- 17.Nauta, A.J., B. Bottazzi, A. Mantovani, G. Salvatori, U. Kishore, W.J. Schwaeble, A.R. Gingras, S. Tzima, F. Vivanco, J. Egido, et al. 2003. Biochemical and functional characterization of the interaction between pentraxin 3 and C1q. Eur. J. Immunol. 33:465–473. [DOI] [PubMed] [Google Scholar]
- 18.Bottazzi, B., V. Vouret-Craviari, A. Bastone, L. De Gioia, C. Matteucci, G. Peri, F. Spreafico, M. Pausa, C. Dettorre, E. Gianazza, et al. 1997. Multimer formation and ligand recognition by the long pentraxin PTX3. Similarities and differences with the short pentraxins C-reactive protein and serum amyloid P component. J. Biol. Chem. 272:32817–32823. [DOI] [PubMed] [Google Scholar]
- 19.Diniz, S.N., R. Nomizo, P.S. Cisalpino, M.M. Teixeira, G.D. Brown, A. Mantovani, S. Gordon, L.F. Reis, and A.A. Dias. 2004. PTX3 function as an opsonin for the dectin-1-dependent internalization of zymosan by macrophages. J. Leukoc. Biol. 75:649–656. [DOI] [PubMed] [Google Scholar]
- 20.Varani, S., J.A. Elvin, C. Yan, J. DeMayo, F.J. DeMayo, H.F. Horton, M.C. Byrne, and M.M. Matzuk. 2002. Knockout of pentraxin 3, a downstream target of growth differentiation factor-9, causes female subfertility. Mol. Endocrinol. 16:1154–1167. [DOI] [PubMed] [Google Scholar]
- 21.Rovere, P., G. Peri, F. Fazzini, B. Bottazzi, A. Doni, A. Bondanza, V.S. Zimmermann, C. Garlanda, U. Fascio, M.G. Sabbadini, et al. 2000. The long pentraxin PTX3 binds to apoptotic cells and regulates their clearance by antigen-presenting dendritic cells. Blood. 96:4300–4306. [PubMed] [Google Scholar]
- 22.Baruah, P., A. Propato, I.E. Dumitriu, P. Rovere-Querini, V. Russo, R. Fontana, D. Accapezzato, G. Peri, A. Mantovani, V. Barnaba, and A.A. Manfredi. 2006. The pattern recognition receptor PTX3 is recruited at the synapse between dying and dendritic cells, and edits the cross-presentation of self, viral, and tumor antigens. Blood. 107:151–158. [DOI] [PubMed] [Google Scholar]
- 23.Souza, D.G., A.C. Soares, V. Pinho, H. Torloni, L.F. Reis, M.M. Teixeira, and A.A. Dias. 2002. Increased mortality and inflammation in tumor necrosis factor-stimulated gene-14 transgenic mice after ischemia and reperfusion injury. Am. J. Pathol. 160:1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeannin, P., B. Bottazzi, M. Sironi, A. Doni, M. Rusnati, M. Presta, V. Maina, G. Magistrelli, J.F. Haeuw, G. Hoeffel, et al. 2005. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity. 22:551–560. [DOI] [PubMed] [Google Scholar]
- 25.Nathan, C. 2006. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 6:173–182. [DOI] [PubMed] [Google Scholar]
- 26.Cassatella, M.A., S. Gasperini, and M.P. Russo. 1997. Cytokine expression and release by neutrophils. Ann. N Y Acad. Sci. 832:233–242. [DOI] [PubMed] [Google Scholar]
- 27.Segal, A.W. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23:197–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottazzi, B., C. Garlanda, G. Salvatori, P. Jeannin, A. Manfredi, and A. Mantovani. 2006. Pentraxins as a key component of innate immunity. Curr. Opin. Immunol. 18:10–15. [DOI] [PubMed] [Google Scholar]
- 29.Theilgaard-Monch, K., L.C. Jacobsen, R. Borup, T. Rasmussen, M.D. Bjerregaard, F.C. Nielsen, J.B. Cowland, and N. Borregaard. 2005. The transcriptional program of terminal granulocytic differentiation. Blood. 105:1785–1796. [DOI] [PubMed] [Google Scholar]
- 30.Lominadze, G., D.W. Powell, G.C. Luerman, A.J. Link, R.A. Ward, and K.R. McLeish. 2005. Proteomic analysis of human neutrophil granules. Mol. Cell. Proteomics. 4:1503–1521. [DOI] [PubMed] [Google Scholar]
- 31.Brinkmann, V., U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D.S. Weiss, Y. Weinrauch, and A. Zychlinsky. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535. [DOI] [PubMed] [Google Scholar]
- 32.Grossman, W.J., and T.J. Ley. 2004. Granzymes A and B are not expressed in human neutrophils. Blood. 104:906–907. [DOI] [PubMed] [Google Scholar]
- 33.Metkar, S.S., and C.J. Froelich. 2004. Human neutrophils lack granzyme A, granzyme B, and perforin. Blood. 104:905–906. [DOI] [PubMed] [Google Scholar]
- 34.Wagner, C., C. Iking-Konert, B. Denefleh, S. Stegmaier, F. Hug, and G.M. Hansch. 2004. Granzyme B and perforin: constitutive expression in human polymorphonuclear neutrophils. Blood. 103:1099–1104. [DOI] [PubMed] [Google Scholar]
- 35.Doni, A., M. Michela, B. Bottazzi, G. Peri, S. Valentino, N. Polentarutti, C. Garlanda, and A. Mantovani. 2006. Regulation of PTX3, a key component of humoral innate immunity in human dendritic cells: stimulation by IL-10 and inhibition by IFN-gamma. J. Leukoc. Biol. 79:797–802. [DOI] [PubMed] [Google Scholar]
- 36.Cowland, J.B., and N. Borregaard. 1999. Isolation of neutrophil precursors from bone marrow for biochemical and transcriptional analysis. J. Immunol. Methods. 232:191–200. [DOI] [PubMed] [Google Scholar]
- 37.Devarajan, P., J.J. Johnston, S.S. Ginsberg, H.E. Van Wart, and N. Berliner. 1992. Structure and expression of neutrophil gelatinase cDNA. Identity with type IV collagenase from HT1080 cells. J. Biol. Chem. 267:25228–25232. [PubMed] [Google Scholar]
- 38.Johnston, J.J., P. Rintels, J. Chung, J. Sather, E.J. Benz Jr., and N. Berliner. 1992. Lactoferrin gene promoter: structural integrity and nonexpression in HL60 cells. Blood. 79:2998–3006. [PubMed] [Google Scholar]
- 39.Kjeldsen, L., D.F. Bainton, H. Sengelov, and N. Borregaard. 1993. Structural and functional heterogeneity among peroxidase-negative granules in human neutrophils: identification of a distinct gelatinase-containing granule subset by combined immunocytochemistry and subcellular fractionation. Blood. 82:3183–3191. [PubMed] [Google Scholar]
- 40.Cassatella, M.A., V. Huber, F. Calzetti, D. Margotto, N. Tamassia, G. Peri, A. Mantovani, L. Rivoltini, and C. Tecchio. 2006. Interferon-activated neutrophils store a TNF-related apoptosis-inducing ligand (TRAIL/Apo-2 ligand) intracellular pool that is readily mobilizable following exposure to proinflammatory mediators. J. Leukoc. Biol. 79:123–132. [DOI] [PubMed] [Google Scholar]
- 41.Dewald, B., U. Bretz, and M. Baggiolini. 1982. Release of gelatinase from a novel secretory compartment of human neutrophils. J. Clin. Invest. 70:518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mocsai, A., E. Ligeti, C.A. Lowell, and G. Berton. 1999. Adhesion-dependent degranulation of neutrophils requires the Src family kinases Fgr and Hck. J. Immunol. 162:1120–1126. [PubMed] [Google Scholar]
- 43.Colotta, F., F. Re, N. Polentarutti, S. Sozzani, and A. Mantovani. 1992. Modulation of granulocyte survival and programmed cell death by cytokines and bacterial products. Blood. 80:2012–2020. [PubMed] [Google Scholar]
- 44.Sabroe, I., R.C. Read, M.K. Whyte, D.H. Dockrell, S.N. Vogel, and S.K. Dower. 2003. Toll-like receptors in health and disease: complex questions remain. J. Immunol. 171:1630–1635. [DOI] [PubMed] [Google Scholar]
- 45.Francois, S., J. El Benna, P.M. Dang, E. Pedruzzi, M.A. Gougerot-Pocidalo, and C. Elbim. 2005. Inhibition of neutrophil apoptosis by TLR agonists in whole blood: involvement of the phosphoinositide 3-kinase/Akt and NF-kappaB signaling pathways, leading to increased levels of Mcl-1, A1, and phosphorylated Bad. J. Immunol. 174:3633–3642. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi, F., T.K. Means, and A.D. Luster. 2003. Toll-like receptors stimulate human neutrophil function. Blood. 102:2660–2669. [DOI] [PubMed] [Google Scholar]
- 47.Inforzato, A., G. Peri, A. Doni, C. Garlanda, A. Mantovani, A. Bastone, A. Carpentieri, A. Amoresano, P. Pucci, A. Roos, et al. 2006. Structure and function of the long pentraxin PTX3 glycosidic moiety: fine-tuning of the interaction with C1q and complement activation. Biochemistry. 45:11540–11551. [DOI] [PubMed] [Google Scholar]
- 48.Lee, G.W., A.R. Goodman, T.H. Lee, and J. Vilcek. 1994. Relationship of TSG-14 protein to the pentraxin family of major acute phase proteins. J. Immunol. 153:3700–3707. [PubMed] [Google Scholar]
- 49.Montagnoli, C., F. Fallarino, R. Gaziano, S. Bozza, S. Bellocchio, T. Zelante, W.P. Kurup, L. Pitzurra, P. Puccetti, and L. Romani. 2006. Immunity and tolerance to Aspergillus involve functionally distinct regulatory T cells and tryptophan catabolism. J. Immunol. 176:1712–1723. [DOI] [PubMed] [Google Scholar]
- 50.Sengelov, H., L. Kjeldsen, and N. Borregaard. 1993. Control of exocytosis in early neutrophil activation. J. Immunol. 150:1535–1543. [PubMed] [Google Scholar]
- 51.Sengelov, H., P. Follin, L. Kjeldsen, K. Lollike, C. Dahlgren, and N. Borregaard. 1995. Mobilization of granules and secretory vesicles during in vivo exudation of human neutrophils. J. Immunol. 154:4157–4165. [PubMed] [Google Scholar]
- 52.Basile, A., A. Sica, E. D'Aniello, F. Breviario, G. Garrido, M. Castellano, A. Mantovani, and M. Introna. 1997. Characterization of the promoter for the human long pentraxin PTX3. J. Biol. Chem. 272:8172–8178. [DOI] [PubMed] [Google Scholar]
- 53.Peri, G., M. Introna, D. Corradi, G. Iacuitti, S. Signorini, F. Avanzini, F. Pizzetti, A.P. Maggioni, T. Moccetti, M. Metra, et al. 2000. PTX3, a prototypic long pentraxin, is an early indicator of acute myocardial infarction in man. Circulation. 102:636–641. [DOI] [PubMed] [Google Scholar]
- 54.Latini, R., A.P. Maggioni, G. Peri, L. Gonzini, D. Lucci, P. Mocarelli, L. Vago, F. Pasqualini, S. Signorini, D. Soldateschi, et al. 2004. Prognostic significance of the long pentraxin PTX3 in acute myocardial infarction. Circulation. 110:2349–2354. [DOI] [PubMed] [Google Scholar]
- 55.Schaffner, A., H. Douglas, and A. Braude. 1982. Selective protection against conidia by mononuclear and against mycelia by polymorphonuclear phagocytes in resistance to Aspergillus. Observations on these two lines of defense in vivo and in vitro with human and mouse phagocytes. J. Clin. Invest. 69:617–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerson, S.L., G.H. Talbot, S. Hurwitz, B.L. Strom, E.J. Lusk, and P.A. Cassileth. 1984. Prolonged granulocytopenia: the major risk factor for invasive pulmonary aspergillosis in patients with acute leukemia. Ann. Intern. Med. 100:345–351. [DOI] [PubMed] [Google Scholar]
- 57.Romani, L., and D.H. Howard. 1995. Mechanisms of resistance to fungal infections. Curr. Opin. Immunol. 7:517–523. [DOI] [PubMed] [Google Scholar]
- 58.Cenci, E., S. Perito, K. Enssle, P. Mosci, J. Latge, L. Romani, and F. Bistoni. 1997. Th1 and Th2 cytokines in mice with invasive aspergillosis. Infect. Immun. 65:564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagai, H., J. Guo, H. Choi, and V. Kurup. 1995. Interferon-gamma and tumor necrosis factor-alpha protect mice from invasive aspergillosis. J. Infect. Dis. 172:1554–1560. [DOI] [PubMed] [Google Scholar]
- 60.Jeannin, P., T. Renno, L. Goetsch, I. Miconnet, J.P. Aubry, Y. Delneste, N. Herbault, T. Baussant, G. Magistrelli, C. Soulas, et al. 2000. OmpA targets dendritic cells, induces their maturation and delivers antigen into the MHC class I presentation pathway. Nat. Immunol. 1:502–509. [DOI] [PubMed] [Google Scholar]
- 61.Bourke, E., D. Bosisio, J. Golay, N. Polentarutti, and A. Mantovani. 2003. The Toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 102:956–963. [DOI] [PubMed] [Google Scholar]
- 62.Kjeldsen, L., H. Sengelov, and N. Borregaard. 1999. Subcellular fractionation of human neutrophils on Percoll density gradients. J. Immunol. Methods. 232:131–143. [DOI] [PubMed] [Google Scholar]
- 63.McDonald, P.P., C. Bovolenta, and M.A. Cassatella. 1998. Activation of distinct transcription factors in neutrophils by bacterial LPS, interferon-gamma, and GM-CSF and the necessity to overcome the action of endogenous proteases. Biochemistry. 37:13165–13173. [DOI] [PubMed] [Google Scholar]
- 64.De Togni, P., G. Cabrini, and F. Di Virgilio. 1984. Cyclic AMP inhibition of fMet-Leu-Phe-dependent metabolic responses in human neutrophils is not due to its effects on cytosolic Ca2+. Biochem. J. 224:629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pruijt, J.F., P. Verzaal, R. van Os, E.J. de Kruijf, M.L. van Schie, A. Mantovani, A. Vecchi, I.J. Lindley, R. Willemze, S. Starckx, et al. 2002. Neutrophils are indispensable for hematopoietic stem cell mobilization induced by interleukin-8 in mice. Proc. Natl. Acad. Sci. USA. 99:6228–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.