Abstract
We previously reported (Bell, D., P. Chomarat, D. Broyles, G. Netto, G.M. Harb, S. Lebecque, J. Valladeau, J. Davoust, K.A. Palucka, and J. Banchereau. 1999. J. Exp. Med. 190: 1417–1426) that breast cancer tumors are infiltrated with mature dendritic cells (DCs), which cluster with CD4+ T cells. We now show that CD4+ T cells infiltrating breast cancer tumors secrete type 1 (interferon γ) as well as high levels of type 2 (interleukin [IL] 4 and IL-13) cytokines. Immunofluorescence staining of tissue sections revealed intense IL-13 staining on breast cancer cells. The expression of phosphorylated signal transducer and activator of transcription 6 in breast cancer cells suggests that IL-13 actually delivers signals to cancer cells. To determine the link between breast cancer, DCs, and CD4+ T cells, we implanted human breast cancer cell lines in nonobese diabetic/LtSz-scid/scid β2 microglobulin–deficient mice engrafted with human CD34+ hematopoietic progenitor cells and autologous T cells. There, CD4+ T cells promote early tumor development. This is dependent on DCs and can be partially prevented by administration of IL-13 antagonists. Thus, breast cancer targets DCs to facilitate its development.
Tumor development depends on the interaction between cancer cells and surrounding nonmalignant stroma. Stroma is composed of nonhematopoietic cells (fibroblasts and endothelial cells) and immune cells from both the innate and the adaptive immune systems (1, 2). These two arms of the immune system are connected by DCs (3–6). DCs induce and maintain immune response and, as opposed to macrophages, are able to prime naive lymphocytes. The outcome of this interaction depends on DC activation/maturation (3). Thus, presentation of antigen by immature (nonactivated) DCs leads to tolerance (7–9), whereas mature DCs are geared toward the launching of antigen-specific immunity (10). Furthermore, vaccination with antigen-loaded DCs in both mice and humans can lead to the break of tolerance to cancer (for review see 11). Therefore, DCs might represent an early target for subversion by developing tumors.
The immunological consequences of DC infiltration are not well understood, although many studies in humans reported infiltration of various tumor types with DCs (for review see 12). Inhibition of DC maturation and function is thought to represent one of the means through which tumors evade the immune system (12). For example, increased production of vascular endothelial growth factor (13) inhibits DC maturation (14), thereby resulting in the induction of tolerance. IL-6 secreted by breast cancer cells can switch monocyte differentiation into macrophages at the expense of DCs (15), thereby skewing antigen presentation toward antigen degradation (16). Finally, certain tumors were shown to promote differentiation of IL-10 and/or TGF-β–secreting DCs that in turn expand CD4+CD25+ regulatory T cells (17–19). These are able to inhibit antitumor effector cells, thereby contributing to tumor escape (20).
We found that human breast cancer tumors are infiltrated with DCs (21), including immature myeloid DCs in tumor beds and mature DC-LAMP+ DCs in peritumoral areas. Mature DCs are often found in clusters with CD4+ T cells, suggesting an ongoing immune response (21). The presence of mature DCs outside lymphoid organs is linked with inflammation and can be observed in the synovia of patients with rheumatoid arthritis (22, 23) (24) or in the blood of patients with systemic autoimmune disease (25, 26). However, the immunological consequences of the presence of mature DCs in tumors remain unknown.
In this paper, we have studied the CD4+ T cells infiltrating human breast cancer tumors. We found the presence of IL-13–secreting CD4+ T cells. We also found IL-13 staining on breast cancer cells. To understand the role of IL-13 in vivo, we used our model of humanized mice (27), which we additionally grafted with breast cancer cell lines (unpublished data). These immunodeficient nonobese diabetic/LtSz-scid/scid β2 microglobulin–deficient (NOD/SCID/β2m−/−) mice transplanted with human CD34+ hematopoietic progenitor cells (HPCs) develop all subsets of human DCs and B cells (27). T cells are adoptively transferred. We found that breast cancer polarizes CD4+ T cells in vivo via DCs. These polarized CD4+ T cells secrete IL-13, which contributes to accelerated tumor development.
RESULTS
Breast cancer tumors are infiltrated with CD4+ T cells secreting IL-13
We measured T cell cytokines, both type 1 (IFN-γ) and type 2 (IL-4 and IL-13), in supernatants of breast cancer tumor fragments activated for 16 h with PMA/ionomycin (n = 19; tumor characteristics are given in Table I). As shown in Table II, high levels of IL-2 (3.3 ± 0.8 ng/ml; 17 out of 19 samples) and IFN-γ (4.1 ± 1.5 ng/ml; 17 out of 19 samples), as well as IL-13 (209 ± 67 pg/ml; 15 out of 19 samples) and IL-4 (33 ± 11 pg/ml; 9 out of 19 samples; mean ± SEM for all), were found. The levels were significantly higher in supernatants from tumor sites compared with supernatants from macroscopically uninvolved surrounding tissue (Table II).
Table I.
Tumor sample characteristics
| Sample no. | Histopathological diagnosis | Grade | Stage |
|---|---|---|---|
| 7 | Invasive duct carcinoma | III | IIA |
| 8 | In-situ and invasive duct carcinoma | III | na |
| 9 | Infiltrating carcinoma, mixed lobular, and ductal | I | IIB |
| 11 | Infiltrating ductal carcinoma, microinvasive | Intermediate | na |
| 12 | Invasive high grade ductal carcinoma | nd | IIB |
| 13 | Infiltrating ductal carcinoma | II | na |
| 15 | Infiltrating ductal carcinoma with lobular feature | II | I |
| 16 | Infiltrating carcinoma, predominantly ductal with lobular feature |
I | I |
| 17 | Invasive duct carcinoma | I | na |
| 18 | Invasive duct carcinoma | I and II | na |
| 19 | In-situ and invasive ductal carcinoma | II | I |
| 20 | In-situ and invasive ductal carcinoma | III | IIA |
| 21 | Infiltrating lobular carcinoma, pleomorphic type | nd | na |
| 22 | Invasive ductal carcinoma | II | na |
| 23 | In-situ and invasive ductal carcinoma | III | IIB |
| 24 | In-situ and invasive ductal carcinoma | III | IIA |
| 25 | Multifocal ductal carcinoma in situ with comedo necrosis | III | I |
| 26 | In-situ and invasive duct carcinoma | III | IIA |
| 27 | Invasive and in-situ ductal carcinoma | III | na |
Characteristics of analyzed breast cancer tumors from patients. Histopathology, tumor grade, and clinical stage are shown. nd, not determined; na, not available.
Table II.
T cell cytokines in supernatants of whole-tumor fragments or surrounding tissue
|
|
IL-2 (pg/ml)
|
IFN-γ (pg/ml)
|
IL-4 (pg/ml)
|
IL-13 (pg/ml)
|
||||
|---|---|---|---|---|---|---|---|---|
| Sample no. | ST | Tumor | ST | Tumor | ST | Tumor | ST | Tumor |
| 7 | 1,041 | 5,489 | 1,030 | 2,929 | 5 | 13 | 0 | 123 |
| 8 | 12 | 7,517 | 0 | 1,311 | 0 | 33 | 0 | 343 |
| 9 | 0 | 588 | 0 | 798 | 0 | 5 | 0 | 85 |
| 11 | 114 | 3,512 | 181 | 2,459 | 0 | 199 | 0 | 310 |
| 12 | 348 | 2,108 | 904 | 3,887 | 22 | 66 | 43 | 395 |
| 13 | 262 | 280 | 83 | 149 | 0 | 0 | 0 | 24 |
| 15 | 750 | 1,456 | 1,013 | 1,769 | 5 | 25 | 0 | 60 |
| 16 | 0 | 236 | 0 | 543 | 0 | 0 | 0 | 0 |
| 17 | 164 | 2,641 | 218 | 3,554 | 0 | 0 | 0 | 19 |
| 18 | 0 | 75 | 0 | 124 | 0 | 0 | 0 | 47 |
| 19 | 172 | 6,281 | 211 | 13,170 | 0 | 43 | 0 | 128 |
| 20 | 8,410 | 7,067 | 7,304 | 6,774 | 0 | 11 | 54 | 87 |
| 21 | 1,436 | 13,022 | 3,087 | 27,599 | 6 | 39 | 6 | 931 |
| 22 | 0 | 5 | 0 | 0 | 2 | 3 | 0 | 0 |
| 23 | 1,665 | 0 | 558 | 0 | 6 | 2 | 6 | 0 |
| 24 | 590 | 870 | 135 | 1,376 | 6 | 6 | 12 | 31 |
| 25 | 81 | 2,371 | 92 | 777 | 7 | 52 | 3 | 263 |
| 26 | 2,035 | 3,052 | 1,352 | 1,855 | 13 | 49 | 63 | 123 |
| 27 | 1,815 | 7,553 | 1,454 | 8,959 | 10 | 77 | 84 | 1,001 |
| p-value (t test) |
0.007 | 0.03 | 0.01 | 0.007 | ||||
The breast cancer microenvironment is rich in type 2 cytokines. Size-comparable (∼10-mm3) fragments of breast cancer tumor or macroscopically uninvolved surrounding tissue (ST) were stimulated for 16 h with PMA/ionomycin, and the cytokines IL-2, IFN-γ, IL-4, and IL-13 were analyzed in the supernatant by multiplex cytokine analysis (pg/ml; n = 19 patients). Values for each analyzed patient are shown. p-values reflect a comparison of cytokine concentration in the supernatants of breast cancer tumor or macroscopically uninvolved surrounding tissue (using the paired t test).
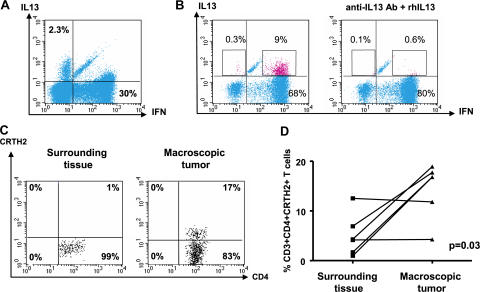
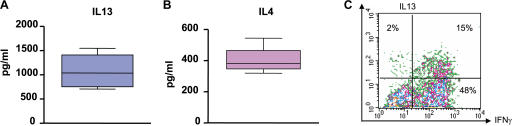
Flow cytometry on single-cell suspensions demonstrated the presence of T cell infiltrate with a prevalence of CD3+CD4+ T cells over CD3+CD8+ T cells (11 ± 3% and 5 ± 1%, respectively; n = 18; P = 0.01). Intracellular staining of single-cell suspensions after 5 h of activation with PMA/ionomycin demonstrated expression of IFN-γ, as well as IL-13, by CD4+ T cells (Fig. 1, A and B). The IL-13 staining was specific, as it could be blocked by excess recombinant IL-13 (Fig. 1 B). The mean frequency of IL-13–expressing CD4+ T cells in 11 samples analyzed was 4 ± 0.6% (range 0.2–9%). Two types of CD4+ T cell staining were observed: double-positive cells expressing both IL-13 and IFN-γ (Fig. 1 B) in a majority of tumors and, in some tumors, single-positive cells expressing either IL-13 or IFN-γ (Fig. 1 A), the latter one consistent with the definition of T cell polarization (28). CD4+ T cells expressing chemoattractant receptor–homologous molecule expressed on Th2 cells (CRTH2) (29, 30) were detected by flow cytometry (14 ± 2%; n = 6; Fig. 1 C). In four out of six analyzed tumors, the infiltration with CRTH2+CD4+ T cells was significantly higher in the tumor sites compared with macroscopically uninvolved surrounding tissue (P = 0.03; Fig. 1 D). Thus, breast cancer tumors from patients are infiltrated with CD4+ T cells secreting IFN-γ and IL-13.[ID]FIG3[/ID][ID]FIG3[/ID]
Figure 1.
Breast cancer is infiltrated with CD4+ T cells secreting type 1 and type 2 cytokines. (A and B) Intracellular staining of IL-13 and IFN-γ on gated CD3+CD4+ T cells infiltrating breast cancer. Flow cytometry plots (A) and (B) represent samples from two different patients. To demonstrate specificity, anti–IL-13 mAb was pretreated with rhIL-13 before staining. (C) Representative flow cytometry analysis of CRTH2 expression by gated CD3+CD4+ T cells. In A–C, numbers indicate the percentage of cells positive for the specified marker. (D) Percentage of CRTH2+CD4+ T cells (ordinate) within breast cancer tumor or corresponding macroscopically not involved surrounding tissue (n = 6 patients). P = 0.03 using the paired t test.
Breast cancer cells express IL-13 and phosphorylated STAT6 (pSTAT6)
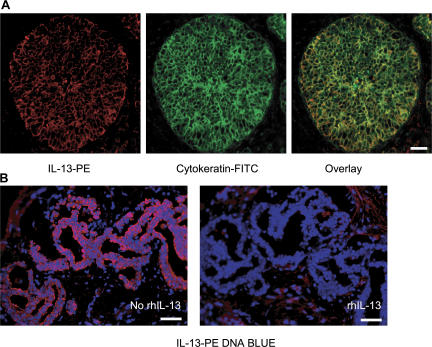
To determine IL-13 expression in situ, breast cancer tissue sections were stained with specific mAb and analyzed by immunofluorescence. As expected based on flow cytometry analysis, T cells staining with IL-13 could be found (unpublished data). However, a major IL-13 staining was found in large cells organized in nests, consistent with the staining on cancer cells (Fig. 2 A, left). To confirm this, the tissue was counterstained with cytokeratin (Fig. 2 A, middle). Indeed, nearly all cytokeratin-expressing cancer cells coexpressed IL-13 (Fig. 2 A, right). This staining was abolished by the excess of recombinant human (rh) IL-13 (Fig. 2 B) demonstrating specificity. IL-13 staining could be detected in 11 out of 11 breast cancer tumor samples analyzed (Fig. 2, A and B).
Figure 2.
IL-13 staining on breast cancer cells. (A) Frozen breast cancer tumor sections were labeled with cytokeratin (green) and IL-13 (red). (B) Section from tumor obtained from a different patient. IL-13 staining of breast cancer cells can be inhibited by rhIL-13. Bars: (A) 20 μm; (B) 50 μm.
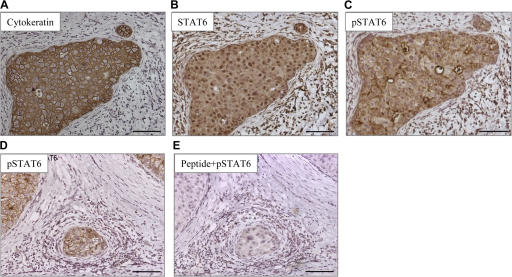
Phosphorylation of STAT6 is considered a signature of IL-13 (and/or IL-4) signaling (31–33). All analyzed breast cancer tumor samples showed expression of STAT6 by cancer cells (Fig. 3, A and B; and not depicted). 8 out of 11 tumors showed cytoplasmic expression of pSTAT6, and three of these tumors showed strong expression (Fig. 3, C and D). The staining was blocked by excess peptide that was used to generate anti-pSTAT6 antibody demonstrating specificity (Fig. 3 E).
Figure 3.
Breast cancer cells express pSTAT6. Immunohistochemistry on paraffin-embedded tissue sections (A–C) and (D and E) represent tumors from two different patients. (A–C) A nest of breast cancer cells expressing cytokeratin (A) can also be stained with antibody recognizing STAT6 (B), as well as with an antibody recognizing pSTAT6 (C). (D and E) pSTAT6 staining is predominantly found on cancer nests (D) and can be blocked by a peptide used to generate the antibody (E). Bars, 100 μm.
Thus, in breast cancer tumors, T cells and breast cancer cells express IL-13. Breast cancer cells also express pSTAT6, suggesting that IL-13 actually delivers signals to cancer cells.
CD4+ T cells promote the development of breast cancer tumors in vivo
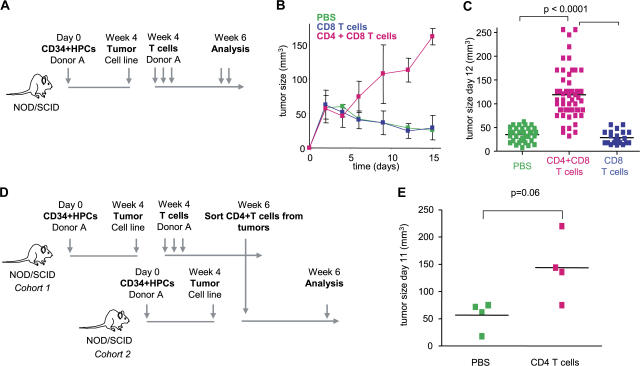
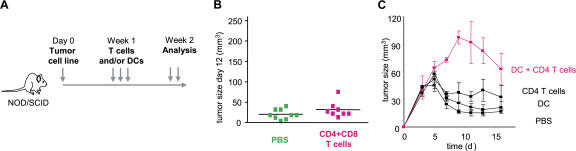
Humanized mice were then used to demonstrate the link between breast cancer and IL-13–secreting CD4+ T cells. The experimental scheme is given in Fig. 4 A. There, sublethally irradiated adult NOD/SCID/β2m−/− mice were transplanted with human G-CSF––mobilized CD34+HPCs from a healthy donor (27). 4 wk later, when these mice display human DCs and B cells, 107 Hs578T breast cancer cells were implanted subcutaneously into the flank (unpublished data). Mice were then reconstituted with T cells autologous to the grafted CD34+HPCs (Fig. 4 A). Repeated injections of 10 × 106 T cells (both CD4+ and CD8+) resulted in accelerated tumor development (Fig. 4 B). Indeed, at day 15, the tumor volume tripled (P < 0.0001; Fig. 4, B and C). This was dependent on CD4+ T cells, as reconstitution with purified CD8+ T cells did not affect early tumor development (Fig. 4, B and C).
Figure 4.
CD4+ T cells promote development of breast cancer tumors. (A) Experimental scheme. (B and C) 100 μl PBS (36 mice analyzed) and 10 × 106 autologous CD8+ T cells/100 μl PBS (21 mice analyzed) alone or together with 10 × 106 CD4+ T cells/100 μl PBS (55 mice analyzed) were transferred into Hs578T breast tumor–bearing humanized mice three times at days 3, 6, and 9 after tumor implantation. Kinetics of tumor development (mean ± SD; B, curves) and tumor size at day 12 (each dot represents one mouse; C) are shown. Horizontal bars represent the mean. (D) Experimental scheme. (E) CD4+ T cells generated as in A were purified from tumors and transferred into Hs578T tumor-bearing humanized mice. Tumor size at day 11 is shown (two experiments with four recipients per Humouse). Horizontal bars represent the mean.
To analyze whether CD4+ T cells actually confer the acceleration of tumor development, CD4+ T cells were sorted from day 15 Hs578T breast cancer tumors pooled from several donor humanized mice. These in vivo primed CD4+ T cells were then injected into humanized mice bearing Hs578T breast cancer tumors but no T cells (Fig. 4 D, experimental scheme). Control mice received PBS. A single transfer of 1.5 × 106 in vivo primed CD4+ T cells per recipient mouse led to acceleration of breast cancer tumor development in three out of four tested mice in two independent experiments (mean tumor volume ± SEM = 51 ± 17 in control mice that received PBS vs. 167 ± 27 in experimental mice that received T cells; P = 0.06; Fig. 4 E).
To determine whether the human DCs had any role in tumor development, we used NOD/SCID/β2m−/− mice without CD34+HPC grafts into which we implanted Hs578T breast cancer cells (Fig. 5 A, experimental scheme). As shown in Fig. 5 B, no change in tumor volume was observed upon injection of T cells isolated from different donors. However, coinjection of autologous DCs (generated by culturing monocytes with GM-CSF and IL-4) and CD4+ T cells led to acceleration of breast cancer tumor development (Fig. 5 C). Thus, CD4+ T cells require DCs to promote the development of breast cancer tumors.
Figure 5.
CD4+ T cells promote development of breast cancer tumors through DCs. (A) Experimental scheme. (B) 100 μl PBS or 10 × 106 CD4+ and CD8+ T cells/100 μl PBS were transferred into Hs578T tumor-bearing mice. The tumor size 12 d after tumor inoculation is shown for two experiments (nine mice per group). Horizontal bars represent the mean. (C) 100 μl PBS, and 106 DCs/100 μl PBS and 10 × 106 autologous CD4+ T cells/100 μl PBS, or both, were transferred into Hs578T tumor-bearing mice. Tumor growth was monitored (three mice per group; mean ± SD).
CD4+ T cells are polarized in vivo to secrete IL-13
To analyze cytokine secretion in vivo, CD4+ T cells were sorted from the tumors of humanized mice 15 d after transfer. IL-2 (>10 ng/ml; not depicted), IFN-γ (>10 ng/ml; not depicted), IL-13 (mean ± SEM = 1,080 ± 200 pg/ml; n = 4; Fig. 6 A), and IL-4 (406 ± 48 pg/ml; n = 4; Fig. 6 B) were detected in supernatants of PMA/ionomycin-activated CD4+ T cells. Up to 17% of CD4+ T cells showed IL-13 expression by flow cytometry (13 ± 3% of IL-13+CD4+ T cells; Fig. 6 C). Most of the IL-13–expressing CD4+ T cells also expressed IFN-γ (Fig. 6 C), as observed in some of the patient tumors. Thus, breast cancer tumors in humanized mice are infiltrated with CD4+ T cells secreting IL-13, as in patient tumor samples.
Figure 6.
CD4+ T cells isolated from tumors secrete IFN-γ as well as type 2 cytokines. 10 × 106 autologous CD4+ and CD8+ T cells /100 μl PBS were transferred into Hs578T breast tumor–bearing humanized mice at days 3, 6, and 9 after tumor inoculation. At day 15, T cells were purified from tumors and restimulated in vitro overnight with PMA/ionomycin. (A and B) Cytokine secretion was measured in the supernatant by Luminex (four humanized mice per group). Error bars represent the mean ± SEM. (C) Intracellular cytokine staining was also performed after restimulation in presence of Brefeldin A (B; two experiments with seven humanized mice). Numbers indicate the percentage of cells positive for the specified marker.
Breast tumors instruct DCs to induce CD4+ T cells secreting IL-13
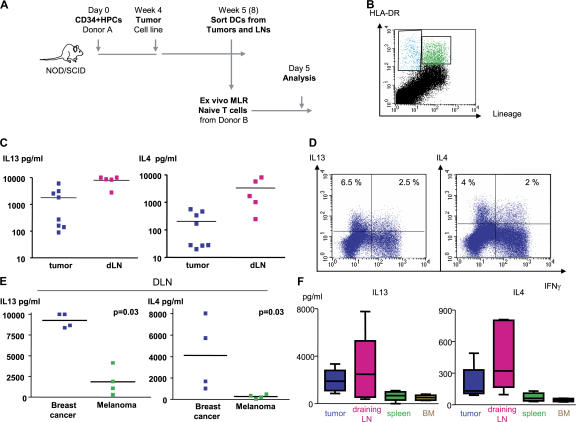
To determine whether DCs represent the link between breast cancer and IL-13–secreting CD4+ T cells, we sorted DCs from Hs587T breast cancer tumors and their draining lymph nodes established in humanized mice without adoptively transferred T cells and analyzed their function in vitro. The experimental scheme is given in Fig. 7 A. As described elsewhere, breast cancer tumors implanted in humanized mice attract human DCs (unpublished data). DCs were sorted at day 4 after tumor implant based on the expression of lineage markers and HLA-DR (Fig. 7 B) and were assessed in vitro for their capacity to trigger allogeneic naive T cell proliferation and cytokine secretion. After 5 d, T cells were activated with PMA and ionomycin, and cytokines were assessed in the supernatants (Fig. 7 A). DCs isolated from both tumors and their draining lymph nodes induced allogeneic CD4+ T cells to proliferate and secrete large amounts (>10 ng/ml) of IL-2 and IFN-γ (unpublished data). Furthermore, high levels of IL-4 and IL-13 were found (mean concentration ± SEM = 205 ± 78.5 and 3,345 ± 1,508 pg/ml IL-4; and 1,775 ± 745 and 7,961 ± 1,342 pg/ml IL-13 for tumor- and, particularly, for draining lymph node–derived DCs, respectively; Fig. 7 C). Flow cytometry confirmed the presence of CD4+ T cells expressing IL-13 with or without IFN-γ (Fig. 7 D). The capacity of DCs to induce large amounts of IL-4 and IL-13 secretion from naive CD4+ T cells was specific to breast cancer, as DCs isolated from the lymph nodes draining melanoma tumors established in the same cohort of mice triggered significantly less IL-13 and IL-4 secretion (mean concentration ± SEM = 269.5 ± 94 and 4,119 ± 1,760 pg/ml IL-4 for melanoma and breast cancer, respectively [P = 0.03]; and 1,848 ± 828.5 and 9,261 ± 430 pg/ml IL-13 for melanoma and breast cancer, respectively [P = 0.03]; Fig. 7 E). Furthermore, although at day 4 after breast tumor implant the capacity of DCs to trigger IL-13 and IL-4 secretion was particularly apparent in draining lymph nodes (Fig. 7 C and not depicted), at day 30 it was clearly confined to DCs isolated from breast cancer tumors and their draining lymph nodes (Fig. 7 F). Thus, breast cancer instructs DCs to prime a fraction of CD4+ T cells to produce IL-4 and IL-13.
Figure 7.
Breast cancer instructs DCs to induce CD4+ T cells to secrete type 2 cytokines. (A) Experimental scheme. (B) Representative FACS analysis of breast cancer tumor cell suspension showing staining with HLA-DR (ordinate) and lineage (abscissa) mAbs. HLA-DR+Lin− DCs are sorted from tumors and draining lymph nodes and co-cultured for 5 d with allogeneic naive CD4+ T cells at a ratio 1:10. (C) Cytokine secretion to co-culture supernatants measured with multiplex bead analysis after overnight PMA/ionomycin restimulation. Each dot represents a separate Humouse tumor. Draining lymph nodes were pooled to obtain a sufficient number of cells for analysis. Horizontal bars represent the mean. (D) Intracellular cytokine expression by CD4+ T cells primed with DCs sorted from day 4 tumors and restimulated for 5 h with PMA/ionomycin in the presence of Brefeldin A. Dot plots are gated on CD3+CD4+ T cells (representative of n = 8 Humouse). Numbers indicate the percentage of cells positive for the specified marker. (E) IL-13 and IL-4 response in breast cancer, but not melanoma, environment. HLA-DR+Lin− DCs were sorted from lymph nodes draining Hs578T breast or Me275 melanoma tumors (day 4; lymph nodes were pooled from a total of 15 Humouse per group in four independent experiments). Horizontal bars represent the mean. P = 0.03 using the paired t test. (F) HLA-DR+Lin− DCs were sorted from tumors, draining lymph nodes, spleen, and BM of Hs578T breast cancer–bearing Humouse (day 30; n = 22 Humouse). Sorted DCs were co- cultured for 5 d with allogeneic naive CD4+ T cells at a ratio 1:10. Cytokine secretion was measured with multiplex bead analysis after overnight PMA/ionomycin restimulation. Box and whiskers data representation show median, range, and SE.
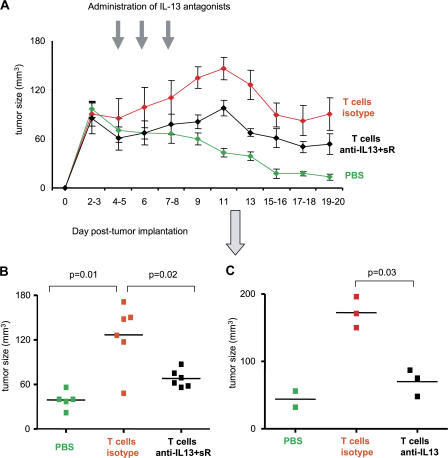
IL-13 antagonists prevent breast cancer tumor development
To establish if IL-13 plays any role in the observed breast cancer tumor development, humanized mice bearing Hs587T breast cancer tumors were treated with an antibody-neutralizing IL-13 and a soluble IL-13R. As shown in Fig. 8 A, mice treated with both IL-13 antagonists showed sustained inhibition of tumor development. Tumor volume at day 13 was 39 ± 5 mm3 in animals without T cells (PBS control) and 128 ± 18 in animals with T cells (mean tumor volume ± SEM; P = 0.01; Fig. 8 B). Neutralizing anti–IL-13 mAb alone was sufficient to prevent acceleration in breast cancer tumor development (mean tumor volume at day 11 = 172 ± 13 in animals treated with isotype control and 70 ± 11.5 in animals treated with IL-13–neutralizing mAb; P = 0.03; Fig. 8 C). Thus, accelerated development of breast cancer tumors in the humanized mouse (Humouse) reconstituted with CD4+ T cells can be partially prevented by treatment with IL-13 antagonists.
Figure 8.
CD4+ T cells promote tumor development via IL-13. 100 μl PBS or 10 × 106 autologous T cells/100 μl PBS were transferred into Hs578T breast tumor–bearing humanized mice at days 3 and 6 after tumor inoculation. Isotype or a mixture of anti–IL-13 antibody and rhIL-13Rα2/Fc chimera (100 μg per injection) were administrated at days 4, 6, and 8 after tumor implantation. (A) Kinetic of tumor size. Error bars represent the mean ± SEM. (B and C) Tumor size 13 d after tumor inoculation is shown (two experiments with six humanized mice per group). Data in C are from one of the experimental cohorts shown in B. Horizontal bars represent the mean.
DISCUSSION
We identified CD4+ T cells secreting IFN-γ and IL-13 in breast cancer tumors. We also found that breast cancer cells express IL-13. The production of IL-13 by CD4+ T cells infiltrating breast cancer suggests the paracrine origin of the IL-13 staining. It is, however, also possible that breast cancer cells might be induced to secrete IL-13, though we did not find expression in breast cancer cell lines in vitro or in vivo in the absence of T cells (unpublished data). Autocrine IL-13 has been shown important in the pathophysiology of Hodgkin's disease (32, 34, 35). There, IL-13 and IL-13R are frequently expressed by Hodgkin and Reed-Sternberg cells (35), and IL-13 stimulates their growth (34, 36). Similar to Hodgkin's cells (32), breast cancer cells express pSTAT6, suggesting that IL-13 actually delivers signals to cancer cells. Accordingly, earlier in vitro studies demonstrated that IL-13 (as well as IL-4) can inhibit estrogen-induced proliferation and favor acquisition of a breast cancer cell differentiation marker, gross cystic disease fluid protein–15 (37, 38). In line with the possible direct effect of IL-13 on breast cancer cells in vivo are recent findings on the expression of IL-13Rα2 in highly aggressive variants of breast cancer with the propensity to form lung metastasis (39).
Humanized mice bearing breast cancer tumors permitted us to conclude that CD4+ T cells and IL-13 are actually involved in breast cancer pathophysiology. There, CD4+ T cells promote early tumor development, which can be partially prevented by IL-13 antagonists. The mechanism of this tumor development needs to be determined. In addition to a possible direct effect on breast cancer cells, indirect mechanisms need also be considered. There is clearly enhanced angiogenesis (unpublished data), which might be caused by activation of type 2 macrophages (40, 41). Furthermore, DCs in such a microenvironment could acquire a suppressive phenotype involving, for example, indoleamine 2,3-dioxygenase expression (42) or, as discussed earlier, TGF-β production (17–19, 43). The role of IL-13 in the indirect suppression of tumor surveillance has been demonstrated in other tumor models (43–46) and is frequently linked with suppression of cytotoxic T cell function. There, blockade of IL-13 permitted inhibition of tumor growth and eventual rejection. Our preliminary results suggest that at least CD8+ T cell priming is not inhibited in our model in the presence of CD4+ T cells. Further studies are needed to establish whether their in vivo function and capacity to reject breast cancer tumors is altered.
Our study further demonstrates that CD4+ T cells facilitate tumor development, provided DCs are present. This ménage a trois generates an environment rich in IL-13 produced by T cells. Indeed, DCs infiltrating breast cancer tumors in humanized mice appear responsible for the induction of IL-13–secreting CD4+ T cells, at least in the initial phase of tumor development. Two patterns of cytokine expression by CD4+ T cells were observed. The first, with the presence of single-positive CD4+ T cells expressing either IL-13 or IFN-γ, is consistent with the classical definition of type 2 polarization (28). This suggests bona fide Th1 and Th2 cells in the breast cancer tumor microenvironment. The second pattern, found in the majority of tumors from patients, is the presence of double-positive IL-13– and IFN-γ–expressing CD4+ T cells, possibly reflecting Th0 cells (28). The presence of double-positive cells could also reflect some degree of plasticity in Th1/Th2 cell polarization, particularly in the presence of high IFN-γ (33). Such plasticity has been demonstrated by genetic reprogramming of human Th1 and Th2 cell clones (47, 48). IL-13 secretion by CD4+ T cells appears independent of the presence of invariant chain–expressing NKT cells, as their depletion does not affect the frequency of IL-13–expressing CD4+ T cells in the in vitro experiments (unpublished data). Finally, they appear restricted at large by MHC class II, as blocking HLA-DR on DCs leads to >90% inhibition of IL-13– secreting CD4+ T cells (unpublished data).
In conclusion, by combining the studies of human cancer using ex vivo analysis of patient samples and in vivo analysis in the model of the human immune system and human cancer, we demonstrated the immune deviation via DCs that breast cancer might use to develop. This immune deviation can be counteracted with IL-13 antagonists, pointing to the role of IL-13 in the pathophysiology of breast cancer.
MATERIALS AND METHODS
Generation of humanized mice.
CD34+HPCs were obtained from apheresis of adult healthy volunteers mobilized with G-CSF and purified as previously described (27). The CD34− fraction of apheresis was Ficoll-purified, and obtained PBMCs were stored frozen and used as a source of autologous T cells. 2.5 × 106 CD34+HPCs were transplanted intravenously into sublethally irradiated (12 cGy/g body weight of 137Cs γ irradiation) NOD/SCID/β2m−/− mice (Jackson ImmunoResearch Laboratories; Institutional Animal Care and Use Committee no. A01-005). 10 × 106 breast cancer cells (Hs578T) or 10 × 106 melanoma cells (Me275) were harvested from long-term cultures and injected subcutaneously into the flanks of the mice. For experiments with NOD/SCID/β2m−/− mice, they were sublethally irradiated the day before tumor implantation. Tumor size was monitored every 2–3 d. Tumor volume (ellipsoid) was calculated as follows: (short diameter)2 × long diameter/2.
Monocyte-derived DCs and T cell purification.
Monocyte-derived DCs were generated from the adherent fraction of PBMCs by culturing with 100 ng/ml GM-CSF (Immunex) and 25 ng/ml IL-4 (R&D Systems). CD4+ and CD8+ T cells were positively selected from thawed PBMCs using magnetic selection according to the manufacturer's instructions (Miltenyi Biotec). The purity was routinely >90%.
Immunofluorescence.
Tissues were frozen in optimal cutting temperature compound (Tissue-Tek; Allegiance), cryosectioned on slides (Superfrost Plus; Fisher Scientific), and fixed with cold acetone. Direct staining was done with HLA-DR FITC (BD Biosciences), CRTH2-PE (Miltenyi Biotec), IL-13–PE (BD Biosciences), and CD3-FITC. Confocal microscopy was performed using a TCS-NT SP (Leica).
Flow cytometry.
Cell suspensions were obtained from tumors by digestion with 2 mg/ml collagenase D (Roche Diagnostics) for 45 min at 37°C. Bone marrow cells were washed out of the harvested bones. Cell suspensions were washed twice and stained in PBS with serum (2 mM EDTA, 5% AB) using the following antibodies: Lin, CD45, HLA-ABC, and CD3-PE (all from BD Biosciences).
T cell cytokines.
To assess cytokine expression by intracellular staining, T cells were restimulated for 5 h with PMA and ionomycin. 10 mg/ml Brefeldin A (BD Biosciences) was added for the last 2.5 h. T cells were labeled with anti-CD3 and antibodies to IL-4, IL-13, TNF, IFN-γ, and IL-2 (BD Biosciences).
In vivo IL-13 blocking.
Mice were injected intratumorally at days 4, 6, and 8 after tumor implantation with anti–IL-13 mAbs and rhIL-13Rα2/Fc chimera or goat IgG isotype control (100 μg/ml each; R&D Systems).
Tumor samples.
Tumor samples from patients diagnosed with breast carcinoma (in situ and invasive duct and/or mucinous carcinoma of the breast, as well as lobular carcinoma) were obtained from the Baylor University Medical Center Tissue Bank (Institutional Review Board no. 005-145). Whole-tissue fragments were placed in culture with 50 ng/ml PMA and 1 μg/ml ionomycin (Sigma-Aldrich) for 16 h. Cytokine production was analyzed in the culture supernatant by Luminex (Beadlyte custom kit; Upstate Biotechnology). For cell suspensions, samples were minced into small fragments and digested in a triple enzyme mix containing 2.5 mg/ml collagenase, 1 mg/ml hyaluronidase, and 20 U/ml DNase for 2–3 h at 37°C. The suspension was filtered and washed, and obtained cells were resuspended at a concentration of 106 cells/ml and activated for 5 h with PMA and ionomycin. 10 mg/ml Brefeldin A was added for the last 2.5 h. Cells were labeled with anti-CD3 and anti-CD4 mAb, and intracellular cytokine staining was performed using antibodies to IL-13 and IFN-γ (BD Biosciences). For inhibition of IL-13 staining, anti–IL-13 mAb was incubated with 5 μg/ml rhIL-13 for 1 h at room temperature before use. Cells were fixed in 1% PFA and analyzed by flow cytometry. A piece of each tissue was frozen for immunofluorescence analysis. Sections were labeled with CD3 Alexa Fluor 488 mAb (BD Biosciences) and mounted with DAPI.
STAT 6 detection in paraffin-embedded breast cancer tissue.
The slides were deparaffinized in two changes of xylene, washed in 100% ethanol, and hydrated in 95% ethanol, 75% ethanol, and PBS. For antigen retrieval, the slides were placed in preheated to boiling temperature 10 mM sodium citrate, 0.05% Tween, pH 6, in a pressure cooker and incubated for 8 min. Rinsed slides were incubated in 0.3% hydrogen peroxide for 8 min, rinsed in 1× PBS followed by a Biotin-Blocking kit (Invitrogen), and blocked with 1% BSA/0.1% saponin. They were then incubated overnight with the primary antibody (cytokeratin [DakoCytomation], STAT6 [BD Biosciences], or pSTAT6 [Cell Signaling]) overnight at room temperature at 0.8 μg/ml. The secondary antibody was goat anti–rabbit biotin from Invitrogen, used at a 1:1,000 dilution for 30 min. Ready-to-use horseradish peroxidase–streptavidin was placed on the slides for 30 min (Vector Laboratories), followed by DAB substrate from BD Biosciences for 20 min, and counterstained with hematoxylin and mounted with Cytoseal XYL (Richard-Allan Scientific). The images were acquired using a digital camera (DXM 1200C; Nikon).
DC–T cell co-cultures.
Naive CD4+ T cells were obtained from buffy coats after magnetic depletion using CD8, CD14, CD19, CD16, CD56, and glycophorine A microbeads (Miltenyi Biotec) and sorted based on the CD4+CCR7+CD45RA+ phenotype. NKT cells were depleted by exclusion of Vα24+ CD4+ T cells from the sort gate. DCs were sorted based on HLA-DR+Lin−CD11c+ and HLA-DR+Lin-CD123+ phenotypes. 5 × 104 naive CD4+ T cells/well were cultured with 5 × 103 DCs/well in RPMI 1640 supplemented with 10% human AB serum (Gemini BioProducts). To assess cytokine secretion by Luminex, T cells were harvested at day 5, washed twice, resuspended at a concentration of 106 cells/ml, and restimulated for 16 h with 50 ng/ml PMA and 1 μg/ml ionomycin (Sigma- Aldrich). To assess cytokine expression by intracellular staining, T cells were harvested on day 6 of the culture, washed twice, and restimulated for 5 h with PMA and ionomycin. 10 mg/ml Brefeldin A was added for the last 2.5 h. T cells were labeled with anti-CD3 and antibodies to IL-4, IL-13, and IFN-γ (BD Biosciences).
Statistics.
The parametric t test and nonparametric Mann-Whitney and Wilcoxon tests were used to assess the significance of observed differences. The parametric Pearson correlation and nonparametric Spearman correlation were used as indicated in the figure legends.
Acknowledgments
We are grateful to Albert Barnes, Sebastien Coquery, Jennifer Shay, and Lynnette Walters for help; Dr. Joseph Fay for help with healthy volunteers; Cindy Samuelsen for continuous support; and Dr. D. Savino at the Department of Pathology at Baylor University Medical Center. We thank Carson Harrod for editorial help. We thank Drs. William Duncan, Ira Mellman, Ralph Steinman, and Gerard Zurawski for critical reading of the manuscript. We thank Dr. Michael Ramsay for continuous support.
This work was supported by the Baylor Health Care Systems Foundation, the Dana Foundation (J. Banchereau), the Defense Advanced Research Projects Agency (J. Banchereau), and the National Institutes of Health (grants RO-1 CA89440 and R21 AI056001 to A. Karolina Palucka; grants U19 AIO57234, RO-1 CA78846, and CA85540 to J. Banchereau). J. Banchereau holds the Caruth Chair for Transplantation Immunology Research. A. Karolina Palucka holds the Ramsay Chair for Cancer Immunology Research.
The authors have no conflicting financial interests.
Abbreviations used: CRTH2, chemoattractant receptor– homologous molecule expressed on Th2 cells; HPC, hematopoietic progenitor cell; Humouse, humanized mouse; NOD/SCID/β2m−/−, nonobese diabetic/LtSz-scid/scid β2 microglobulin–deficient; pSTAT6, phosphorylated STAT6; rh, recombinant human.
References
- 1.Joyce, J.A. 2005. Therapeutic targeting of the tumor microenvironment. Cancer Cell. 7:513–520. [DOI] [PubMed] [Google Scholar]
- 2.Coussens, L.M., and Z. Werb. 2002. Inflammation and cancer. Nature. 420:860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinman, R.M., D. Hawiger, and M.C. Nussenzweig. 2003. Tolerogenic dendritic cells. Annu. Rev. Immunol. 21:685–711. [DOI] [PubMed] [Google Scholar]
- 4.Banchereau, J., and R.M. Steinman. 1998. Dendritic cells and the control of immunity. Nature. 392:245–252. [DOI] [PubMed] [Google Scholar]
- 5.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767–811. [DOI] [PubMed] [Google Scholar]
- 6.Shortman, K., and Y.J. Liu. 2002. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2:151–161. [DOI] [PubMed] [Google Scholar]
- 7.Albert, M.L., B. Sauter, and N. Bhardwaj. 1998. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature. 392:86–89. [DOI] [PubMed] [Google Scholar]
- 8.Albert, M.L., S.F. Pearce, L.M. Francisco, B. Sauter, P. Roy, R.L. Silverstein, and N. Bhardwaj. 1998. Immature dendritic cells phagocytose apoptotic cells via αvβ5 and CD36 and cross-present antigens to cytotoxic T lymphocytes. J. Exp. Med. 188:1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath, W.R., and F.R. Carbone. 2001. Cross-presentation, dendritic cells, tolerance and immunity. Annu. Rev. Immunol. 19:47–64. [DOI] [PubMed] [Google Scholar]
- 10.Finkelman, F.D., A. Lees, R. Birnbaum, W.C. Gause, and S.C. Morris. 1996. Dendritic cells can present antigen in vivo in a tolerogenic or immunogenic fashion. J. Immunol. 157:1406–1414. [PubMed] [Google Scholar]
- 11.Banchereau, J., and A.K. Palucka. 2005. Dendritic cells as therapeutic vaccines against cancer. Nat. Rev. Immunol. 5:296–306. [DOI] [PubMed] [Google Scholar]
- 12.Gabrilovich, D. 2004. Mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat. Rev. Immunol. 4:941–952. [DOI] [PubMed] [Google Scholar]
- 13.Wang, T., G. Niu, M. Kortylewski, L. Burdelya, K. Shain, S. Zhang, R. Bhattacharya, D. Gabrilovich, R. Heller, D. Coppola, et al. 2004. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 10:48–54. [DOI] [PubMed] [Google Scholar]
- 14.Gabrilovich, D.I., H.L. Chen, K.R. Girgis, H.T. Cunningham, G.M. Meny, S. Nadaf, D. Kavanaugh, and D.P. Carbone. 1996. Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat. Med. 2:1096–1103. [DOI] [PubMed] [Google Scholar]
- 15.Chomarat, P., J. Banchereau, J. Davoust, and A.K. Palucka. 2000. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat. Immunol. 1:510–514. [DOI] [PubMed] [Google Scholar]
- 16.Delamarre, L., M. Pack, H. Chang, I. Mellman, and E.S. Trombetta. 2005. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 307:1630–1634. [DOI] [PubMed] [Google Scholar]
- 17.Enk, A.H., H. Jonuleit, J. Saloga, and J. Knop. 1997. Dendritic cells as mediators of tumor-induced tolerance in metastatic melanoma. Int. J. Cancer. 73:309–316. [DOI] [PubMed] [Google Scholar]
- 18.Ghiringhelli, F., P.E. Puig, S. Roux, A. Parcellier, E. Schmitt, E. Solary, G. Kroemer, F. Martin, B. Chauffert, and L. Zitvogel. 2005. Tumor cells convert immature myeloid dendritic cells into TGF-β–secreting cells inducing CD4+CD25+ regulatory T cell proliferation. J. Exp. Med. 202:919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levings, M.K., S. Gregori, E. Tresoldi, S. Cazzaniga, C. Bonini, and M.G. Roncarolo. 2005. Differentiation of Tr1 cells by immature dendritic cells requires IL-10 but not CD25+CD4+ Tr cells. Blood. 105:1162–1169. [DOI] [PubMed] [Google Scholar]
- 20.Terabe, M., and J.A. Berzofsky. 2004. Immunoregulatory T cells in tumor immunity. Curr. Opin. Immunol. 16:157–162. [DOI] [PubMed] [Google Scholar]
- 21.Bell, D., P. Chomarat, D. Broyles, G. Netto, G.M. Harb, S. Lebecque, J. Valladeau, J. Davoust, K.A. Palucka, and J. Banchereau. 1999. In breast carcinoma tissue, immature dendritic cells reside within the tumor, whereas mature dendritic cells are located in peritumoral areas. J. Exp. Med. 190:1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas, R., K.P. MacDonald, A.R. Pettit, L.L. Cavanagh, J. Padmanabha, and S. Zehntner. 1999. Dendritic cells and the pathogenesis of rheumatoid arthritis. J. Leukoc. Biol. 66:286–292. [DOI] [PubMed] [Google Scholar]
- 23.Radstake, T.R., A.W. van Lieshout, P.L. van Riel, W.B. van den Berg, and G.J. Adema. 2005. Dendritic cells, Fc{gamma} receptors, and Toll-like receptors: potential allies in the battle against rheumatoid arthritis. Ann. Rheum. Dis. 64:1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon, S., and P.R. Taylor. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953–964. [DOI] [PubMed] [Google Scholar]
- 25.Blanco, P., A.K. Palucka, M. Gill, V. Pascual, and J. Banchereau. 2001. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 294:1540–1543. [DOI] [PubMed] [Google Scholar]
- 26.Banchereau, J., V. Pascual, and A.K. Palucka. 2004. Autoimmunity through cytokine-induced dendritic cell activation. Immunity. 20:539–550. [DOI] [PubMed] [Google Scholar]
- 27.Palucka, A.K., J. Gatlin, J.P. Blanck, M.W. Melkus, S. Clayton, H. Ueno, E.T. Kraus, P. Cravens, L. Bennett, A. Padgett-Thomas, et al. 2003. Human dendritic cell subsets in NOD/SCID mice engrafted with CD34+ hematopoietic progenitors. Blood. 102:3302–3310. [DOI] [PubMed] [Google Scholar]
- 28.Mosmann, T.R., and R.L. Coffman. 1989. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 7:145–173. [DOI] [PubMed] [Google Scholar]
- 29.Nagata, K., K. Tanaka, K. Ogawa, K. Kemmotsu, T. Imai, O. Yoshie, H. Abe, K. Tada, M. Nakamura, K. Sugamura, and S. Takano. 1999. Selective expression of a novel surface molecule by human Th2 cells in vivo. J. Immunol. 162:1278–1286. [PubMed] [Google Scholar]
- 30.Cosmi, L., F. Annunziato, M.I.G. Galli, R.M.E. Maggi, K. Nagata, and S. Romagnani. 2000. CRTH2 is the most reliable marker for the detection of circulating human type 2 Th and type 2 T cytotoxic cells in health and disease. Eur. J. Immunol. 30:2972–2979. [DOI] [PubMed] [Google Scholar]
- 31.Wurster, A.L., T. Tanaka, and M.J. Grusby. 2000. The biology of Stat4 and Stat6. Oncogene. 19:2577–2584. [DOI] [PubMed] [Google Scholar]
- 32.Skinnider, B.F., A.J. Elia, R.D. Gascoyne, B. Patterson, L. Trumper, U. Kapp, and T.W. Mak. 2002. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 99:618–626. [DOI] [PubMed] [Google Scholar]
- 33.Ansel, K.M., I. Djuretic, B. Tanasa, and A. Rao. 2006. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24:607–656. [DOI] [PubMed] [Google Scholar]
- 34.Kapp, U., W.C. Yeh, B. Patterson, A.J. Elia, D. Kagi, A. Ho, A. Hessel, M. Tipsword, A. Williams, C. Mirtsos, et al. 1999. Interleukin 13 is secreted by and stimulates the growth of Hodgkin and Reed-Sternberg cells. J. Exp. Med. 189:1939–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skinnider, B.F., A.J. Elia, R.D. Gascoyne, L.H. Trumper, F. von Bonin, U. Kapp, B. Patterson, B.E. Snow, and T.W. Mak. 2001. Interleukin 13 and interleukin 13 receptor are frequently expressed by Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 97:250–255. [DOI] [PubMed] [Google Scholar]
- 36.Trieu, Y., X.Y. Wen, B.F. Skinnider, M.R. Bray, Z. Li, J.O. Claudio, E. Masih-Khan, Y.X. Zhu, S. Trudel, J.A. McCart, et al. 2004. Soluble interleukin-13Ralpha2 decoy receptor inhibits Hodgkin's lymphoma growth in vitro and in vivo. Cancer Res. 64:3271–3275. [DOI] [PubMed] [Google Scholar]
- 37.Blais, Y., S. Gingras, D.E. Haagensen, F. Labrie, and J. Simard. 1996. Interleukin-4 and interleukin-13 inhibit estrogen-induced breast cancer cell proliferation and stimulate GCDFP-15 expression in human breast cancer cells. Mol. Cell. Endocrinol. 121:11–18. [DOI] [PubMed] [Google Scholar]
- 38.Serve, H., E. Oelmann, A. Herweg, D. Oberberg, S. Serve, B. Reufi, C. Mucke, A. Minty, E. Thiel, and W.E. Berdel. 1996. Inhibition of proliferation and clonal growth of human breast cancer cells by interleukin 13. Cancer Res. 56:3583–3588. [PubMed] [Google Scholar]
- 39.Minn, A.J., G.P. Gupta, P.M. Siegel, P.D. Bos, W. Shu, D.D. Giri, A. Viale, A.B. Olshen, W.L. Gerald, and J. Massague. 2005. Genes that mediate breast cancer metastasis to lung. Nature. 436:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinha, P., V.K. Clements, and S. Ostrand-Rosenberg. 2005. Interleukin-13-regulated M2 macrophages in combination with myeloid suppressor cells block immune surveillance against metastasis. Cancer Res. 65:11743–11751. [DOI] [PubMed] [Google Scholar]
- 41.Mantovani, A., A. Sica, and M. Locati. 2005. Macrophage polarization comes of age. Immunity. 23:344–346. [DOI] [PubMed] [Google Scholar]
- 42.Munn, D.H., and A.L. Mellor. 2004. IDO and tolerance to tumors. Trends Mol. Med. 10:15–18. [DOI] [PubMed] [Google Scholar]
- 43.Terabe, M., S. Matsui, J.M. Park, M. Mamura, N. Noben-Trauth, D.D. Donaldson, W. Chen, S.M. Wahl, S. Ledbetter, B. Pratt, et al. 2003. Transforming growth factor–β production and myeloid cells are an effector mechanism through which CD1d-restricted T cells block cytotoxic T lymphocyte–mediated tumor immunosurveillance: abrogation prevents tumor recurrence. J. Exp. Med. 198:1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terabe, M., S. Matsui, N. Noben-Trauth, H. Chen, C. Watson, D.D. Donaldson, D.P. Carbone, W.E. Paul, and J.A. Berzofsky. 2000. NKT cell-mediated repression of tumor immunosurveillance by IL-13 and the IL-4R-STAT6 pathway. Nat. Immunol. 1:515–520. [DOI] [PubMed] [Google Scholar]
- 45.Park, J.M., M. Terabe, L.T. van den Broeke, D.D. Donaldson, and J.A. Berzofsky. 2005. Unmasking immunosurveillance against a syngeneic colon cancer by elimination of CD4+ NKT regulatory cells and IL-13. Int. J. Cancer. 114:80–87. [DOI] [PubMed] [Google Scholar]
- 46.Terabe, M., J. Swann, E. Ambrosino, P. Sinha, S. Takaku, Y. Hayakawa, D.I. Godfrey, S. Ostrand-Rosenberg, M.J. Smyth, and J.A. Berzofsky. 2005. A nonclassical non-Vα14Jα18 CD1d-restricted (type II) NKT cell is sufficient for down-regulation of tumor immunosurveillance. J. Exp. Med. 202:1627–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundrud, M.S., S.M. Grill, D. Ni, K. Nagata, S.S. Alkan, A. Subramaniam, and D. Unutmaz. 2003. Genetic reprogramming of primary human T cells reveals functional plasticity in Th cell differentiation. J. Immunol. 171:3542–3549. [DOI] [PubMed] [Google Scholar]
- 48.Messi, M., I. Giacchetto, K. Nagata, A. Lanzavecchia, G. Natoli, and F. Sallusto. 2003. Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nat. Immunol. 4:78–86. [DOI] [PubMed] [Google Scholar]