Abstract
Mouse cytomegalovirus (MCMV) susceptibility often results from defects of natural killer (NK) cell function. Here we describe Jinx, an N-ethyl-N-nitrosourea–induced MCMV susceptibility mutation that permits unchecked proliferation of the virus, causing death. In Jinx homozygotes, activated NK cells and cytotoxic T lymphocytes (CTLs) fail to degranulate, although they retain the ability to produce cytokines, and cytokine levels are markedly elevated in the blood of infected mutant mice. Jinx was mapped to mouse chromosome 11 on a total of 246 meioses and confined to a 4.60–million basepair critical region encompassing 122 annotated genes. The phenotype was ascribed to the creation of a novel donor splice site in Unc13d, the mouse orthologue of human MUNC13-4, in which mutations cause type 3 familial hemophagocytic lymphohistiocytosis (FHL3), a fatal disease marked by massive hepatosplenomegaly, anemia, and thrombocytopenia. Jinx mice do not spontaneously develop clinical features of hemophagocytic lymphohistiocytosis (HLH), but do so when infected with lymphocytic choriomeningitis virus, exhibiting hyperactivation of CTLs and antigen-presenting cells, and inadequate restriction of viral proliferation. In contrast, neither Listeria monocytogenes nor MCMV induces the syndrome. In mice, the HLH phenotype is conditional, which suggests the existence of a specific infectious trigger of FHL3 in humans.
Mouse cytomegalovirus (MCMV) is a β-herpesvirus that is contained by the host through the action of NK cells before the onset of the adaptive immune response. Mice of the C57BL/6 or C57BL/10 background show robust resistance to MCMV due to the expression of the NK cell–activating receptor Ly49H, whereas BALB/c mice, lacking Ly49H, are highly susceptible (1–4).
We have previously described a genetic screen for susceptibility to MCMV, performed in C57BL/6 mice homozygous for random N-ethyl-N-nitrosourea (ENU)-induced germline mutations (5). Based on the frequency of transmissible susceptibility mutations, we have estimated that ∼300 genes comprise the MCMV resistome: that set of genes with nonredundant function in early resistance to this pathogen (6).
Among the known components of the resistome are genes required to sense virally encoded nucleic acids and proteins (e.g., Tlr9, Tlr3, Myd88, Trif, Unc93b1, Ly49h, and Dap12 genes; references 1 and 7–12). Also within the resistome are genes coding for several cytokine mediators (13–16), their receptors (13, 15), and their transducers (5, 17–20). In addition, several genes code for components of the cellular machinery required for NK cell granule exocytosis, or components of the granules themselves. These include Lyst (21), Prf1 (22), and genes defective in Griscelli syndrome type II (23) or the Hermansky-Pudlak syndrome type II (unpublished data). Notable in this context is the fact that among proteins involved in granule exocytosis, many contribute to melanosome and/or neuronal exocytosis; hence, complex phenotypes are observed in which mutations that affect pigmentation may also have immunological or neurological consequences (24).
As described previously (5), the Jinx mutation [MGI: 3626342], one of eight defects identified by screening 3,500 G3 mutant mice for MCMV susceptibility, is associated with exaggerated cytokine production after MCMV inoculation, consistent with the preservation of innate immune sensing function and inadequate effector function. Jinx does not cause aberrant pigmentation or obvious neurological dysfunction. Here we report the detailed phenotypic characterization and positional cloning of Jinx, which represents the first animal model of type 3 familial hemophagocytic lymphohistiocytosis (FHL3) in humans. We show that in the mouse, the hemophagocytic lymphohistiocytosis (HLH) phenotype is conditional, in that it depends upon a specific infectious trigger.
RESULTS
The Jinx phenotype
When inoculated with 105 PFU of Smith strain MCMV, WT C57BL/6 mice normally survive infection, showing no sign of illness, and when killed after 5 d, show very few PFU in the spleen. The Jinx mutation was detected in a G3 mouse that showed severe illness after inoculation with 105 PFU of MCMV. It was retrieved by recrossing the corresponding G1 sire and G2 dam, and then brought to homozygosity by repeated sibling inbreeding. All Jinx mice were normally pigmented and showed normal cage activities, and their primary and secondary lymphoid organs were grossly normal in appearance. No abnormalities of lymphoid subsets were evident on CD4, CD8, B220, and NK1.1 typing, nor was there evidence of anemia or a bleeding diathesis (not depicted).
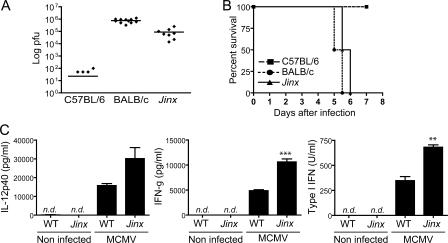
5 d after MCMV infection, viral titers in BALB/c mice and Jinx homozygotes are four to five orders of magnitude higher than in WT C57BL/6 mice (Fig. 1 A). Although Jinx homozygotes do not usually die after challenge with 105 PFU of MCMV, an inoculum of 2.5 × 105 PFU is uniformly lethal to both Jinx and BALB/c mice within the same time frame (Fig. 1 B). Jinx homozygotes show exaggerated production of IL-12, IFN-γ, and IFN-α/β (type I IFN) 36 h after inoculation with the virus (Fig. 1 C). This finding is consistent with normal sensing by APCs in the context of an inadequate NK cell effector response, permitting unfettered accumulation of the virus and therefore a stronger stimulus for cytokine production.
Figure 1.
Jinx mutants show high susceptibility and an increase in cytokine production after MCMV infection. (A) PFU were measured in spleens from C57BL/6, BALB/c, and Jinx/Jinx mice on day 5 after the inoculation with 105 PFU of MCMV. BALB/c mice were used as controls for susceptibility. Each point represents an individual animal, and lines refer to means. (B) Time-dependent death of C57BL/6 mice, BALB/c mice, and Jinx/Jinx mutants when challenged with 2.5 × 105 PFU of MCMV. For each genotype, n = 6. The experiment was concluded after 7 d, but no additional deaths were observed for at least 10 additional days. (C) IL-12p40, IFN-γ, and IFN-α/β levels in serum measured 36 h after MCMV infection. n.d., not detected.
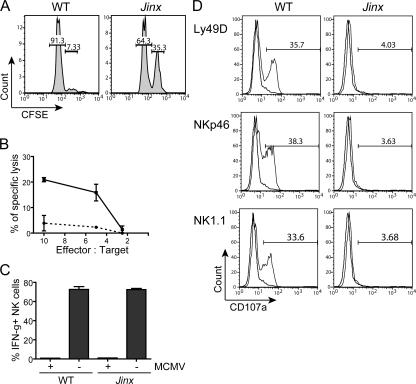
Because NK cells are pivotal in resistance to acute infection with MCMV, we sought to analyze NK cytolytic activity and IFN-γ production because defects in either of these processes would suggest an explanation for the observed failure to restrict viral proliferation within infected tissues. In Jinx mutants, NK cell–mediated cytotoxicity against β2-microglobulin (β2m)-deficient target cells in vivo (Fig. 2 A) and against YAC-1 cells in vitro (Fig. 2 B) is abolished. However, NK cells from MCMV-infected Jinx mice are able to be activated as indicated by their ability to secrete normal levels of IFN-γ (Fig. 2 C). Accordingly, we suspected a problem with NK cell degranulation and used the CD107a surface translocation method (25, 26) to assess degranulation in Jinx and WT NK cells. A gross abnormality of NK cell granule exocytosis was observed, as reflected by the failure to transfer CD107a to the cell surface in response to stimulation by NK cell–activating receptors Ly49D, NKp46, or NK1.1 (Fig. 2 D).
Figure 2.
Jinx NK cells produce IFN-γ after MCMV infection but fail to kill target cells due to a defect in degranulation. (A) In vivo assay of NK cell killing. WT, cells were injected into C57BL/6 mouse; Jinx, cells injected into a Jinx/Jinx homozygote. Numbers refer to the percentage of cells in each gate. (B) Killing of YAC-1 cells in vitro by NK cells purified from MCMV-infected Jinx homozygotes (dashed line) or C57BL/6 cells (solid line). (C) IFN-γ production by NK cells obtained from MCMV-infected C57BL/6 (WT) and Jinx/Jinx homozygotes 2 d after inoculation. Data show the percentage of IFN-γ+ cells among the gated NK1.1+ CD3ɛ− population. (D) Stimulus-induced degranulation of NK cells measured by CD107a surface expression. Plate-bound antibodies specific for some NK cell receptors (solid line) or their respective isotype controls (faded line) were used for induction. Numbers indicate the percentage of NK1.1+ CD3ɛ− cells expressing CD107a at their surface.
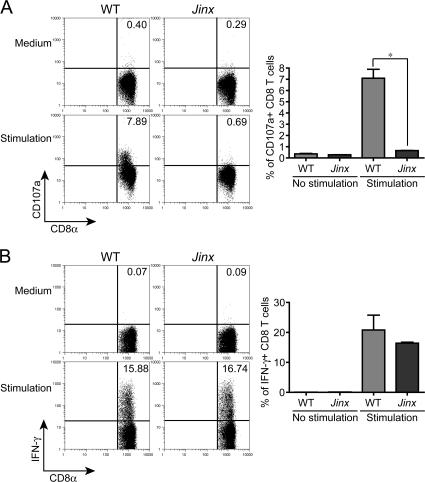
When we examined CTL function in Jinx homozygotes, a similar phenotype was evident. Polyclonal stimulation of CTLs from Jinx homozygotes revealed a profound defect in their ability to degranulate (Fig. 3 A), but normal production of IFN-γ was apparent (Fig. 3 B). We therefore inferred that the Jinx defect likely involved a component of machinery for exocytosis and, further, concluded that the protein in question had nonredundant function in lymphoid cells but was not required for an analogous function in melanocytes or neurons because melanosome exocytosis to the hair shaft and neurological function was at least grossly intact. Moreover, Jinx homozygotes showed no obvious enhancement of susceptibility to Listeria monocytogenes (not depicted), consistent with the conclusion that the protein affected has no essential role in neutrophil function.
Figure 3.
Jinx CD8+ T cells produce a normal amount of IFN-γ upon polyclonal stimulation with PMA/ionomycin but fail to degranulate. (A) Surface expression of CD107a. Inset numbers indicate percentage of cells with induced expression. (B) Up-regulation of intracellular IFN-γ. Graphs beneath each FACS illustration show data for three mice.
Positional cloning of Jinx
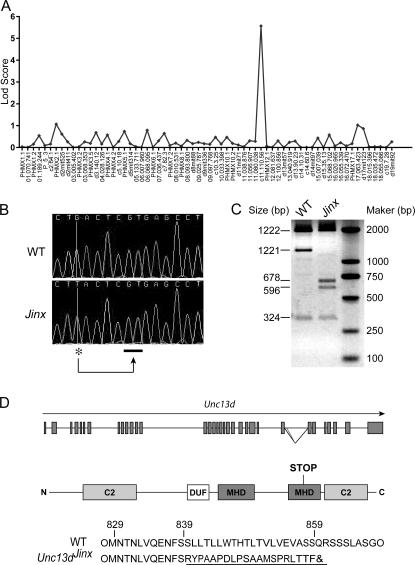
The Jinx mutation was mapped by outcrossing the mutant stock to mice of the C57BL/10 and C3H/HeN strains and backcrossing F1 animals to mice of the mutant stock. Using a panel of 68 informative single nucleotide polymorphisms and microsatellite markers to examine 30 meioses derived from the C57BL/10 cross and phenotyping based on MCMV susceptibility, the mutation was mapped to the distal end of chromosome 11 with a log odds distance score of 5.5 (Fig. 4 A). On 216 additional meioses derived from the C3H/HeN cross and phenotyping based on MCMV susceptibility, the mutation was confined to a region circumscribed by markers at 113.03 million basepairs (Mb) and 117.63 Mb removed from the centromere (Ensembl Build 41; Table I). This 4.60-Mb region encompassed 122 annotated genes, 22 of which were excluded from consideration by DNA sequencing, most of them at the cDNA level (Table II).
Figure 4.
Mapping and positional cloning of Jinx (A) Genome-wide confinement of the phenotype on 30 meioses using a panel of 68 informative markers (bottom). Strongest linkage observed with a marker on chromosome 11, 110.56 Mb. (B) Genomic sequence from a part of intron 26 of Unc13d reveals a G→T transversion (*), which causes splicing to occur distally (underscore), causing incorporation of 53 bp of intronic sequence into exon 26. (C) Effect of the Jinx mutation at the mRNA level. KpnI cuts the WT cDNA twice with a 3,467-bp amplification fragment leading to 1,922-, 1,221-, and 324-bp bands. In Jinx, the 53-bp insertion contains an additional KpnI site, giving 1,922-, 596-, 678-, and 324-bp bands. In the mutant cDNA pool, no WT transcript is detectable. (D) Location of Jinx mutation in the genomic sequence of Unc13d and the structure of the truncated protein predicted from the Jinx mutation. C2, Ca2+-binding domain; MHD, MUNC homology domain; DUF, DUF1041 domain.
Table I.
Markers used for Jinx fine mapping
| Markers | Position (Mb) | Susceptible | Susceptible | Resistant | Resistant |
|---|---|---|---|---|---|
| c11.110.56 | 110.56 | D | S-B | S-B | S-B |
| d11mit291 | 112.59 | D | S-B | S-B | S-B |
| c11.113.1 | 113.03 | S-B | S-B | S-B | D |
| c11.115.2 | 115.46 | S-B | S-B | D | D |
| d11mit203 | 116.18 | S-B | S-B | D | D |
| c11.117.1 | 117.63 | S-B | D | D | D |
The marker positions are derived from Ensembl build 41 database. S-B, single-B6 (homozygous B6 allele); D, double (heterozygous B6-C3H allele).
Table II.
List and positions of the sequenced genes localized in the Jinx critical region
| Start position (Mb) | Ensembl gene ID | Description | Level sequenced |
|---|---|---|---|
| 113.55 | ENSMUSG00000041598 | CDC42 effector protein 4 | cDNA |
| 114.71 | ENSMUSG00000034652 | CD300A antigen | cDNA and Genomic |
| 114.75 | ENSMUSG00000063193 | CD300 antigen like family member B | cDNA and Genomic |
| 114.78 | ENSMUSG00000058728 | CD300C antigen | cDNA and Genomic |
| 114.80 | ENSMUSG00000034641 | RIKEN cDNA 4732429D16 | Genomic |
| 114.82 | ENSMUSG00000044811 | CD300A antigen | cDNA and Genomic |
| 114.83 | ENSMUSG00000069610 | cDNA and Genomic | |
| 114.84 | ENSMUSG00000069609 | cDNA and Genomic | |
| 114.85 | ENSMUSG00000069608 | cDNA and Genomic | |
| 114.86 | ENSMUSG00000069607 | CMRF-35-like molecule 3 | cDNA and Genomic |
| 114.87 | ENSMUSG00000048498 | CD300 antigen like family member E | cDNA and Genomic |
| 114.91 | ENSMUSG00000020732 | RAB37, member of RAS oncogene | cDNA |
| 114.94 | ENSMUSG00000047798 | CD300 antigen like family member F | cDNA and Genomic |
| 115.41 | ENSMUSG00000020740 | ARF binding protein 3 | cDNA |
| 115.46 | ENSMUSG00000059923 | growth factor receptor bound protein 2 | cDNA |
| 115.75 | ENSMUSG00000020755 | transcriptional regulator protein | cDNA |
| 115.90 | ENSMUSG00000057948 | unc-13 homolog D (C. elegans) | cDNA and Genomic |
| 115.96 | ENSMUSG00000020776 | Fas (TNFRSF6) binding factor 1 | cDNA |
| 116.11 | ENSMUSG00000020792 | exocyst complex component 7 | cDNA |
| 116.15 | ENSMUSG00000034227 | forkhead box J1 | cDNA |
| 116.93 | ENSMUSG00000061878 | Sphingosine kinase 1 | cDNA |
| 116.93 | ENSMUSG00000020823 | Sec14-like 1 | cDNA |
Out of 22 sequenced genes, Unc13d was the only gene to be mutated (bold type). All positions and descriptions are derived from the Ensembl Build 41 database (www.ensembl.org).
The Unc13d gene was considered as a candidate because mutations in the human orthologue MUNC13-4 cause FHL3, a disease in which CTL and NK cells from affected patients show a defect of degranulation (27, 28). When the Unc13d cDNA was amplified from splenocyte mRNA derived from Jinx mice and sequence, a 53-bp insertion (degeneration to double sequence) was observed (GenBank accession no. EF127645) in a portion of the cDNA corresponding to exon 26 of the gene.
Further sequencing at the genomic level disclosed a G→T transversion at position 4,538 downstream from the distal end of exon 26 (Fig. 4 B), which created a new donor splice site, preferred to the complete exclusion of the normal donor site (Fig. 4 C). As confirmed by genomic sequencing (GenBank accession no. EF127646), this splicing defect causes the aberrant incorporation of the 53 intronic nucleotides that normally follow exon 26 into the Unc13dJinx-derived mRNA, which in turn leads to the incorporation of 20 aberrant amino acids into the UNC-13D protein, followed by premature termination of the polypeptide chain due to an in-frame TGA codon after amino acid 859 (Fig. 4 D). Normally, 1,085 amino acids in length, the UNC-13D protein features two Ca2+-binding (C2) domains separated by two “MUNC homology” domains (MHDs) and a domain of unknown function (DUF1041; Fig. 4 D). The Jinx mutation is predicted to eliminate the second of the C2 domains and part of the second MHD. C2 domains are also found in phospholipases, protein kinases C, and in other members of the MHD-defined family, and extend across many eukaryotic species, including Caenorhabditis elegans and Drosophila melanogaster. C2 domains appear to bind phospholipids, inositol polyphosphates, and intracellular proteins, consistent with a possible role in signal transduction.
Conditional induction of an FHL3-like phenotype by lymphocytic choriomeningitis virus (LCMV) infection
FHL3 is a severe disease that occurs in children with mutations of MUNC13-4, the human orthologue of Unc13d (27). Although the pathogenesis of the disease is not well understood, the clinical features of the disorder include massive hepatosplenomegaly, thrombocytopenia, neutropenia, and anemia. The two principal hallmarks of the disease are an overwhelming proliferation of CD8+ T cells and macrophages, associated with sustained production of cytokines. Histologically, macrophages are seen to ingest erythroid precursors as well as erythrocytes and platelets. Other forms of FHL occur as a result of mutations in the perforin-encoding gene (29) and the syntaxin-11 gene (30). Some (but not all) features of FHL can be observed in patients with Griscelli syndrome type II (caused by mutations of RAB27A; reference 31) and in patients with Chediak-Higashi syndrome (caused by mutations in LYST; reference 32).
It has been proposed that an infectious trigger—either viruses such as EBV or intracellular bacteria (33)—is required for expression of the FHL phenotype, a possibility supported by the observation that in Prf1 mutant mice, L. monocytogenes causes increased CD8+ T cell activation (34) and LCMV infection causes both uncontrolled CD8+ T cell expansion and hepatosplenomegaly (35–37). In Unc13dJinx/Jinx mice, hepatosplenomegaly is neither observed in uninfected animals maintained under standard conditions of housing nor in animals deliberately infected with L. monocytogenes, sustaining 7 d of documented infection and followed through 2 wk, whereon all mice recovered (not depicted). Moreover, when 2 × 104 PFU of MCMV are administered to Unc13dJinx/Jinx mice, sickness develops by day 5, but through 12 d of observation, when sickness has fully resolved, there is no evidence of HLH. At day 7, there is neutrophilia (fourfold higher than in WT mice) and monocytosis (threefold higher than in WT mice). The neutrophilia resolves by day 14, whereas the monocytosis does not (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20062447/DC1). The liver, spleen, and salivary glands all remain normal in size and appearance.
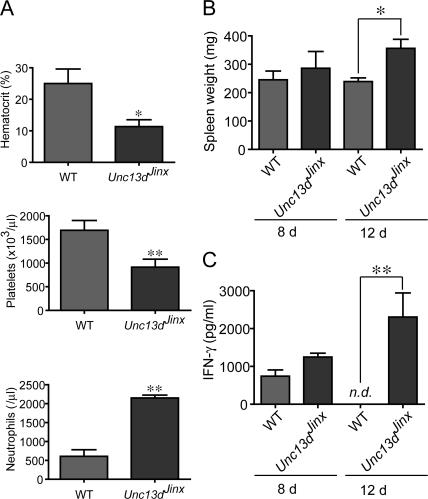
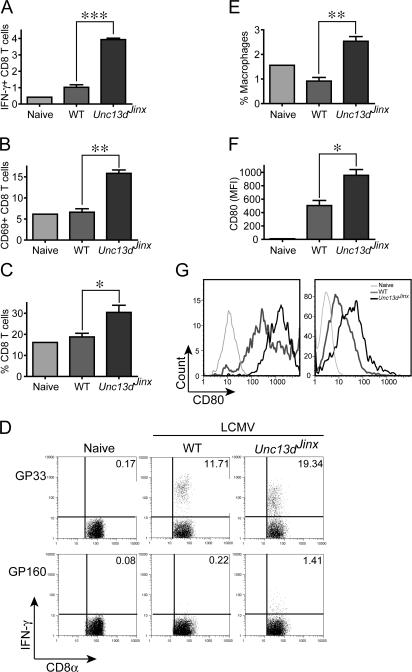
However, HLH can be induced by at least one infectious agent. When infected with LCMV (Armstrong strain) and examined after 12 d, mice have severe anemia and very significant thrombocytopenia, but neutrophilia rather than neutropenia (Fig. 5 A). Splenomegaly (Fig. 5 B) and sustained elevation of IFN-γ in serum (Fig. 5 C) were also present, both hallmarks of the human disease process. Exaggerated production of IFN-γ is evident in splenic CD8+ T cells from infected Unc13dJinx/Jinx mice (Fig. 6 A) as is the activation marker CD69 (Fig. 6 B). The percentage of CD8+ T cells is approximately doubled in the spleen (Fig. 6 C). Similarly doubled with respect to the response observed in WT control mice are the number of Unc13dJinx/Jinx splenic CD8+ T cells that respond to GP33 peptide, derived from LCMV (Fig. 6 D). In Unc13dJinx/Jinx mice, an approximately 2.5-fold increase in the number of F4/80+/CD11b+ cells were present in the spleen (Fig. 6 E). Among the cells of this population, the APC activation marker CD80 is abnormally up-regulated in Unc13dJinx/Jinx mice (Fig. 6 F). This up-regulation applied equally in both macrophage and DC populations (Figs. 6 H).
Figure 5.
Unc13dJinx/Jinx mice develop an FHL-like phenotype when infected with LCMV. (A) Hematocrit, platelet count, and neutrophil count in the blood of LCMV-infected WT and Unc13dJinx/Jinx mice 12 d after infection. (B) IFN-γ production in the serum of LCMV-infected WT and Unc13dJinx/Jinx mice 8 and 12 d after infection. n.d., not detected. (C) Spleen weight in WT and Unc13dJinx/Jinx mice 12 d after infection. n = 3 mice per group.
Figure 6.
Phenotype of CD8+ T cells and APCs in Unc13dJinx/Jinx mutants after LCMV infection. (A–D) Splenocytes from Unc13dJinx/Jinx or WT mice infected with LCMV 12 d previously or uninfected controls were stained for CD8α, CD3ɛ, CD69, and IFN-γ. (A) The percentage of IFN-γ+ CD8+ T cells. (B) Percentage of CD69+ CD8α+ T cells. (C) The percentage of CD8α+ CD3ɛ+ T cells among all splenocytes. (D) Splenocytes were stimulated for 4 h in the presence of LCMV-specific peptide GP33 or the irrelevant peptide GP160. Inset numbers represent the percentage of CD8α+ CD3ɛ+ IFN-γ+ cells. (E) The percentage of macrophages (F4/80+ CD11b+ cells) among all splenocytes. (F) The expression of CD80 (mean fluorescence intensity) by splenic macrophages (gated on the F4/80+ and CD11b+ population). (G) CD80 up-regulation in macrophages (left) and dendritic cells (right) in uninfected mice and in infected WT and Unc13dJinx/Jinx mice. A single uninfected (naive) mouse was used in all experiments, and a representative set of individual mice were used in D and G. For all other panels, n = 3.
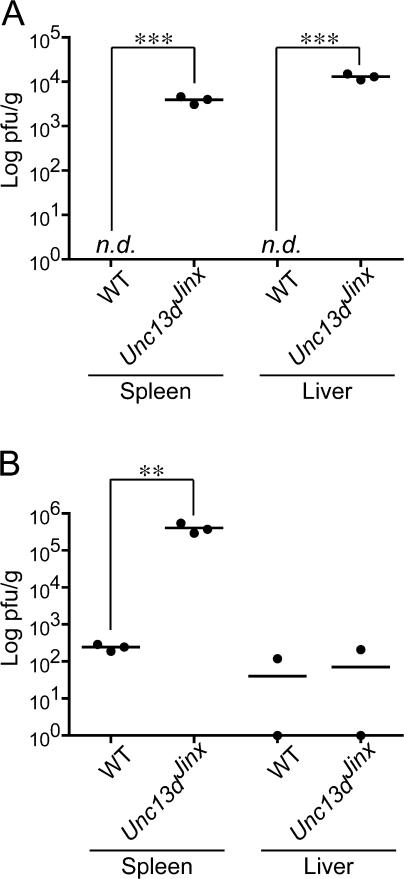
Despite evidence of exaggerated innate and adaptive immune responses to infection, Unc13dJinx/Jinx mice were not able to eradicate the LCMV infection, which grew out of control so that plaque assays of spleen homogenates showed viral titers >1,000-fold higher in Unc13dJinx/Jinx mice than in WT mice at 12 d after inoculation. Interestingly, some containment was achieved in the liver, where titers declined between days 8 and 12 after inoculation (Fig. 7, A and B).
Figure 7.
LCMV titre in the liver and spleen after infection in WT and Unc13dJinx/Jinx mice. Standard plaque assays were performed on spleens and livers 8 (A) and 12 d (B) after LCMV infection. n.d., not detected. n = 3 mice for each group.
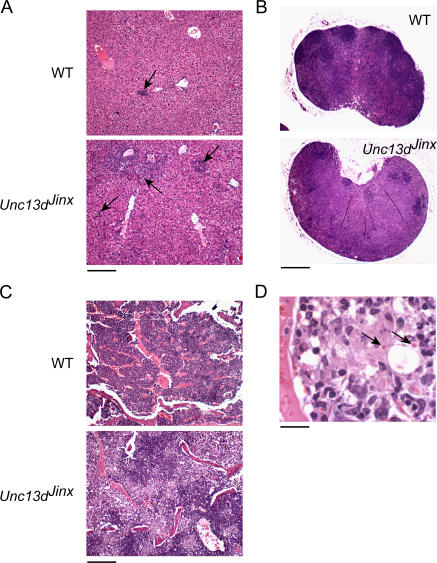
Histologically at day 12, Unc13dJinx/Jinx mice showed abundant hepatic granulomata compared with WT mice (Fig. 8 A), and depletion of germinal centers with replacement by macrophages in peripheral lymph nodes (Fig. 8 B). In the bone marrow as well, macrophage infiltration was evident in Unc13dJinx/Jinx mice (Fig. 8 C), and at high magnification, hemophagocytosis could clearly be observed (Fig. 8 D).
Figure 8.
Histologic appearance of the liver and spleen in LCMV-infected Unc13dJinx/Jinx and WT mice displays features of FHL in different organs. Hematoxylin and eosin staining of sections of livers from LCMV-infected Unc13dJinx/Jinx mice (A) showed an increased number of granulomas of larger size (arrow) compared with livers from infected controls (bar, 200 μm). (B) Infiltration of macrophages is observed in lymph nodes in Unc13dJinx/Jinx mice (arrow), along with a paucity of germinal centers (bar, 0.4 mm). (C) Bone marrow from LCMV-infected Unc13dJinx/Jinx mice is heavily infiltrated by macrophages compared with LCMV-infected WT marrow (bar, 200 μm), and at high magnification (D) RBCs are seen to reside within vesicles in bone marrow macrophages (arrows, hemophagocytosis). Bar, 20 μm.
DISCUSSION
In this paper we have reported the first phenovariant allele of Unc13d in mice, detected because it caused severe immunocompromise in the setting of MCMV infection. Although mutations of the human orthologue MUNC13-4 have been identified as the cause of FHL3, no animal model of this disorder has previously been identified, and hence, there has been little chance for insight into the environmental “trigger” of this disease, if such exists.
EBV, a γ-herpesvirus, has been suggested to trigger FHL (33), but assignment of cause and effect has been elusive because EBV infection is relatively common and because data on the penetrance of this relatively rare human disorder have not been available. We attempted to induce FHL-like disease in Unc13dJinx/Jinx mice using the β-herpesvirus MCMV. The acute lethal effect of MCMV infection in Unc13dJinx/Jinx mice made the experiment impossible to perform with the highest doses normally tolerated by C57BL/6J controls. However, we note that low doses of the virus, sufficient to produce visible sickness in Unc13dJinx/Jinx mice, were ultimately cleared without producing an HLH-like disease.
LCMV was investigated as an alternative pathogen because NK cells do not play a prominent role in early host defense against this agent. The LCMV-driven HLH phenotype of Unc13dJinx/Jinx mice is consistent with a model in which the infection is not effectively cleared in the absence of effective CTL degranulation. Viral infection then drives the expansion of APCs (which are themselves actively infected) and CD8+ T cells, which may be infected as well, and which respond to the ongoing antigenic stimulus that the APCs present. We consider that like the arenavirus LCMV, herpesviruses and possibly many other agents might ultimately initiate a comparable cycle of APC and CTL proliferation that is clinically interpreted as FHL. However, not all microbes will readily cause FHL, and in our initial studies of Unc13dJinx/Jinx mice, L. monocytogenes infection was found to be incapable of doing so.
It is interesting to note that the Unc13dJinx/Jinx mice infected with LCMV present one important discrepancy from humans with FHL3: the former show fourfold elevation in peripheral neutrophil counts, whereas FHL3 patients are neutropenic (27, 33, 38, 39). This discrepancy may reflect a different role of the UNC-13D protein in neutrophils in mice as compared with humans, or may alternatively be a property of the microbial inducer that is most commonly culpable in humans. Of interest in this regard, Prf −/− mice do develop neutropenia when infected with LCMV (37).
Host resistance mechanisms often incorporate proteins that are multifunctional, not only in the sense that they confer resistance to multiple infectious agents, but also in the sense that they are required for diverse biological processes. Many of the proteins required for NK cell or neutrophil granule exocytosis are also required for pigmentation or have neurological functions. UNC-13D, the protein affected by the Jinx mutation, appears to be dedicated to resistance and does not noticeably affect pigmentation or neurological function. Essential in NK cells and CTLs, it is probably dispensable for neutrophil function because defects of neutrophil function usually cause severe immunocompromise during L. monocytogenes infection.
UNC-13D is required for priming (fusion competence) of cytoplasmic vesicles. Although we cannot formally exclude the possibility that some functions of UNC-13D might be intact in Unc13dJinx/Jinx homozygotes, it appears most likely that Unc13dJinx/Jinx encodes a completely nonfunctional protein based on deletion/complementation studies of the UNC-13 homologue in C. elegans, which have shown that deletion or point mutation within the second MHD abolishes fusion competence and complementation of the motility phenotype (40).
The molecular ancestry of UNC-13D has been traced by homology searches focused on individual domains, such as the C2 domain (41), which is widely distributed in cellular proteins involved in signal transduction or membrane trafficking, and serves as a Ca2+-dependent membrane-targeting module, enabling proteins to bind phospholipids within various biological membranes. Different C2 domains exhibit lipid selectivity, favoring phosphatidylserine or phosphatidylcholine binding, and the C2 domain of UNC13 from C. elegans can engage phorbol esters (42). The DUF1041 motif is often found in conjunction with C2 domains and is represented in several proteins in addition to members of the UNC-13 family, including the brain-specific angiogenesis inhibitor-1–associated protein 3 and the calcium-dependent secretion activator-1 and -2 proteins. The MHD is the signature domain of the MUNC family of proteins and is regarded as an interaction motif based on studies in C. elegans, in which it was shown that a double amino acid substitution within the MHD could abolish interaction between UNC-13 and syntaxin (40).
MUNC13-1, MUNC13-2, and MUNC13-3, the closest relatives of MUNC13-4 (UNC13D), are expressed in the brain, and single knockout mutations each produce a CNS phenotype (43–45). Although MUNC13-1 knockouts are often born dead and show relatively severe neurological impairment (43), individual MUNC13-2 and MUNC13-3 mice, and MUNC13-2 and MUNC13-3 double knockouts, are viable (46), consistent with the conclusion that these paralogues serve at least partially redundant functions. Deletion of the more distantly related MUNC18-1–encoding gene Stxbp1 (syntaxin binding protein-1) causes perinatal lethality related to neurological dysfunction as well (47).
MUNC13-4 (and by implication, UNC-13D) is believed to associate with RAB27A on the basis of immunoprecipitation studies performed with human platelets (48), and is believed to be required for the priming of platelet-dense granules. However, we are unable to find evidence of platelet dysfunction in Unc13dJinx/Jinx homozygotes, suggesting that alternative mechanisms for dense granule priming, independent of UNC-13D, must exist in mice. The strong NK cell phenotype suggests nonredundancy of function, and although it may not apply to all classes of NK cell granules, it is clear that the LAMP1 (CD107a) compartment is forbidden exocytosis in Unc13dJinx/Jinx homozygotes. Fas ligand, granzymes, and perforin colocalize within the same class of vesicles in NK cells (49, 50), and it is therefore likely that in Unc13dJinx/Jinx mice, a defect of Fas ligand release also exists. This may contribute to the proliferative syndrome that is observed, to the extent that Fas ligand initiates apoptosis in the target cell population. However, the absence of spontaneous lymphoproliferative disease in Unc13dJinx/Jinx mice (as compared with Gld mice, for example) indicates that the NK cells and CTLs do not act as the essential source of Fas ligand required for homeostatic control of T cells in vivo.
MATERIALS AND METHODS
Mice, ENU mutagenesis, MCMV susceptibility screen, and Jinx mapping.
C57BL/6 (also referred to as WT), C57BL/10, C3H/HeN, BALB/c, β2m −/−, and Jinx/Jinx mice (MMRRC:016137) were maintained under specific pathogen-free conditions in the TSRI vivarium, and all studies involving mice were performed in accordance with institutional regulations governing animal care and use. ENU mutagenesis was performed as described previously (51) in a C57BL/6 background, naturally resistant to MCMV infection. Because most inbred strains show either a strong or moderate susceptibility to MCMV infection, the chromosome location of Jinx mutation was obtained by outcrossing Jinx homozygotes to C57BL/10 mice, and then backcrossing to Jinx stock and infecting F2 mice with MCMV at 6 wk of age. For mapping against the C57BL/10 background, 31 informative single nucleotide polymorphisms and 37 microsatellite polymorphisms were used (Table S1, available at http://www.jem.org/cgi/content/full/jem.20062447/DC1), distributed across the genome at ∼50-Mb intervals. For fine mapping, C57BL/10 and C3H/HeN strains were used using the microsatellite markers shown in Table I. Four informative markers were identified in our laboratory and are defined in Table S2).
Viruses, in vivo susceptibility screen, PFU assay, and serum cytokine detection.
The generation of the MCMV Smith strain stock from the salivary glands of 3-wk-old MCMV-infected BALB/c mice, as well as the conditions of the in vivo MCMV susceptibility screen of ENU germline mutants, was described previously (5). MCMV was administrated i.p. The Armstrong strain of LCMV (provided by E. Zuniga and M.B.A. Oldstone, TSRI, La Jolla, CA) was injected i.p. at 105 PFU per mouse. Viral loads were determined after organ homogenization in DMEM, 3% FCS, by standard plaque assays on 3T3-NIH cells for MCMV (52) and on VERO cells for LCMV (53).
Serum cytokine detection and complete blood counts.
36 h after MCMV infection or at the times indicated after LCMV infection, mice were bled from the retroorbital sinus, and the concentration of IL-12p40, TNF-α, IL-6, and IFN-γ in the serum was assayed by ELISA (eBioscience). The bioactivity of serum type I IFN was measured by luciferase assay using L929-ISRE cells as described previously (5). Complete blood cells counts were performed by Antech Diagnostics.
In vivo assay for NK cell cytotoxic activity.
Splenic suspensions from β2m-deficient mice and C57BL/6 controls were resuspended at 107 cells/ml in PBS. Control and target splenocytes were, respectively, labeled with a low and high concentration of CFSE at room temperature for 10 min. The labeling was stopped by dumping cold FCS on cell suspensions. Cells were washed twice, counted, and resuspended at 5 × 107 cells/ml. The two populations were mixed at a 1:1 ratio and injected i.v. into recipient mice. Recipients were bled the next day, and PBMCs were analyzed for CFSE staining by flow cytometry.
Leukocyte preparations and purification of NK cells and CD8+ T cells.
Noninfected or infected mice were killed, and spleens were removed in RPMI, FCS 10%, minced, and filtered through 70-μm nylon mesh. RBCs were lysed with RBC lysis buffer (Sigma-Aldrich), and splenocytes were resuspended in complete medium. The enrichment of NK cells or CD8+ T cells from splenocytes was performed using the NK cell isolation kit or the CD8+ T cell isolation kit, respectively (Miltenyi Biotec). Cells of each type were counted, resuspended in RPMI, FCS 5%, and used in subsequent experiments.
In vitro assay for NK cell function and CD8+ T cell function.
To determine their cytolytic function, enriched NK cells were isolated from splenocytes 48 h after MCMV infection and incubated with YAC-1 cells at 37°C for 6 h. The percent-specific lysis was measured according to the release of lactate dehydrogenase into the supernatant, using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega). The degranulation capacity of NK cells was assayed by incubating NK cells with plate-bound antibodies directed against NK cell receptors (NK1.1, NKp46, and Ly49D, as indicated) for 4 h in the presence of FITC-conjugated anti–mouse CD107a (BD Biosciences) and monensin (Golgi-Stop; BD Biosciences). Enriched CD8+ T cells were stimulated with PMA and ionomycin in the presence of brefeldin A (Golgi-Plug; BD Biosciences) for 4 h to measure IFN-γ synthesis, or with PMA and ionomycin in the presence of FITC-antiCD107a for 4 h to measure their degranulation. Splenocytes from LCMV-infected mice were stimulated with the LCMV peptide GP33 or with the irrelevant HIV-derived peptide GP160 for 4 h in the presence of brefeldin A (Golgi-Plug; BD Biosciences), after which IFN-γ production was measured by intracellular staining.
Antibodies, intracellular staining, and statistical analysis.
Antibodies used in this study included the following: NK1.1 (PK136), IFN-γ (XMG1.2), CD8α (53-6.7), CD80 (B7-1), CD11b (M1/70), CD69 (H1.2F3; eBioscience), CD3ɛ (145-2C11), Ly49D (4E5), CD107a (1D4B; BD Biosciences), NKp46 (R&D Systems), F4/80 (BM8; Caltag laboratories). Intracellular staining was performed after classical staining for surface markers. Cells were then fixed and permeabilized using Cytofix/Cytoperm solution (BD Biosciences) for 20 min at 4°C, and then stained with IFN-γ antibody diluted in permeabilization buffer provided by BD Biosciences. All statistics were calculated using the Student's t test (two tail). *, P < 0.05; **, P < 0.01; ***, P < 0.001 in all figures. Error bars show SEM.
Online supplemental material.
Table S1 shows a list and positions of the markers used for Jinx mapping using the C57BL/6 and C57BL/10 strains. Table SII shows a list and positions of markers identified in our laboratory and used for Jinx fine mapping on C57BL/6, C57BL/10, and C3H/HeN strains. In Fig. S1, mice (Jinx homozygotes and WT C57BL/6J controls) were infected with 2 × 104 PFU of MCMV and observed over a period of 14 d. Although the control animals did not appear ill at any time during the infection, the Jinx mice did show signs of sickness at day 5, with resolution by day 12. Mice were bled at days 7 and 14 for complete blood counts. Mice were killed at day 14 for visual inspection and weighing for the liver, spleen, and salivary glands. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20062447/DC1.
Supplemental Material
Acknowledgments
The authors would like to thank Dr. Elina Zuniga and Prof. Michael B.A. Oldstone (TSRI) for providing cells, virus, and reagents, and for technical advice, as well as Dr. Thierry Walzer (CIML, Marseille, France) for helpful discussions.
This work was supported by National Institutes of Health grant number AI054523. S. Rutschmann was supported by a Human Frontier Science Program long-term fellowship. This is TSRI manuscript number 18610.
The authors have no conflicting financial interests.
Abbreviations used: β2m, β2-microglobulin; C2, Ca2+-binding; ENU, N-ethyl-N-nitrosourea; FHL3, type 3 familial hemophagocytic lymphohistiocytosis; HLH, hemophagocytic lymphohistiocytosis; LCMV, lymphocytic choriomeningitis virus; Mb, million basepair; MCMV, mouse cytomegalovirus; MHD, MUNC homology domain.
S. Rutschmann's present address is Dept. of Immunology, Faculty of Medicine, Imperial College London SW7 2AZ, London, UK.
References
- 1.Arase, H., E.S. Mocarski, A.E. Campbell, A.B. Hill, and L.L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326. [DOI] [PubMed] [Google Scholar]
- 2.Lee, S.H., S. Girard, D. Macina, M. Busa, A. Zafer, A. Belouchi, P. Gros, and S.M. Vidal. 2001. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat. Genet. 28:42–45. [DOI] [PubMed] [Google Scholar]
- 3.Lee, S.H., A. Zafer, Y. de Repentigny, R. Kothary, M.L. Tremblay, P. Gros, P. Duplay, J.R. Webb, and S.M. Vidal. 2003. Transgenic expression of the activating natural killer receptor Ly49H confers resistance to cytomegalovirus in genetically susceptible mice. J. Exp. Med. 197:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M.G., A.O. Dokun, J.W. Heusel, H.R. Smith, D.L. Beckman, E.A. Blattenberger, C.E. Dubbelde, L.R. Stone, A.A. Scalzo, and W.M. Yokoyama. 2001. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 292:934–937. [DOI] [PubMed] [Google Scholar]
- 5.Crozat, K., P. Georgel, S. Rutschmann, N. Mann, X. Du, K. Hoebe, and B. Beutler. 2006. Analysis of the MCMV resistome by ENU mutagenesis. Mamm. Genome. 17:398–406. [DOI] [PubMed] [Google Scholar]
- 6.Beutler, B., K. Crozat, J.A. Koziol, and P. Georgel. 2005. Genetic dissection of innate immunity to infection: the mouse cytomegalovirus model. Curr. Opin. Immunol. 17:36–43. [DOI] [PubMed] [Google Scholar]
- 7.Delale, T., A. Paquin, C. Asselin-Paturel, M. Dalod, G. Brizard, E.E. Bates, P. Kastner, S. Chan, S. Akira, A. Vicari, et al. 2005. MyD88-dependent and -independent murine cytomegalovirus sensing for IFN-alpha release and initiation of immune responses in vivo. J. Immunol. 175:6723–6732. [DOI] [PubMed] [Google Scholar]
- 8.Krug, A., A.R. French, W. Barchet, J.A. Fischer, A. Dzionek, J.T. Pingel, M.M. Orihuela, S. Akira, W.M. Yokoyama, and M. Colonna. 2004. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 21:107–119. [DOI] [PubMed] [Google Scholar]
- 9.Tabeta, K., P. Georgel, E. Janssen, X. Du, K. Hoebe, K. Crozat, S. Mudd, L. Shamel, S. Sovath, J. Goode, et al. 2004. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. USA. 101:3516–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabeta, K., K. Hoebe, E.M. Janssen, X. Du, P. Georgel, K. Crozat, S. Mudd, N. Mann, S. Sovath, J. Goode, et al. 2006. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat. Immunol. 7:156–164. [DOI] [PubMed] [Google Scholar]
- 11.Hoebe, K., X. Du, P. Georgel, E. Janssen, K. Tabeta, S.O. Kim, J. Goode, P. Lin, N. Mann, S. Mudd, et al. 2003. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 424:743–748. [DOI] [PubMed] [Google Scholar]
- 12.Sjolin, H., E. Tomasello, M. Mousavi-Jazi, A. Bartolazzi, K. Karre, E. Vivier, and C. Cerboni. 2002. Pivotal role of KARAP/DAP12 adaptor molecule in the natural killer cell–mediated resistance to murine cytomegalovirus infection. J. Exp. Med. 195:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Presti, R.M., J.L. Pollock, A.J. Dal Canto, A.K. O'Guin, and H.W. Virgin IV. 1998. Interferon γ regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels. J. Exp. Med. 188:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pien, G.C., A.R. Satoskar, K. Takeda, S. Akira, and C.A. Biron. 2000. Cutting edge: selective IL-18 requirements for induction of compartmental IFN-gamma responses during viral infection. J. Immunol. 165:4787–4791. [DOI] [PubMed] [Google Scholar]
- 15.Salazar-Mather, T.P., C.A. Lewis, and C.A. Biron. 2002. Type I interferons regulate inflammatory cell trafficking and macrophage inflammatory protein 1alpha delivery to the liver. J. Clin. Invest. 110:321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salazar-Mather, T.P., and K.L. Hokeness. 2003. Calling in the troops: regulation of inflammatory cell trafficking through innate cytokine/chemokine networks. Viral Immunol. 16:291–306. [DOI] [PubMed] [Google Scholar]
- 17.Strobl, B., I. Bubic, U. Bruns, R. Steinborn, R. Lajko, T. Kolbe, M. Karaghiosoff, U. Kalinke, S. Jonjic, and M. Muller. 2005. Novel functions of tyrosine kinase 2 in the antiviral defense against murine cytomegalovirus. J. Immunol. 175:4000–4008. [DOI] [PubMed] [Google Scholar]
- 18.Durbin, J.E., R. Hackenmiller, M.C. Simon, and D.E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 84:443–450. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C.K., D.T. Rao, R. Gertner, R. Gimeno, A.B. Frey, and D.E. Levy. 2000. Distinct requirements for IFNs and STAT1 in NK cell function. J. Immunol. 165:3571–3577. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen, K.B., T.P. Salazar-Mather, M.Y. Dalod, J.B. Van Deusen, X.Q. Wei, F.Y. Liew, M.A. Caligiuri, J.E. Durbin, and C.A. Biron. 2002. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J. Immunol. 169:4279–4287. [DOI] [PubMed] [Google Scholar]
- 21.Shellam, G.R., J.E. Allan, J.M. Papadimitriou, and G.J. Bancroft. 1981. Increased susceptibility to cytomegalovirus infection in beige mutant mice. Proc. Natl. Acad. Sci. USA. 78:5104–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh, J., D.T. Chu, A.K. O'Guin, W.M. Yokoyama, and H.W. Virgin IV. 2005. Natural killer cells utilize both perforin and gamma interferon to regulate murine cytomegalovirus infection in the spleen and liver. J. Virol. 79:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haddad, E.K., X. Wu, J.A. Hammer III, and P.A. Henkart. 2001. Defective granule exocytosis in Rab27a-deficient lymphocytes from Ashen mice. J. Cell Biol. 152:835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stinchcombe, J., G. Bossi, and G.M. Griffiths. 2004. Linking albinism and immunity: the secrets of secretory lysosomes. Science. 305:55–59. [DOI] [PubMed] [Google Scholar]
- 25.Rubio, V., T.B. Stuge, N. Singh, M.R. Betts, J.S. Weber, M. Roederer, and P.P. Lee. 2003. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat. Med. 9:1377–1382. [DOI] [PubMed] [Google Scholar]
- 26.Alter, G., J.M. Malenfant, and M. Altfeld. 2004. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods. 294:15–22. [DOI] [PubMed] [Google Scholar]
- 27.Feldmann, J., I. Callebaut, G. Raposo, S. Certain, D. Bacq, C. Dumont, N. Lambert, M. Ouachee-Chardin, G. Chedeville, H. Tamary, et al. 2003. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3). Cell. 115:461–473. [DOI] [PubMed] [Google Scholar]
- 28.Marcenaro, S., F. Gallo, S. Martini, A. Santoro, G.M. Griffiths, M. Arico, L. Moretta, and D. Pende. 2006. Analysis of natural killer-cell function in familial hemophagocytic lymphohistiocytosis (FHL): defective CD107a surface expression heralds Munc13-4 defect and discriminates between genetic subtypes of the disease. Blood. 108:2316–2323. [DOI] [PubMed] [Google Scholar]
- 29.Stepp, S.E., R. Dufourcq-Lagelouse, F. Le Deist, S. Bhawan, S. Certain, P.A. Mathew, J.I. Henter, M. Bennett, A. Fischer, G. de Saint Basile, and V. Kumar. 1999. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 286:1957–1959. [DOI] [PubMed] [Google Scholar]
- 30.zur Stadt, U., S. Schmidt, B. Kasper, K. Beutel, A.S. Diler, J.I. Henter, H. Kabisch, R. Schneppenheim, P. Nurnberg, G. Janka, and H.C. Hennies. 2005. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type-4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum. Mol. Genet. 14:827–834. [DOI] [PubMed] [Google Scholar]
- 31.Menasche, G., E. Pastural, J. Feldmann, S. Certain, F. Ersoy, S. Dupuis, N. Wulffraat, D. Bianchi, A. Fischer, F. Le Deist, and G. de Saint Basile. 2000. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat. Genet. 25:173–176. [DOI] [PubMed] [Google Scholar]
- 32.Rubin, C.M., B.A. Burke, R.W. McKenna, K.L. McClain, J.G. White, M.E. Nesbit Jr., and A.H. Filipovich. 1985. The accelerated phase of Chediak-Higashi syndrome. An expression of the virus-associated hemophagocytic syndrome? Cancer. 56:524–530. [DOI] [PubMed] [Google Scholar]
- 33.Fisman, D.N. 2000. Hemophagocytic syndromes and infection. Emerg. Infect. Dis. 6:601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Badovinac, V.P., A.R. Tvinnereim, and J.T. Harty. 2000. Regulation of antigen-specific CD8+ T cell homeostasis by perforin and interferon-gamma. Science. 290:1354–1358. [DOI] [PubMed] [Google Scholar]
- 35.Matloubian, M., M. Suresh, A. Glass, M. Galvan, K. Chow, J.K. Whitmire, C.M. Walsh, W.R. Clark, and R. Ahmed. 1999. A role for perforin in downregulating T-cell responses during chronic viral infection. J. Virol. 73:2527–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kagi, D., B. Odermatt, and T.W. Mak. 1999. Homeostatic regulation of CD8+ T cells by perforin. Eur. J. Immunol. 29:3262–3272. [DOI] [PubMed] [Google Scholar]
- 37.Jordan, M.B., D. Hildeman, J. Kappler, and P. Marrack. 2004. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood. 104:735–743. [DOI] [PubMed] [Google Scholar]
- 38.Henter, J.I., G. Elinder, and A. Ost. 1991. Diagnostic guidelines for hemophagocytic lymphohistiocytosis. The FHL Study Group of the Histiocyte Society. Semin. Oncol. 18:29–33. [PubMed] [Google Scholar]
- 39.Henter, J.I., A. Horne, M. Arico, R.M. Egeler, A.H. Filipovich, S. Imashuku, S. Ladisch, K. McClain, D. Webb, J. Winiarski, and G. Janka. 2007. HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr. Blood Cancer. 48:124–131. [DOI] [PubMed] [Google Scholar]
- 40.Madison, J.M., S. Nurrish, and J.M. Kaplan. 2005. UNC-13 interaction with syntaxin is required for synaptic transmission. Curr. Biol. 15:2236–2242. [DOI] [PubMed] [Google Scholar]
- 41.Brose, N., K. Hofmann, Y. Hata, and T.C. Sudhof. 1995. Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J. Biol. Chem. 270:25273–25280. [DOI] [PubMed] [Google Scholar]
- 42.Ahmed, S., I.N. Maruyama, R. Kozma, J. Lee, S. Brenner, and L. Lim. 1992. The Caenorhabditis elegans unc-13 gene product is a phospholipid-dependent high-affinity phorbol ester receptor. Biochem. J. 287:995–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Augustin, I., C. Rosenmund, T.C. Sudhof, and N. Brose. 1999. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 400:457–461. [DOI] [PubMed] [Google Scholar]
- 44.Varoqueaux, F., A. Sigler, J.S. Rhee, N. Brose, C. Enk, K. Reim, and C. Rosenmund. 2002. Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc. Natl. Acad. Sci. USA. 99:9037–9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Augustin, I., S. Korte, M. Rickmann, H.A. Kretzschmar, T.C. Sudhof, J.W. Herms, and N. Brose. 2001. The cerebellum-specific Munc13 isoform Munc13-3 regulates cerebellar synaptic transmission and motor learning in mice. J. Neurosci. 21:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varoqueaux, F., M.S. Sons, J.J. Plomp, and N. Brose. 2005. Aberrant morphology and residual transmitter release at the Munc13-deficient mouse neuromuscular synapse. Mol. Cell. Biol. 25:5973–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verhage, M., A.S. Maia, J.J. Plomp, A.B. Brussaard, J.H. Heeroma, H. Vermeer, R.F. Toonen, R.E. Hammer, T.K. van den Berg, M. Missler, et al. 2000. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 287:864–869. [DOI] [PubMed] [Google Scholar]
- 48.Shirakawa, R., T. Higashi, A. Tabuchi, A. Yoshioka, H. Nishioka, M. Fukuda, T. Kita, and H. Horiuchi. 2004. Munc13-4 is a GTP-Rab27-binding protein regulating dense core granule secretion in platelets. J. Biol. Chem. 279:10730–10737. [DOI] [PubMed] [Google Scholar]
- 49.Bossi, G., and G.M. Griffiths. 1999. Degranulation plays an essential part in regulating cell surface expression of Fas ligand in T cells and natural killer cells. Nat. Med. 5:90–96. [DOI] [PubMed] [Google Scholar]
- 50.Blott, E.J., and G.M. Griffiths. 2002. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 3:122–131. [DOI] [PubMed] [Google Scholar]
- 51.Du, X., K. Tabeta, K. Hoebe, H. Liu, N. Mann, S. Mudd, K. Crozat, S. Sovath, X. Gong, and B. Beutler. 2004. Velvet, a dominant Egfr mutation that causes wavy hair and defective eyelid development in mice. Genetics. 166:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orange, J.S., B. Wang, C. Terhorst, and C.A. Biron. 1995. Requirement for natural killer cell–produced interferon γ in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin 12 administration. J. Exp. Med. 182:1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dutko, F.J., and M.B. Oldstone. 1983. Genomic and biological variation among commonly used lymphocytic choriomeningitis virus strains. J. Gen. Virol. 64:1689–1698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.