Abstract
Adaptive immunity is initiated by T cell recognition of foreign peptides presented on dendritic cells (DCs) by major histocompatibility molecules. These interactions take place in secondary lymphoid tissues, such as lymph nodes (LNs) and spleen, and hence the anatomical structure of these tissues plays a crucial role in the development of immune responses. Two-photon microscopy (2PM) imaging in LNs suggests that T cells walk in a consistent direction for several minutes, pause briefly with a regular period, and then take off in a new, random direction. Here, we construct a spatially explicit model of T cell and DC migration in LNs and show that all dynamical properties of T cells could be a consequence of the densely packed LN environment. By means of 2PM experiments, we confirm that the large velocity fluctuations of T cells are indeed environmentally determined rather than resulting from an intrinsic motility program. Our simulations further predict that T cells self-organize into microscopically small, highly dynamic streams. We present experimental evidence for the presence of such turbulent streams in LNs. Finally, the model allows us to estimate the scanning rates of DCs (2,000 different T cells per hour) and T cells (100 different DCs per hour).
The T cell repertoire is comprised of at least 25 million receptors, each with different antigen specificity (1, 2). During an immune response, only a small fraction (10−5–10−6) of the T cells will recognize foreign antigen, activate, and undergo proliferation (3). In LNs, these antigen-specific T cells face the daunting task of first finding a DC presenting their cognate antigen. This seems especially difficult because the LN is densely packed with millions of competing T cells having irrelevant specificity, DCs presenting noncognate peptide–MHC complexes, and many solid obstacles, such as the reticular network (RN). Recently, it has become possible to visualize the in vivo motility of different immune cells through the application of two-photon microscopy (2PM; references 4–15). The resulting vivid movies and the measurements of the events occurring in LNs (4, 6, 8) suggest that T cells achieve their aim by moving around at high velocities of, on average, more than one cell diameter per minute (9–12 μm min−1). They walk in a consistent direction for several minutes, reaching peak velocities of >25 μm min−1, but crawl along random trajectories in the long term (however, note that it was recently shown that chemokines help in guiding CD8+ T cells to sites of CD4+ T cell–DC interaction; reference 15). The large velocity fluctuations and sudden directional changes give the impression that T cells pause once in every 1–3 min and are then likely to start off in a new direction. This “stop and go” fashion of walking has been suggested to be part of a program of intrinsic rhythmicity (6, 16), but the existence of such a program has until now only been examined in one study (6). Large velocity fluctuations and sharp directional changes in the tracks of T cells, however, are consistently observed (4, 8, 17–19) and seem to point in the direction of an inherent motility program. However, two-photon imaging studies reveal neither the underlying mechanism of the observed behaviors nor the consequences of the densely packed LN environment on T cell motility.
The visualization of dynamic processes in lymphoid tissues by 2PM opens up possibilities for combined modeling and experimental efforts (20, 21). Here, we construct a spatial model of the T cell area in LNs to investigate the default dynamics of naive T cells in the absence of cognate antigen. We use a three-dimensional Cellular Potts Model (CPM; references 22 and 23), which is lattice based, can describe the formation of patterns, and has previously been used to simulate cellular dynamics in a large variety of organisms (24–28). Because in cellular dynamics inertia is dominated by viscosity, the CPM describes cell motion in terms of local energy changes by means of a Hamiltonian (see Materials and methods) rather than by explicit forces (29).
In our three-dimensional model of the LN, each in silico T cell has a preferred direction of movement, which is updated according to its recent displacement. This appears to be sufficient to explain all dynamical properties of T cells, i.e., persistent motion on a short timescale, random motion on a long timescale, and large velocity fluctuations with apparent periodic pausing. Our results suggest that these complicated behaviors could be determined by the dense LN environment rather than resulting from an autonomous T cell clock. To test this hypothesis generated by the model, we perform a 2PM experiment studying T cell velocity fluctuations over a long time period and confirm the prediction that the velocity fluctuations are environmentally determined. The model further predicts the presence of microscopically small, highly dynamic streams of T cells. Indeed, an analysis of new experimental data demonstrates the presence of such turbulent streams in LNs.
RESULTS
The LN T cell area in silico
In our model we consider T cells and DCs because they are the main players in the LN T cell area. For simplicity we do not model influx or egress of cells from LNs, but assume that cells are confined to a limited, wrapped space. In silico T cells have a small preference to adhere to DCs. In experiments only fluorescently labeled cells can be visualized, whereas our model allows us to study the behavior of both “labeled” and “nonlabeled” cells. Note that these cells are assumed to differ only in their labeling and not in any other characteristics.
In LNs, fibroblastic reticular cells construct the RN (30), which we model as a static network that consists of thin, long rods with a random position and orientation (Video S1, which is available at http://www.jem.org/cgi/content/full/jem.20061278/DC1). The flexibility of the CPM allows us to choose whether T cells adhere preferentially to the RN or not. We studied both scenarios in our simulations and obtained similar results regarding the migratory behavior of T cells (see Discussion). The lattice sites that do not belong to cells, or to the RN, represent extracellular matrix (ECM) that cells can move freely into.
DCs are named after their dendritic shape, and they continuously extend and retract long dendrites (7). Our simulated DCs are assigned a large surface area to volume ratio that results in small, dynamic tentacles. Stochastic membrane fluctuations result in natural-looking shape changes.
For a T cell to move around, it needs to acquire a polarity (31, 32), which determines the formation of a leading edge and hence its approximate direction of movement. Abrupt directional changes are not possible because it takes time to displace the complicated internal structure that brings about motion. We describe this mechanism of motility in a simplified way by endowing each simulated T cell with a “target direction.” This preferred direction of travel is assumed to change over a period of seconds according to its recent displacement, which results in a natural, self-adjusting type of motility.
Motility of simulated T cells
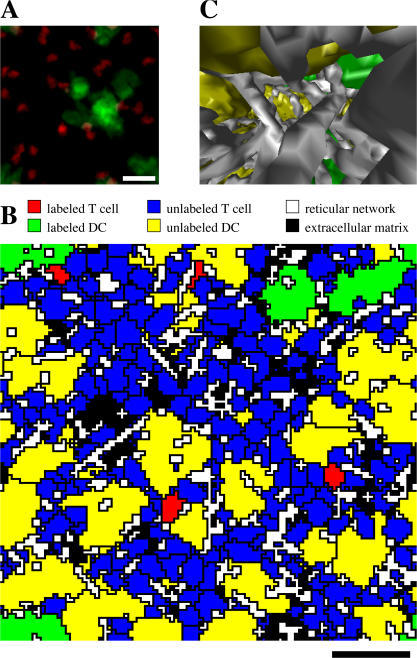
This minimal set of assumptions is sufficient to describe the basic properties of T cell motility found in experiments. As is the case in (the snapshots of) the experimental movies, the space appears to be relatively empty when one visualizes only the labeled cells in a maximum intensity top-view projection (Fig. 1 A and Video S2, available at http://www.jem.org/cgi/content/full/jem.20061278/DC1, which is strikingly similar to Movie 2 in Miller et al. [18]) or in a three-dimensional representation (Video S3). These movies give a false impression of emptiness, where in reality the space is densely packed. This becomes clear when all cells and RN are visualized in a cross section with a thickness of a single pixel (Fig. 1 B and Video S4), or when one considers what a T cell would perceive during its voyage through the simulated LN (Fig. 1 C and Videos S5 and S6).
Figure 1.
Simulated T cells and DCs in a densely packed space. (A) Snapshot with compression along the z direction (top-view) at time 00:00 showing labeled T cells and DCs. (B) Cross section through space at time 18:00 showing all cell types. (C) Three-dimensional snapshot at time 21:39 taken from the viewpoint of a T cell looking in its direction of motion showing RN (gray), labeled DCs (green), and nonlabeled DCs (yellow). Snapshots and cross sections along different directions yield similar pictures. Bars, 20 μm.
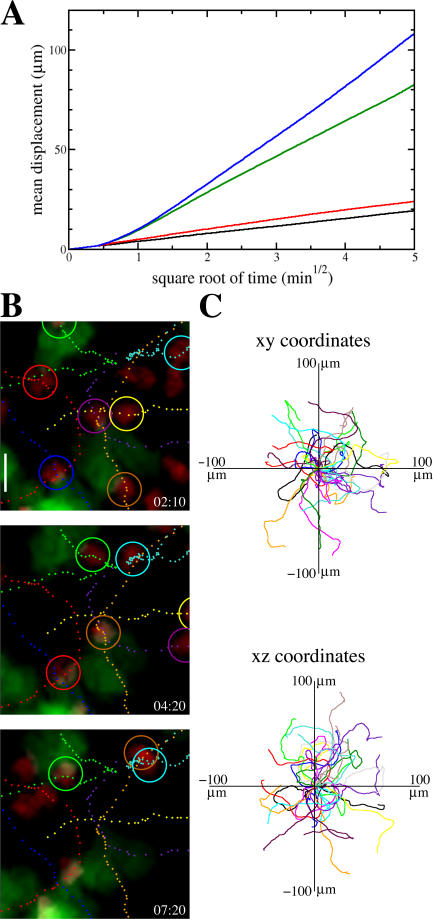
The dynamical properties of simulated T cells are in close correspondence with the experimental results (compare all panels in Fig. 2 with figures in Miller et al. [6]). The relation between mean displacement of simulated T cells and square root of time shows that in the short term (up to several minutes) they move in a consistent direction, but in the long term they perform a random walk (Fig. 2 A). Both small, random fluctuations in the T cell path and an environment that prevents T cells from keeping a consistent direction for a long time causes the random walk behavior. The persistence time and motility coefficient can be scaled by changing how often T cells update their target direction. Realistic motility coefficients (M) of 50−100 μm2 min−1 and persistence times of ∼2 min (6, 8, 18) are obtained when the target direction is updated every 15−20 s (Fig. 2 A). The random movement pattern of simulated T cells is confirmed by an analysis of the tracks along which they crawl. Individual tracks reveal persistent motion in the short term (Fig. 2 B), whereas an overlay of normalized T cell tracks shows that there is no preferred direction in the long run (Fig. 2 C).
Figure 2.
T cell motion. (A) Mean displacement plots for different update times of the target direction Δt (black line, Δt = 5 s, M ≈ 3 μm2 min−1; red line, Δt = 10 s, M ≈ 4 μm2 min−1; green line, Δt = 15 s, M ≈ 55 μm2 min−1; blue line, Δt = 20 s, M ≈ 102 μm2 min−1). (B) Sequence of compressions along the z direction (top-view) showing T cell movement over a period of several minutes (time shown in min:s). Tracks of 7 T cells (encircled) are shown at 10-s intervals (DCs green, T cells red). Bar, 10 μm. (C) Overlay of individual T cell tracks from a 10-min period in xy and xz coordinates after aligning their starting positions.
Simulated T cells self-organize into streams
Although the motility pattern exhibited by fluorescently labeled T cells justifies the conclusion that there is no large-scale bulk motion of these cells (6), we observe bulk motion on a local level in our simulations. This self-organization occurs because nearby T cells that travel in approximately the same direction will continue doing so, whereas nearby T cells that travel in alternative directions will often collide, having to adjust their preferred route.
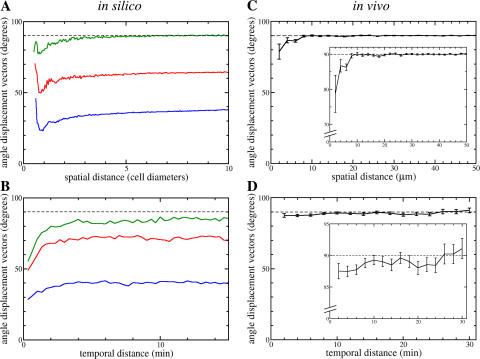
One of the advantages of our modeling approach is that we can study the behavior of all T cells. To study the presence of local streams of T cells, we plot the mean angle between the vectors of recent displacement of all T cell pairs versus the distance between the cells of the pairs (Fig. 3 A, green line). The angle between the displacement vectors of T cells that are far apart is on average 90°, which corresponds to uncorrelated motion. The average angle between the displacement of nearby T cells is <90°. Hence, nearby T cells are more likely to travel in the same direction than T cells that are far apart. Note that for T cells whose mean positions are clearly closer than the diameter of a single cell, the average angle between their displacement vectors increases again. This is because T cells approaching each other close enough to change each other's shape are likely to be involved in a collision.
Figure 3.
T cells travel in streams. The average angle between the displacement vectors of all possible T cell pairs in our simulations (A and B) and in 2PM experiments (C and D) as a function of the spatial (A and C) and temporal (B and D) distance between the members of pairs. Data shown are the average angle per group (bin sizes per panel: 0.05 cell diameters [A], 10 s [B], 2 μm [C], and 2 min [D]), and error bars represent standard error of the mean. Colored lines in in silico data: blue, no rods, 220 DCs, μmax = 4 × 106; red, 1,000 rods, 195 DCs, μmax = 4.5 × 106; green, 3,000 rods, 145 DCs, μmax = 6 × 106.
The presence of small streams of T cells is also visible when one considers temporal changes in the direction of movements at fixed spatial positions. Other T cells that pass through these positions at later time points are likely to originate from a similar direction as the T cells that were there before (Fig. 3 B, green line). This temporal correlation is almost completely lost in the longer term. Hence, the local T cell streams are extremely dynamic and turbulent, with constant changes in their direction of motion. The fact that the temporal correlation fails to attain 90° suggests the presence of locations that do not allow travel in all directions. This is a result of certain configurations of the rods that make up the in silico RN.
In our default simulation, bulk motion is still detectable two cells away and for up to 2 min (Fig. 3, A and B, green line). In simulations with a less dense RN (while keeping the average velocity of T cells approximately the same by adjusting the directional persistence of cells), the T cell streams become more global, which is visible both on the spatial and temporal scale (Fig. 3, A and B, red and blue line). Moreover, a decrease in RN density leads to increased displacement of T cells (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20061278/DC1).
Small, dynamic T cell streams are present in real LNs
To test if the turbulent streams predicted by our simulations are present in LNs, we next designed new 2PM experiments and performed the same spatial and temporal correlation analyses on the new data. In these experiments, large numbers of T cells (Video S7, available at http://www.jem.org/cgi/content/full/jem.20061278/DC1) were injected into mice to obtain as many data points as possible for the correlation analysis. This strategy enabled us to detect evidence for small, dynamic streams (Fig. 3, C and D). The effect of spatial and temporal distance on correlations in direction of movement is small but significant.
The average angle between the displacement of T cells <6 μm apart is significantly different from that of cells >6 μm apart (Fig. 3 C; P = 7.31 × 10−7). Hence, the spatial scale at which correlated movement occurs is approximately one cell diameter. Regarding the temporal scale (Fig. 3 D), a correlation in travel direction through fixed spatial positions is detectable for up to at least 6 min (in comparison with longer periods; P = 0.013). Collectively, these data demonstrate that T cell streams exist in real LNs and that they are extremely dynamic, as predicted by our simulations.
Contact duration and scanning rates
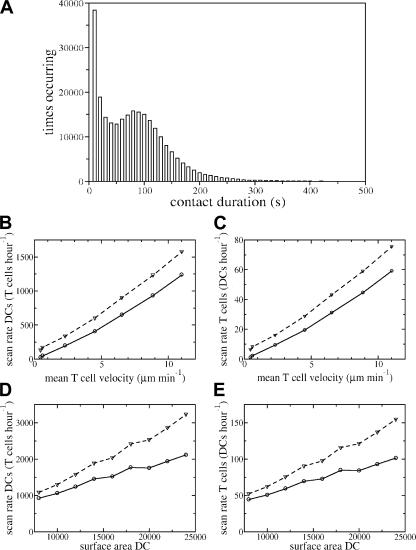
Knowing the exact whereabouts of all simulated cells at all times also allows one to determine precisely how long the contacts between simulated DCs and T cells last, as well as how many DCs each T cell scans per hour, and vice versa. Contacts take up to 10 min but most are much briefer (Fig. 4 A), with an average of 1.3 min (1.9 min when only interactions >1 min are included, as in Mempel et al. [8]). This is of the same order as the 3-min contacts found in experiments (7, 8, 18). Changing the adhesion between simulated T cells and DCs or the target surface area of DCs hardly affects the duration of contacts (not depicted). Note that this is because the adhesion between all T cells and all DCs is changed; the resulting fierce competition between T cells for DC contact explains this result.
Figure 4.
Contact duration and scanning rates. (A) Distribution of contact times measured over all T cells and all DCs. (B–E) Number of T cells that DCs scan per hour (B and D), and number of DCs that T cells scan per hour (C and E) as a function of the T cell velocity (B and C) and of the DC surface area (D and E). Simulations are performed for 15 min. T cell velocity is increased by changing μ from 0 to 6 × 106 with a step size 106 (B and C). In D and E, we change the target surface area of DCs. Dashed lines, total number of cells scanned; solid lines, number of unique cells scanned.
As hypothesized previously (7, 17), the scanning rate of both DCs and T cells becomes larger when we increase the average T cell velocity (Fig. 4, B and C) or the DC surface area (Fig. 4, D and E). Indeed, the number of different cells scanned is extremely low if T cells are not motile (Fig. 4, D and E). From experimental data, DCs were estimated to scan 500–5,000 T cells per hour (7, 17) and to be in contact with 250 T cells at any instant (7). When using a realistic surface area to volume ratio for a DC that is approximately threefold larger than a spherical cell of the same volume (7), a simulated DC is in contact with 75 T cells at each moment, occupying ∼40% of its surface area. Our DCs scan in total 3,000 T cells per hour, and 2,000 of these contacts are unique. This means that if antigen-specific T cells are present at a precursor frequency of 10−5, a population of 100 antigen-bearing DCs each scanning 2,000 different T cells per hour would detect specific T cells within 2 h with a probability of 95% (see Materials and methods). In silico T cells scan 150 DCs per hour, of which 100 contacts are unique (see Discussion for implications on thymic negative selection).
Periodic velocity fluctuations
The three-dimensional velocities of simulated T cells exhibit fluctuations from 0 to 25 μm min−1 (Fig. S2 A, available at http://www.jem.org/cgi/content/full/jem.20061278/DC1), i.e., are as large as those observed in experiments (data from Miller et al. [6] are replotted in Fig. S2 B). In the model, this is a consequence of the obstacles (RN, DCs, and other T cells) that T cells encounter on their preferred path, resulting in brief pauses. Miller et al. (6) performed a Fourier analysis on the velocity traces from single cells, using a time series of 64 data points. They found that T cells “stop” approximately every 2 min and suggested that this is regulated by an intrinsic control mechanism (our reanalysis of the velocity data is plotted in Fig. S2 B). We performed Fourier and autocorrelation analyses on velocities of simulated T cells and found that apparent periodicity can also result from pausing due to random obstacles (Fig. S2 A, most clearly visible in the autocorrelation analysis). This quasi-periodicity is a spurious result and is due to a combination of large velocity fluctuations (which is caused by obstacles; see Fig. S1 B) and a brief time series used for the analysis. This became clear when we analyzed longer time series, compared analyses of two- and three-dimensional velocities, and studied a modified model in which we describe T cells with a true internal clock (see Supplemental Results and Figs. S4 and S5). From this, we found that the velocity fluctuations in our simulations are random, but appear to have a characteristic period when one analyzes time series that are too brief. Hence, the time series used for analysis of experimental velocity data performed thus far were too brief to establish whether T cells have an internal clock that regulates velocity fluctuations over time.
Long time series in 2PM experiments
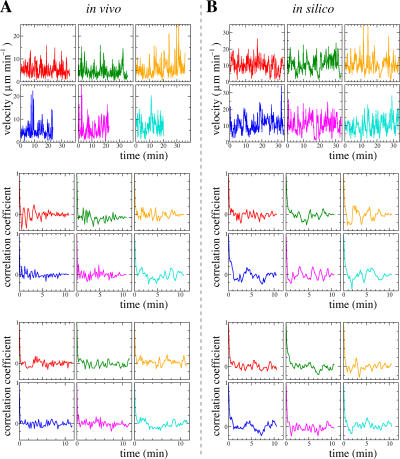
To find out if T cell stops are due to an intrinsic motility program or obstacles, i.e., to test our model-generated hypothesis, we performed new 2PM experimental work (Video S8, which is available at http://www.jem.org/cgi/content/full/jem.20061278/DC1, and Fig. 5 A; see Fig. 5 B for comparison with simulation results) with frequent (each 11 s) scanning of a large field. This enabled us to follow velocity fluctuations of individual T cells for 20–40 min (Fig. 5 A, top), which corresponds to 100–200 data points. Our simulations predict that longer time series should result in correlations fading out slowly when the environment causes the large velocity fluctuations, whereas they should become stronger when there is an internal clock in T cells. The number of data points is too low to extinguish the correlations completely (compare experimental and simulation results from the bottom panels of Fig. 5). However, our new experimental data clearly demonstrate that the evidence for periodic pausing does not increase when an autocorrelation analysis of only the first 64 data points of each T cell (Fig. 5 A, middle) is compared with the analysis for all available data points of the same T cells (Fig. 5 A, bottom). Hence, these results are consistent with our hypothesis that velocity fluctuations are environmentally determined rather than due to an intrinsic rhythmicity.
Figure 5.
T cell velocity fluctuations are environmentally determined. Velocity fluctuations (top) of six real T cells over a 20−40-min period (A) and of six simulated T cells (B). Autocorrelation analyses of the first 64 velocity data points (middle) and of all data points (bottom two panels) are shown.
DISCUSSION
We demonstrated that the dynamical properties of T cells (random walk, large velocity fluctuations) do not require T cells to pause regularly and subsequently choose a new random direction. In our three-dimensional model of the LN T cell area, both behaviors follow automatically from the simple assumption that T cells have a preferred direction of motion that is adjusted according to their recent displacement. Our 2PM experiments confirm the model prediction that the complicated T cell behaviors are environmentally determined phenomena: T cells seem to walk in a more or less consistent direction until they see obstacles on their path. Note that our simulated T cells still exhibit sharp turns if they move in an environment that is not densely packed with other cells or obstacles (see Video S9, available at http://www.jem.org/cgi/content/full/jem.20061278/DC1), which is due to stochastic fluctuations. However, this occurs less frequently than in a densely packed space.
The idea that T cell–persistent motion in the short term automatically leads to random walk behavior in the longer term, without the need for separate processes to explain these phenomena, is analogous to ideas on polymer dynamics (for an introduction see references 33 and 34). Two classical ideal chain models of polymers are directly relevant for this analogy: the freely jointed chain and the worm-like chain. In the freely jointed chain, segments of fixed length are randomly placed on top of each other. This would be similar to an ideal stop and go motion, in which T cells move perfectly straight for a fixed period of time, and subsequently choose a new, random direction. It results in a linear relation between squared displacement and polymer length (the latter is analogous to time in the T cell case). The worm-like chain considers a continuously flexible rod, whose orientation correlation diminishes exponentially with polymer length. Our description of T cell motion is similar to the worm-like chain model. For short polymers, this results in a linear relation between displacement and length, but it changes to a linear relation between squared displacement and length for longer polymers. This is because the orientation correlation is completely lost in the long run. For sufficiently long polymers, the worm-like chain is equivalent to a freely jointed chain with segments of size twice the persistence length. This analogy demonstrates that one cannot infer the mechanism underlying T cell behavior from the displacement versus square root of time relation alone.
Our model predicts the presence of extremely turbulent local T cell streams. We therefore performed new 2PM experiments and confirmed the presence of such streams in real LNs. The direction of travel through fixed positions is more or less maintained for at least 6 min, which demonstrates how dynamic this phenomenon is. Furthermore, small correlations in the direction of travel are detectable between cells that are approximately one cell diameter apart. In our simulations, the local T cell streams turn into global bulk motion when there are few inhomogeneities present (RN, DCs). Such strong global bulk motion may be unwelcome in areas of the LN with high DC density (around high endothelial venules; references 35 and 36) because T cells of appropriate antigen specificity need to be retained in that area for a sufficiently long time for activation. Hence, a function of the RN may be to prevent global bulk motion of T cells. The mean T cell velocity in deep areas of the T cell zone is higher than that in superficial areas (8). A possible explanation for this finding is that T cell bulk motion is more global in deep areas of LNs due to a lower density of obstacles (for instance-different DC density, as shown in Bajénoff et al. [35]).
Our simulations allow more precise estimates of DC scanning rates of T cells than can be obtained from experiments (although our estimates of course depend on the correctness of the chosen parameters). Experimental estimates (7, 17) remain imprecise because they are based on the small number of contacts between fluorescently labeled T cells and DCs, and the percentage of labeled T cells. Our scanning estimates are more direct because we know the whereabouts of all cells and can easily detect brief interactions of small contact area. Importantly, we are also able to count which contacts are unique, a property currently inaccessible to experimental techniques.
During negative selection in the thymic medulla, maturing T cells with a high affinity for self-antigens are deleted. Based on an average contact time between T cells and DCs of 5 min, Müller et al. (37) estimated that T cells scan maximally 4,000 DCs in 14 d. This, as well as other estimates related to the selection threshold of thymocytes (number of self-peptide–MHC molecules on DCs required for recognition), led them to conclude that there is not enough time for negative selection by all self-peptides. In our simulations, we find that T cells are able to scan 100 different DCs per hour. Hence, under the circumstances in our model we expect that T cells scan ∼34,000 different DCs in 14 d. The almost 10-fold difference between these estimates can be explained by the high number of very brief contacts (see Fig. 4 A), a brief inter-contact period, and the regular occurrence of contacts with multiple DCs in our simulations. It is important to realize that such scanning estimates will depend on the ratio of DCs and T cells (shown in Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20061278/DC1), a factor not taken into account by Müller et al. (37). Our new scanning estimate helps to solve the problem identified by Müller et al. (37); there may be enough time for T cells to see all self-peptides (see also Bandeira et al. [38]).
In recent 2PM experiments, the fibroblastic reticular cells that construct and ensheath the RN have been imaged (39), leading to very detailed images of the complete RN. The movies and analysis of the T cell behavior in these experiments demonstrate that T cells preferentially move along the RN fibers. In our simulations we studied scenarios with and without preferential adhesion of T cells to the RN, and found that the migratory behavior of T cells (random walk, large velocity fluctuations, and contact time between DCs and T cells; not depicted) was similar in both cases (compare Videos S2 and S10, available at http://www.jem.org/cgi/content/full/jem.20061278/DC1). This similarity can be explained by the fact that T cells are equal in their competition to adhere to the very dense and random RN. Combining our new experimental and modeling evidence for T cells moving in small, dynamic streams through the LN with the recent data by Bajénoff et al. (39), showing that T cells preferentially walk along the RN, our conjecture is that the micro-streams occur primarily alongside the RN fibers. We expect that a larger preference of T cells to adhere to the RN will lead to a stronger orientation of the T cell streams along the RN. This is one of the issues that will be interesting to address in the future by means of a combined experimental and modeling approach.
MATERIALS AND METHODS
Model description.
In the CPM (22, 23), several connected sites (with three-dimensional coordinates i, j, and k) on a lattice together comprise a cell that is of type τ(σ), where σ is the identification number of a cell. One of the important advantages of the assumption that cells occupy several sites on the lattice is that they have a shape (e.g., roundish or elongated), which can change over time. Sites that are in contact with other cells, RN, or ECM have a surface energy with their direct surroundings. Changes in cell configurations and movements over time occur because cells are assumed to minimize their surface energies. During a simulation it is constantly being considered how an extension of a lattice site into a random neighbor would change the surface energy. This is described in the so-called Hamiltonian, which consists of the sum of all surface energies J and a term λ(v−V)2 (v is the actual volume, V is the target volume, and λ is the inelasticity). The latter term is required to keep cells at a nonzero volume because otherwise the system could minimize its total surface energy by shrinking all cells to zero volume.
To describe DCs, which have a large surface area to volume ratio because of their dendrites, we extend the Hamiltonian with target surface area P and actual surface area p:
 |
(1) |
where δ is the Kronecker delta, and σi′j′k′ sums over all 26 neighbors (i.e., up to third order neighbors). The third term in the Hamiltonian is only applied to DCs. The probability that a lattice site is copied to a neighboring site is one if ΔH<−θ and e −(ΔH+θ)/T otherwise, where ΔH is the change in H due to the considered modification, θ represents a yield energy (the resistance to deform) and T represents the membrane fluctuation amplitude of cells. We made no special assumptions on the movement of DCs. Hence, their changes in shape and displacement are a result of membrane fluctuations alone.
To describe motile T cells, they are assumed to exhibit a target direction. Extensions of lattice sites that are approximately in this direction are more likely than other extensions; we implemented this by extending ΔH for T cells as follows:
 |
(2) |
where μ is the “directional propensity” of cells, and α is the angle between target direction and the considered displacement vector (the vector given by the coordinate of the position whose modification is considered and the mean position of the cell). The initial target direction of T cells is random, but it is updated each Δt seconds to become the displacement vector of the previous period. This gives rise to a self-adjusting type of motility. To describe cell turning in a realistic way, i.e., abrupt directional changes are not possible, the actual μ value of a T cell is also adjusted according to its recent displacement: μ = μmax e −ρ(1−cosβ), where μmax is the maximum directional propensity T cells can obtain, β is the angle between the displacement and target vector of the previous period, and ρ determines how rapidly the directional propensity declines when a turn is occurring. We use the directional propensity of T cells to adjust the average velocity of T cells to realistic values. The model as well as the measurements on cellular behavior were implemented in the C programming language.
Default model parameters.
When not otherwise noted, we use the default parameters as described in this subsection. We consider a wrapped cubic space (three-dimensional torus) with a length of 100 μm in all three dimensions. To set the spatial scale of the simulations, we choose one site of the lattice to equal 1 μm3. The RN consists of 3,000 rods with a radius of 1 μm and a height of 20 μm (in total taking up ∼17% of the space). We vary the density of the RN to investigate its impact on T cell behavior. In imaging experiments, fluorescently labeled T cells typically comprise 1−2% of all T cells (4, 7, 8, 17), whereas labeled DCs make up 10−30% of all DCs and 0.5% of total cellularity (7). According to this estimate, 2−5% of cells in LNs are DCs. Because DCs are not homogeneously spread in the LN, but preferentially reside around high endothelial venules (35, 36), we take the upper limit (5%) for calculating the number of DCs in our simulations. For simplicity, we consider the other 95% of cells in our simulations to be T cells. Together, this leads to the following numbers of cells in the space we consider: 30 fluorescently labeled T cells, 3,000 nonlabeled T cells, 10 fluorescently labeled DCs, and 135 nonlabeled DCs. Cells are initialized at a random position, as a 27 μm3 block, and subsequently grow to their target volumes (150 μm3 for T cells and 2,200 μm3 for DCs, corresponding to diameters of, respectively, 6.6 μm and 16.1 μm if they attain a spherical shape). The surface area of a DC is equal to the number of three-dimensional Moore-neighbors (this includes all direct diagonal neighbors) that do not belong to the DC, summed over all DC sites. Its default target surface area is 12,000, which is ∼1.58 times the value it would have if it were a sphere (larger values result in tentacles breaking off frequently). We investigate the effect of target surface area on scanning rates. The mentioned cell and RN density leaves ∼5% of the space for ECM, which fits well with the impression of a densely packed environment one gets from looking at histology slides. Decreasing the space for ECM to 0% does not influence the results qualitatively (not depicted). We assume that T cells have a small preference to adhere to DCs. Further, we perform simulations with and without preferential adhesion of T cells to the RN, which results in similar migratory behavior (compare Videos S2 and S10; refer to Discussion). All other movies and results shown in the paper are for the case without preferential adhesion between T cells and RN. Other than T cell–DC and T cell–RN interactions, we assume that there is no differential adhesion between DCs, T cells, RN, and ECM. The default surface energy parameters and resulting adhesion strengths are shown in Table I (surface tension between cell types x and y, γx,y, is calculated from the surface energies as follows [23]: γx,y = J x,y − (J x,x +J y,y)/2). Other default parameters used: μmax= 6 × 106, Δt = 20 s, ρ = 3, T = 5 × 105, θ = 0, λv = 5 × 105, λp = 600. Results shown are representative of several simulations.
Table I.
Default surface energies and surface tensions
| T cell | DC | RN | ECM | |
|---|---|---|---|---|
| T cell | JT,T = 106 | γT,DC =− 2 × 103 | γT,RN = 0 | γT,ECM = 0 |
| DC | JDC,T = 5.03 × 106 | JDC,DC = 104 | γDC,RN = 0 | γDC,ECM = 0 |
| RN | JRN,T = 5 × 105 | JRN,DC = 5 × 103 | JRN,RN = 0 | γRN,ECM = 0 |
| ECM | JECM,T = 5 × 105 | JECM,DC = 5 × 103 | JECM,RN = 0 | JECM,ECM = 0 |
Surface tensions (γ) are shown in bold font, and surface energies (J) are shown in normal font.
Scanning of simulated T cells by DCs.
To calculate the consequences of DC scanning rates, we consider the probability with which a population of n antigen–bearing DCs fails to detect antigen-specific T cells that are present with precursor frequency fp within t hours. Given that DCs scan s different T cells per hour, this probability is (1−sfp)nt, which, for low sfp, approximately equals  . Hence, with a probability of 95%, the DC population will detect such T cells within t = ln(20)/(sfpn) hours (this calculation is similar to the one performed by Miller et al. [7]).
. Hence, with a probability of 95%, the DC population will detect such T cells within t = ln(20)/(sfpn) hours (this calculation is similar to the one performed by Miller et al. [7]).
Simulation measurements.
One Monte Carlo time step in the simulations (after which all positions in the lattice have been considered for updating) corresponds to 1 s in real time. Measurements start after an initial 400 s, defined as time 00:00 (min:s). The mean positions of T cells are registered each 10 s and are used to calculate displacements and velocities. Motility coefficients are estimated according to the formula  (explained in Sumen et al. [40]). Interactions between T cells and DCs are counted as contacts when they have at least one direct neighbor.
(explained in Sumen et al. [40]). Interactions between T cells and DCs are counted as contacts when they have at least one direct neighbor.
Imaging experiments.
CD4+ T cells were isolated by magnetic negative selection (Miltenyi Biotec) from the spleens and LNs of C57BL6 mice (The Jackson Laboratories). The T cell purity of this method is typically >90% CD4+ CD3+ as assessed by flow cytometry. T cells were labeled with 5 μM CFSE or 10 μM CMTMR (Invitrogen) for 30 min at 37°C and washed once. 10−50 million cells (depending on the experiment) were transferred into B6 mice by tail vein injection. 18 h after T cell transfer, mice were killed by CO2 asphyxiation and cervical LNs were removed for microscopy. LNs were secured in the flow chamber with a thin film of VetBond (3M) and maintained at 37°C by perfusing the chamber with warm RPMI bubbled with a mixture of 95% O2 and 5% CO2 (Carbagen gas; reference 4). Time-lapse imaging was performed using a custom built two-photon microscope at the Washington University School of Medicine. CFSE- and CMTMR-labeled T cells were excited by a Chameleon Ti:sapphire laser (Coherent) tuned to 780−800 nm. CFSE fluorescence was collected at 490−540 nm and CMTMR fluorescence was collected above 540 nm. ImageWarp (A&B software) was used to control the various hardware devices during real-time acquisition and to process and archive the image data. Each time point consists of a 200 × 220 × 40−60 μm volume (x and y = 2 pixels μm−1). Z stacks were acquired by taking sequential steps at 2−2.25-μm spacing with an automated z-stepper motor (Prior). For long-term tracking, z stacks of 21 steps were acquired at 11-s intervals (10−15 million CFSE-labeled T cells were injected in this experiment). For T cell streams, three trials were obtained in two experiments. One of these trials consisted of z stacks of 21 steps at 24-s intervals (25 million CFSE-labeled T cells were injected in this trial). The other two trials consisted of z stacks of 31 steps at 42-s intervals (25 million CFSE- and 25 million CMTMR-labeled T cells were injected in these two trials). To increase signal contrast, we averaged 10 video frames for each z-slice. Multidimensional rendering and three- dimensional cell tracking were performed with Volocity (Improvision) and Imaris (Bitplane). The automated tracking was checked manually for all cases where cells approach each other more closely than 2 μm (this ruled out the risk of tracking cells twice, which could be due to interference between red and green fluorescence). Because the continuous flow of medium over and through the LN results in small global drift, we corrected the three- dimensional coordinates in each experiment for this artifact by using the average displacement of all (or a subset of low velocity) cell movements. All experiments were approved by the Washington University School of Medicine Animal Studies Committee, and veterinary care was provided by the Division of Comparative Medicine.
Autocorrelation and Fourier analysis.
Autocorrelation and Fourier analyses (using the Welch window) of both simulated and experimental data are performed using the program Xmgrace (available at http://plasma-gate.weizmann.ac.il/Grace) with the average velocity of each T cell shifted to 0. In our 2PM experiments with long-term tracking, cells that had an average velocity of <4 μm min−1 or had a gradual velocity decrease over a prolonged period (>15 min) were excluded from the analysis. To reanalyze the T cell velocity data from Miller et al. (6), we read out the data using the program Plot Digitizer (available at http://plotdigitizer.sourceforge.net). We padded the 61 data points of the first T cell with three zeroes.
Analysis to measure correlated movement.
To investigate the spatiotemporal scale of bulk motion, T cell pairs are grouped according to either the spatial or the temporal distance between their members. The average angle is calculated per group and plotted in Fig. 3. For the spatial scale at which bulk motion occurs in our simulations, all cell pairs at a single time step are considered. For the temporal scale, only cells whose mean positions come within a 3-μm radius of fixed locations (taken as mean positions of cells at the start of measurement) are considered.
For our analysis on the presence of T cell streams in real LNs, we combined data from three trials. Tracks of <4 min from all trials were excluded. Other than that, each time point of each cell was included in the analysis. We performed two-tailed Mann-Whitney tests to investigate whether the average angle between direction of travel for cells with small spatial or temporal distance differs significantly from that of cells that are a large (spatial or temporal) distance apart.
Online supplemental material.
The supplemental material contains additional results on periodicity of T cell velocity fluctuations. Fig. S1 demonstrates the effect of density of the RN on T cell displacement and velocity fluctuations. Fig. S2 shows a comparison of velocity fluctuations of simulated and real T cells for brief time series. Fig. S3 shows velocity fluctuations of in silico T cells for a long time series and for a model with programmed T cell stop. Fig. S4 illustrates the importance of studying three-dimensional instead of two-dimensional velocities for analyses of periodicity. Fig. S5 shows the effect of the T/DC ratio on the scanning rate of T cells and DCs. Video S1 shows what the RN looks like in our model. Videos S2, S3, and S4 demonstrate the behavior of in silico T cells and DCs in a maximum intensity top-view projection, three-dimensional representation, and cross section, respectively. Videos S5 and S6 show the LN from the perspective of a T cell during its voyage. Video S7 and S8 show maximum intensity top-view projections of two-photon experiments with different numbers of injected T cells. Video S9 shows a simulation of 30 T cells traveling through ECM in the absence of other cells and RN. Video S10 shows a simulation where T cells have a strong preference to adhere to the RN. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20061278/DC1.
Supplemental Material
Acknowledgments
We thank V. Grieneisen for suggesting how to model directional propensity of T cells, and C. Keşmir, V. Ganusov, and three anonymous referees for critical comments on a previous version of the paper. This work was supported by a Human Frontier Science Program (HFSP) grant (J.B. Beltman) and by Netherlands Organisation for Scientific Research (NWO) grants (A.F.M. Marée and R.J. de Boer).
R.J. de Boer instigated the research for this work. A.F.M. Marée developed the CPM simulation environment. J.B. Beltman, A.F.M. Marée, and R.J. de Boer devised the model to analyze LN processes, and J.B. Beltman and A.F.M. Marée implemented these algorithms. J.B. Beltman performed the simulations and analyzed their outcome. M.J. Miller and J.N. Lynch performed the 2PM experiments and analyzed the raw data. J.B. Beltman analyzed the experimental data for periodicity and for T cell streams. J.B. Beltman wrote the paper, and all authors commented extensively on the manuscript.
The authors have no conflicting financial interests.
Abbreviations used: 2PM, two-photon microscopy; CPM, Cellular Potts Model; ECM, extracellular matrix; RN, reticular network.
J.B. Beltman and A.F.M. Marée contributed equally to this work.
References
- 1.Arstila, T.P., A. Casrouge, V. Baron, J. Even, J. Kanellopoulos, and P. Kourilsky. 1999. A direct estimate of the human αβ T cell receptor diversity. Science. 286:958–961. [DOI] [PubMed] [Google Scholar]
- 2.Keşmir, C., J.A.M. Borghans, and R.J. de Boer. 2000. Diversity of human αβ T cell receptors. Science. 288:1135. [DOI] [PubMed] [Google Scholar]
- 3.Blattman, J.N., R. Antia, D.J. Sourdive, X. Wang, S.M. Kaech, K. Murali-Krishna, J.D. Altman, and R. Ahmed. 2002. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 195:657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller, M.J., S.H. Wei, I. Parker, and M.D. Cahalan. 2002. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 296:1869–1873. [DOI] [PubMed] [Google Scholar]
- 5.Bousso, P., N.R. Bhakta, R.S. Lewis, and E. Robey. 2002. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science. 296:1876–1880. [DOI] [PubMed] [Google Scholar]
- 6.Miller, M.J., S.H. Wei, M.D. Cahalan, and I. Parker. 2003. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc. Natl. Acad. Sci. USA. 100:2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller, M.J., S.H. Wei, M.D. Cahalan, and I. Parker. 2004. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc. Natl. Acad. Sci. USA. 101:998–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mempel, T.R., S.E. Henrickson, and U.H. von Andrian. 2004. T cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 427:154–159. [DOI] [PubMed] [Google Scholar]
- 9.Hugues, S., L. Fetler, L. Bonifaz, J. Helft, F. Amblard, and S. Amigorena. 2004. Distinct T cell dynamics in lymph nodes during the induction of tolerance and immunity. Nat. Immunol. 5:1235–1242. [DOI] [PubMed] [Google Scholar]
- 10.Shakhar, G., R.L. Lindquist, D. Skokos, D. Dudziak, J.H. Huang, M.C. Nussenzweig, and M.L. Dustin. 2005. Stable T cell-dendritic cell interactions precede the development of both tolerance and immunity in vivo. Nat. Immunol. 6:707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witt, C.M., S. Raychaudhuri, B. Schaefer, A.K. Chakraborty, and A.E. Robey. 2005. Directed migration of positively selected thymocytes visualized in real time. PLoS Biol. 3:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celli, S., Z. Garcia, and P. Bousso. 2005. CD4 T cells integrate signals delivered during successive DC encounters in vivo. J. Exp. Med. 202:1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinselmeyer, B.H., J. Dempster, A.M. Gurney, D. Wokosin, M. Miller, H. Ho, O.R. Millington, K.M. Smith, C.M. Rush, I. Parker, et al. 2005. In situ characterization of CD4+ T cell behavior in mucosal and systemic lymphoid tissues during the induction of oral priming and tolerance. J. Exp. Med. 201:1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bajénoff, M., B. Breart, A.Y.C. Huang, H. Qi, J. Cazareth, V.M. Braud, R.N. Germain, and N. Glaichenhaus. 2006. Natural killer cell behavior in lymph nodes revealed by static and real-time imaging. J. Exp. Med. 203:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellino, F., A.Y. Huang, G. Altan-Bonnet, S. Stoll, C. Scheinecker, and R.N. Germain. 2006. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 440:890–895. [DOI] [PubMed] [Google Scholar]
- 16.Wei, S.H., I. Parker, M.J. Miller, and M.D. Cahalan. 2003. A stochastic view of lymphocyte motility and trafficking within the lymph node. Immunol. Rev. 195:136–159. [DOI] [PubMed] [Google Scholar]
- 17.Bousso, P., and E. Robey. 2003. Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nat. Immunol. 4:579–585. [DOI] [PubMed] [Google Scholar]
- 18.Miller, M.J., O. Safrina, I. Parker, and M.D. Cahalan. 2004. Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J. Exp. Med. 200:847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei, S.H., H. Rosen, M.P. Matheu, M.G. Sanna, S.-K. Wang, E. Jo, C.-H. Wong, I. Parker, and M.D. Cahalan. 2005. Sphingosine 1-phosphate type 1 receptor antagonism inhibits transendothelial migration of medullary T cells to lymphatic sinuses. Nat. Immunol. 6:1228–1235. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty, A.K., M.L. Dustin, and A.S. Shaw. 2003. In silico models for cellular and molecular immunology: successes, promises and challenges. Nat. Immunol. 4:933–936. [DOI] [PubMed] [Google Scholar]
- 21.Witt, C., S. Raychaudhuri, and A. Chakraborty. 2005. Movies, measurement, and modeling: the three Ms of mechanistic immunology. J. Exp. Med. 201:501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graner, F., and J.A. Glazier. 1992. Simulation of biological cell sorting using a two-dimensional extended Potts model. Phys. Rev. Lett. 69:2013–2016. [DOI] [PubMed] [Google Scholar]
- 23.Glazier, J.A., and F. Graner. 1993. Simulation of the differential adhesion driven rearrangement of biological cells. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 47:2128–2154. [DOI] [PubMed] [Google Scholar]
- 24.Mombach, J.C., J.A. Glazier, R.C. Raphael, and M. Zajac. 1995. Quantitative comparison between differential adhesion models and cell sorting in the presence and absence of fluctuations. Phys. Rev. Lett. 75:2244–2247. [DOI] [PubMed] [Google Scholar]
- 25.Hogeweg, P. 2000. Evolving mechanisms of morphogenesis: on the interplay between differential adhesion and cell differentiation. J. Theor. Biol. 203:317–333. [DOI] [PubMed] [Google Scholar]
- 26.Marée, A.F.M., A.V. Panfilov, and P. Hogeweg. 1999. Phototaxis during the slug stage of Dictyostelium discoideum: a model study. Proc. R. Soc. Lond. B. Biol. Sci. 266:1351–1360. [Google Scholar]
- 27.Marée, A.F.M., and P. Hogeweg. 2001. How amoeboids self-organize into a fruiting body: multicellular coordination in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 98:3879–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keşmir, C., and R.J. de Boer. 2003. A spatial model of germinal center reactions: cellular adhesion based sorting of B cells results in efficient affinity maturation. J. Theor. Biol. 222:9–22. [DOI] [PubMed] [Google Scholar]
- 29.Graner, F. 1993. Can surface adhesion drive cell-rearrangement? Part I: biological cell-sorting. J. Theor. Biol. 164:455–476. [Google Scholar]
- 30.Katakai, T., T. Hara, M. Sugai, H. Gonda, and A. Shimizu. 2004. Lymph node fibroblastic reticular cells construct the stromal reticulum via contact with lymphocytes. J. Exp. Med. 200:783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard, T.D., and G.G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- 32.Friedman, R.S., J. Jacobelli, and M.F. Krummel. 2005. Mechanisms of T cell motility and arrest: deciphering the relationship between intra- and extracellular determinants. Semin. Immunol. 17:387–399. [DOI] [PubMed] [Google Scholar]
- 33.Doi, M., and S.F. Edwards. 1986. The Theory of Polymer Dynamics. Oxford University Press, Oxford. 391 pp.
- 34.Rubinshtein, M., and R.H. Colby. 2003. Polymer Physics. Oxford University Press, Oxford. 454 pp.
- 35.Bajénoff, M., S. Granjeaud, and S. Guerder. 2003. The strategy of T cell antigen-presenting cell encounter in antigen-draining lymph nodes revealed by imaging of initial T cell activation. J. Exp. Med. 198:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katakai, T., T. Hara, J. Lee, H. Gonda, M. Sugai, and A. Shimizu. 2004. A novel reticular stromal structure in lymph node cortex: an immuno-platform for interactions among dendritic cells, T cells and B cells. Int. Immunol. 16:1133–1142. [DOI] [PubMed] [Google Scholar]
- 37.Müller, V., and S. Bonhoeffer. 2003. Quantitative constraints on the scope of negative selection. Trends Immunol. 24:132–135. [DOI] [PubMed] [Google Scholar]
- 38.Bandeira, A., and J. Faro. 2003. Quantitative constraints on the scope of negative selection: robustness and weaknesses. Trends Immunol. 24:172–173. [DOI] [PubMed] [Google Scholar]
- 39.Bajénoff, M., J. Egen, L. Koo, J. Laugier, F. Brau, N. Glaichenhaus, and R. Germain. 2006. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 25:989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sumen, C., T.R. Mempel, I.B. Mazo, and U.H. von Andrian. 2004. Intravital microscopy: visualizing immunity in context. Immunity. 21:315–329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.