Abstract
The Philadelphia chromosome (Ph) encoding the oncogenic BCR-ABL1 kinase defines a subset of acute lymphoblastic leukemia (ALL) with a particularly unfavorable prognosis. ALL cells are derived from B cell precursors in most cases and typically carry rearranged immunoglobulin heavy chain (IGH) variable (V) region genes devoid of somatic mutations. Somatic hypermutation is restricted to mature germinal center B cells and depends on activation-induced cytidine deaminase (AID). Studying AID expression in 108 cases of ALL, we detected AID mRNA in 24 of 28 Ph+ ALLs as compared with 6 of 80 Ph− ALLs. Forced expression of BCR-ABL1 in Ph− ALL cells and inhibition of the BCR-ABL1 kinase showed that aberrant expression of AID depends on BCR-ABL1 kinase activity. Consistent with aberrant AID expression in Ph+ ALL, IGH V region genes and BCL6 were mutated in many Ph+ but unmutated in most Ph− cases. In addition, AID introduced DNA single-strand breaks within the tumor suppressor gene CDKN2B in Ph+ ALL cells, which was sensitive to BCR-ABL1 kinase inhibition and silencing of AID expression by RNA interference. These findings identify AID as a BCR-ABL1–induced mutator in Ph+ ALL cells, which may be relevant with respect to the particularly unfavorable prognosis of this leukemia subset.
Somatic hypermutation (SHM) and class-switch recombination (CSR) represent physiological processes that modify variable (V) and constant regions of Ig genes in mature germinal center B cells (1). Both SHM and CSR critically depend on expression of activation-induced cytidine deaminase (AID), which introduces single-strand breaks into target DNA (2). AID-mediated DNA single-strand breaks (DNA-SSB) leading to SHM and CSR are specifically introduced into V or constant regions of Ig genes, respectively. At much lower frequency, however, AID can also target non-Ig genes in germinal center B cells (3, 4) and may even act as a genome-wide mutator (5). Such targeting errors carry the risk of oncogenic mutation leading to the transformation of a germinal center B cell, which may give rise to B cell lymphoma. For instance, aberrant SHM or CSR may lead to chromosomal translocation of protooncogenes, including MYC, BCL2, BCL6, and CCND1, and cause various types of B cell lymphoma (3). Therefore, tight regulation of AID expression in germinal center B cells and control of DNA strand breaks related to SHM and CSR are critical to prevent B cell malignancy. In fact, previous work demonstrated that Myc-Igh chromosome translocations as they occur in human Burkitt's lymphoma are caused by Aid (6). The emergence of Myc-Igh gene rearrangements is not only prevented by tight regulation of Aid expression; the activation of DNA damage– induced checkpoints during physiological AID- dependent CSR may eventually lead to the activation of the tumor suppressors ATM, NBS1, CDKN2D (INK4D, P19/ARF), and TP53 and is indeed critical to prevent oncogenic Myc-Igh gene rearrangements (7).
RESULTS
Aberrant AID expression correlates with the Philadelphia chromosome (Ph) in acute lymphoblastic leukemia (ALL)
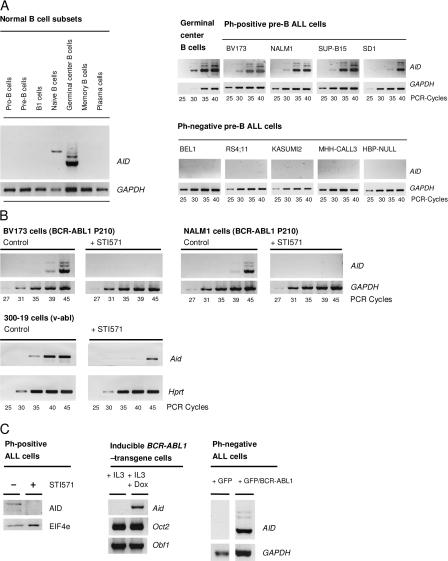
ALL cells are typically derived from pro– or pre–B cells. These B cell precursors do not express AID (Table I and Fig. 1 A, left) and carry Ig genes that have neither undergone SHM nor CSR (8). Therefore, it was unexpected that AID is expressed in a subset of ALL cell lines (Table I and Fig. 1 A, right). Interestingly, AID expression correlates with the presence of t(9;22)(q34;q11), resulting in the so-called Philadelphia chromosome (Ph). Ph encodes the oncogenic BCR-ABL1 kinase and defines a subgroup of ALL with a particularly unfavorable prognosis (9). Studying AID mRNA expression in 108 cases of ALL, AID mRNA was detected in 24 of 28 cases of Ph+ ALL, but only in 6 of 80 cases of Ph− ALL (Table I). Compared with normal germinal center B cells, mRNA levels for AID are lower in most but similar in some Ph+ ALL cell lines (Fig. 1 A, right).
Table I.
Aberrant SHM of Ig- and non-Ig genes in Ph+ ALL cells
| Cell type | IGHV a | BCL6 | MYC | AID mRNA expression |
|||
|---|---|---|---|---|---|---|---|
| Mutated clones/ cases |
Mutations per 103 bp |
Mutated clones/ cases |
Mutations per 103 bp |
Mutated clones/ cases |
Mutations per 103 bp |
||
| Pre–B cells (clones) | 1/36 | 4.4 ± 4 | n.d. | n.d. | No | ||
| Naive B cells (clones) | 0/12 | 3.1 ± 1 | 1/21b | 0.05b | 7/179c | 0.05c | No |
| Germinal center B cells (clones) |
12/14 | 40.2 ± 12 | 5/15b | 1.0b | n.d. | Yes | |
| Memory B cells (clones) | 52/54 | 45.2 ± 9 | 26/71c | 1.38 ± 0.34c | 9/178c | 0.09c | No |
| Ph− ALL (cases) | 6/60 | 4.85 ± 1.23 | 1/5d | 0.11 ± 0.11 | 1/5c | 0.16 ± 0.11 | 6/80 cases |
| Ph+ ALL (cases) | 30/46 | 34.48 ± 4.42 | 7/10d | 1.07 ± 0.23 | 3/9d | 0.51 ± 0.15 | 24/28 cases |
|
MYC-IGH Burkitt's lymphoma (cases) |
12/12b | 69b | 11/30b | 0.4b | 12/12b | 1.9b | Yes |
For normal B cell subsets, rearranged VH gene segments were amplified, cloned, and sequenced from bulk populations. For leukemia and lymphoma cells, individual cases were analyzed. Mutation frequencies are given as means (mutations per 103 bp) ± standard error of the mean. VH gene rearrangements were amplified using Taq DNA polymerase. VH gene rearrangements amplified from ALL cases were considered mutated if the average mutation frequency of all amplified sequences was significantly (P < 0.01) above the error rate of Taq DNA polymerase.
Data from reference 3.
Data from reference 4.
BCL6 and MYC alleles were amplified using PFU DNA polymerase and sequenced from both DNA strands. BCL6 or MYC alleles amplified from ALL cases were considered mutated if at least two mutated sequences were amplified per case with an average mutation frequency of all amplified sequences significantly (P < 0.01) above the error rate of PFU DNA polymerase. Only mutations confirmed on both DNA strands were counted.
Figure 1.
AID expression in Ph+ ALL cells. mRNA expression of AID was measured in normal human pro–B, pre–B, B1, naive, germinal center, and memory B cells as well as plasma cells by RT-PCR (A). In a semiquantitative RT-PCR analysis, AID mRNA expression in Ph+ ALL cells was compared with germinal center B cells and Ph− ALL cells. GAPDH was used for normalization of cDNA amounts (A). Ph+ ALL cell lines (BV173 and Nalm1; 10 μmol/l STI571) and v-abl–transformed mouse pre–B cells (300-19; 1 μmol/l STI571) were treated with or without STI571 for 24 h and subjected to semiquantitative RT-PCR analysis for human AID and GAPDH or murine Aid and Hprt mRNA expression (B). Protein lysates from STI571-treated or untreated Ph+ ALL cells (BV173) were used for Western blotting (C) together with antibodies against AID and EIF4E (used as a loading control). IL-3–dependent murine pro–B cells carrying a doxycycline-inducible BCR-ABL1 transgene were incubated with or without 1 μg/ ml doxycycline for 24 h and subjected to RT-PCR analysis of Aid, Oct2, and Obf1 mRNA expression (C). Ph− ALL cells were transiently transfected with a pMIG vector encoding GFP and/or GFP and BCR-ABL1. After 24 h, GFP-expressing cells were sorted and subjected to RT-PCR analysis for AID expression (C).
SHM and CSR of Ig genes in Ph+ ALL
This raises the possibility that Ig V region genes might aberrantly be targeted by SHM in Ph+ ALL. Therefore, we analyzed the sequence of VH region genes in 60 Ph− and 46 Ph+ cases of ALL. Among Ph− ALL, 6 of 60 cases carry somatically mutated VH region genes (Table I and Table S1, which is available at http://www.jem.org/cgi/content/full/jem.20062662/DC1). In contrast, 30 of 46 Ph+ ALL cases harbor mutated VH gene rearrangements (Table I and Table S2). Counting only Ph+ and Ph− leukemia cases, for which information on both AID expression and VH region sequence was available, this correlation was even more conspicuous: 16 of 18 Ph+ leukemia cases, all expressing AID, also carried mutated VH region genes, whereas 10 Ph− leukemia cases, all lacking AID expression, also all carried unmutated VH gene rearrangements. The average mutation frequency was 34.5 ± 4 mutations/103 bp in Ph+ and 4.9 ± 1 mutations/103 bp in Ph− ALL among all sequences (means based on data from 70 and 76 sequences ± SEM, P < 0.05; Tables S1 and S2). Also among the six Ph− cases carrying mutated VH region genes, the mutation frequency was high, suggesting that in a small subset of Ph− ALL, aberrant SHM may also be induced by other factors that are not related to BCR-ABL1.
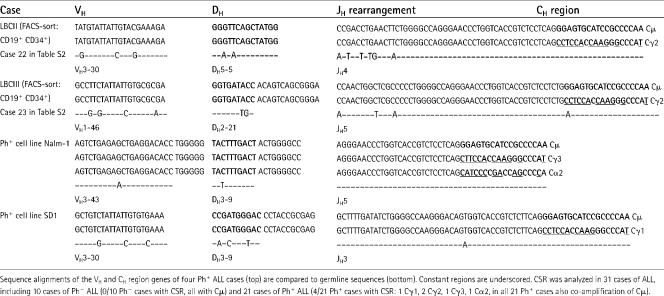
Analyzing CSR of Ig CH region genes in Ph+ and Ph− ALL, we identified switched Cγ1, Cγ2, Cγ3, and Cα2 transcripts in 4 of 21 Ph+ but not in any of 10 Ph− ALL cases (Table II). Based on the small number of cases studied, we cannot give an estimate of the overall frequency of CSR in Ph+ ALL. Collectively, we conclude that Ig genes in Ph+ ALLs can be targeted by SHM and in rare cases even undergo CSR, which is consistent with specific expression of AID in this ALL subset.
Table II.
CSR in Ph+ ALL cells
Aberrant SHM of non-Ig genes in Ph+ ALL
Given that previous studies demonstrated that aberrant SHM also involves mutation of non-Ig genes (3, 4), we also studied known potential target regions of SHM within the BCL6 and MYC genes (3, 4) in Ph− and Ph+ cases. 7 of 10 Ph+ ALL cases and 1 of 5 Ph− ALL cases harbored a mutated BCL6 gene (Table I and Table S3). In both Ph− and Ph+ ALL, the average mutation frequency was above the error rate of the PFU DNA polymerase used in this experiment. Comparing Ph+ and Ph− cases, the average mutation frequency was significantly higher in Ph+ ALL (Table I and Table S3; P < 0.05). Likewise, MYC gene mutations were found in both Ph− and Ph+ ALL cells. In this case, however, the difference between Ph+ and Ph− cells was not significant (Table I; P = 0.13).
Besides rearranged IGH V region genes, CD19+ B cell lineage ALL cells frequently carry TCRB (10) and TCRG (11) gene rearrangements. Consistent with these previous findings, we were able to amplify TCRB and TCRG gene rearrangements from four of eight (TCRB) and seven of seven (TCRG) cases of Ph+ ALL (Table S3, available at http://www.jem.org/cgi/content/full/jem.20062662/DC1). Given that transgenic expression of Aid in murine T cell lymphomas also induces SHM of rearranged TCRB genes (12), we performed sequence analysis of TCRB and TCRG gene rearrangements in Ph+ ALL, as well as normal peripheral blood T cells (Table S3). TCRB gene rearrangements amplified from Ph+ ALL were somatically mutated in all four cases with an average mutation frequency of 6.2 mutations per 103 bp (±2.3 mutations per 103 bp, SEM), which was substantially above the mutation frequency of rearranged TCRB alleles amplified from normal T cells. Only 2 of 30 TCRB gene rearrangements amplified from normal T cells harbored a single mutation with an average mutation frequency of 0.2 ± 0.1 mutations per 103 bp (P < 0.01; Table S3). Likewise, five of seven TCRG gene rearrangements amplified from Ph+ ALL cases carried somatic mutations with an average mutation frequency of 5.7 ± 1.0 mutations per 103 bp. In contrast, only 1 in 16 TCRG gene rearrangements amplified from normal T cells harbored a single point mutation (average mutation frequency: 0.2 ± 0.2 mutations per 103 bp, P < 0.01; Table S3). Collectively, these data indicate that besides rearranged Ig genes, non-Ig genes, namely BCL6, TCRB, and TCRG, can also be targeted by aberrant SHM in Ph+ ALL, which is consistent with aberrant expression of AID in this leukemia subset.
BCR-ABL1–induced AID expression in Ph+ ALL
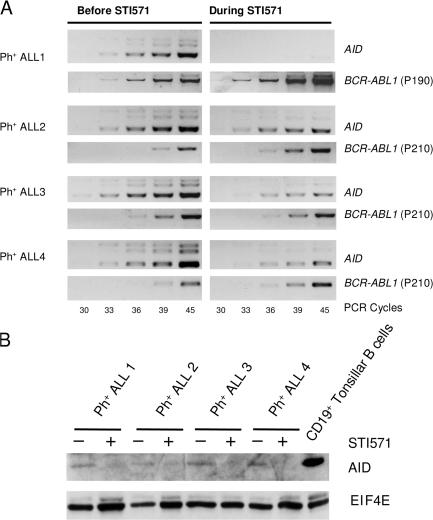
We next investigated whether the Ph-encoded BCR-ABL1 kinase contributes to aberrant AID expression in Ph+ ALL. As shown in Fig. 1, inhibition of BCR-ABL1 kinase activity by STI571 down-regulates AID expression in Ph+ ALL at the mRNA (Fig. 1 B) and protein levels (Fig. 1 C, left). Activation of transgenic expression of BCR-ABL1 in a murine pro–B cell line and forced transient expression of BCR-ABL1 in a Ph− ALL (Fig. 1 C, middle and right) induce de novo expression of AID in these cells. To validate the causative link between BCR-ABL1 kinase activity and aberrant AID expression also in patients suffering from Ph+ ALL, we compared four matched sample pairs of Ph+ ALL before the onset and during continued therapy with the BCR-ABL1 kinase inhibitor STI571 (Fig. 2). Confirming in vitro observations, primary patient–derived Ph+ ALL cells down-regulate AID mRNA (Fig. 2 A) and protein (Fig. 2 B) levels under extended treatment with the BCR-ABL1 kinase inhibitor STI571. We conclude that BCR-ABL1 kinase activity is essential for aberrant AID expression in Ph+ ALL cells.
Figure 2.
AID expression in patient-derived Ph+ ALL cells depends on BCR-ABL1 kinase activity in vivo. Matched sample pairs from four patients with Ph+ ALL before the onset and during continued treatment with the BCR-ABL1 kinase inhibitor STI571 were analyzed for AID mRNA levels by semiquantitative RT-PCR (A). The content of Ph+ ALL cells in all samples was normalized by BCR-ABL1 fusion transcripts (A), with “p190” and p210" indicating the two different breakpoints. Protein lysates from the same Ph+ ALL cases were also subjected to Western blot analysis for AID expression using EIF4E as a loading control (B). Protein lysates from CD19+ tonsillar B cells were used as a positive control.
Given that STI571 inhibits both oncogenic BCR-ABL1 as well as physiologic ABL1 kinase activity, we investigated whether ABL1 kinase activity contributes to AID expression in normal germinal center–derived B cells. To this end, we isolated splenic B cells from C57/BL6 mice and cultured them in the presence or absence of 10 μmol/l STI571 in the presence or absence of 1 ng/ml IL-4 and 25 μg/ml LPS, or both IL-4/LPS and STI571 (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20062662/DC1). To compare the effect of ABL1 kinase inhibition in normal B cells and BCR-ABL1–transformed B cells, splenocytes from BCR-ABL1–transgenic C57/BL6 litter mates were also cultured in the presence or absence of 10 μmol/l STI571. After 3 d, Aid mRNA levels were measured by real-time PCR as a ratio of Aid and Hprt mRNA levels. As described previously (13), activation of splenic B cells resulted in a dramatic increase of Aid mRNA expression. Aid mRNA levels in BCR-ABL1–transgenic B cells were also constitutively higher than in B cells isolated from wild-type littermates (Fig. S1). Interestingly, ABL1 kinase inhibition through STI571 not only diminished Aid mRNA levels in BCR-ABL1–transgenic B cells, but also effectively prevented the up-regulation of Aid in normal B cells upon stimulation by IL-4 and LPS. These findings are paralleled by the blastoid morphology and the formation of large cell aggregates of the Aid-expressing B cells. In the presence of STI571, however, IL-4 and LPS failed to induce a blastoid morphology and formation of cell aggregates (Fig. S1). Although we cannot exclude that the high concentrations of STI571 used in this experiment may inhibit other signaling molecules besides normal ABL1 in the splenic B cells, these findings suggest that Aid induction in normal B cells also requires ABL1 kinase activity. Further experiments that address this possibility are currently under way. Of note, the same concentration of STI571 had no significant effect on Aid mRNA levels in a number of B cell lymphoma cell lines that exhibit constitutive expression of Aid (Fig. S2).
Our finding that BCR-ABL1 induces aberrant AID expression in Ph+ ALL cells is in agreement with a recent study demonstrating that murine B cell precursors infected with the Abelson murine leukemia virus (Abelson-MuLV) also exhibit aberrant expression of Aid (14). The authors of this study attribute aberrant Aid expression induced by the Abelson-MuLV to retroviral infection and interpret aberrant Aid expression as an innate defense mechanism against the transforming retrovirus. Because both BCR-ABL1 and the transforming oncogene of the Abelson-MuLV, v-abl, share ABL1 kinase activity, the results we present here suggest that v-abl kinase activity may also contribute to aberrant expression of Aid in the murine B cell precursors infected by Abelson-MuLV. To test this hypothesis, we analyzed Aid expression in an Abelson-MuLV–transformed murine pre–B cell line (300-19) in the presence or absence of STI571, which inhibits both BCR-ABL1 and v-abl kinase activity (Fig. 1 B). In five repeat experiments, inhibition of v-abl kinase activity resulted in substantial down-regulation of Aid expression in the Abelson-MuLV–transformed 300-19 cells (Fig. 1 B). We conclude that v-abl kinase activity also contributes to Aid expression in Abelson-MuLV–transformed pre–B cells. However, inhibition of v-abl did not abolish Aid expression entirely, which indicates that other factors leading to the up-regulation of Aid (e.g., the anti-retroviral host defense proposed; reference 16) may indeed contribute to Aid expression in these cells as well.
BCR-ABL1–induced up-regulation of AID involves repression of ID2, a negative regulator of AID
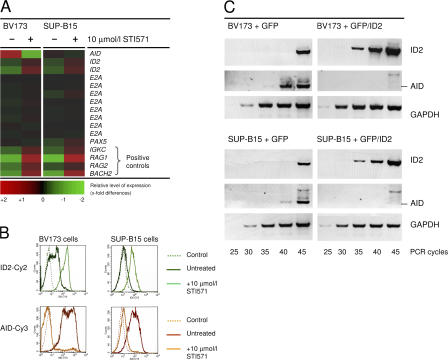
Previous studies demonstrated that AID expression is tightly regulated by the transcription factor pair PAX5 and E2A and the E2A inhibitor ID2 (15, 16). Interestingly, previous work showed that ID2 is among the genes that are transcriptionally activated by STI571-induced ABL kinase inhibition in murine Abelson-MuLV–transformed pre–B cells (17). We therefore investigated the relationship between BCR-ABL1 kinase activity, E2A/PAX5, and their inhibitor ID2 with respect to regulation of AID expression in Ph+ ALL. Analyzing Affymetrix U133A 2.0 microarray data on two Ph+ ALL cell lines (BV173 and SUP-B15; the full dataset is available through GEO accession no. GSE7182) that were cultured in the presence or absence of 10 μmol/l STI571 for 16 h, we confirmed that inhibition of BCR-ABL1 kinase activity by STI571 increased ID2 mRNA levels, whereas mRNA levels for PAX5 and E2A did not change significantly (Fig. 3 A). In addition, the two Ph+ ALL cell lines were treated with STI571 for 48 h, and AID and ID2 protein levels were measured in the surviving cells (annexin V−, propidium iodide−) by flow cytometry (Fig. 3 B). Although BCR-ABL1 kinase inhibition by STI571 decreased AID protein expression, levels of ID2 were clearly increased in the two Ph+ ALL cell lines (Fig. 3 B).
Figure 3.
BCR-ABL1–mediated up-regulation of AID involves repression of ID2, a negative regulator of AID. Two Ph+ ALL cell lines (BV173 and SUP-B15) were incubated in the presence or absence of 10 μmol/l STI571 for 16 h and subjected to microarray analysis using the Affymetrix U133A 2.0 platform as described in Materials and methods (A). mRNA levels of ID2 were compared with those of AID and its positive regulators E2A and PAX5. As controls, known STI571-inducible genes (IGKC, RAG1, RAG2, and BACH2) are shown. (B) The two Ph+ ALL cell lines were cultured for 48 h in the presence or absence of STI571, and protein levels of ID2 (top) and AID (bottom) were measured by flow cytometry. (C) To test the functional relevance of BCR-ABL1–mediated down-regulation of ID2 in Ph+ ALL cells, the effect of ID2 overexpression on AID mRNA levels was measured in Ph+ ALL cells. Therefore, the two Ph+ ALL cell lines were stably transduced with a vector encoding only GFP (left) or both GFP and ID2 (right). Overexpression of ID2 was monitored together with mRNA levels of AID. GAPDH mRNA levels were used for normalization of cDNA amounts.
To test whether up-regulation of ID2 (as observed upon BCR-ABL1 kinase inhibition by STI571) leads to transcriptional repression of AID in Ph+ ALL cells, we transduced two AID-expressing Ph+ ALL cell lines with a lentiviral vector encoding ID2 and GFP or GFP alone as a control. GFP+ cells were sorted and analyzed for mRNA levels of ID2 and AID using GAPDH as a reference gene (Fig. 3 C). Lentiviral overexpression of ID2 indeed resulted in transcriptional inactivation of AID in both Ph+ ALL cell lines. These findings indicate that up-regulation of AID by BCR-ABL1 involves transcriptional repression of ID2, which would act as a negative regulator of AID in the absence of BCR-ABL1 kinase activity.
AID-induced DNA-SSB in Ph+ leukemia cells
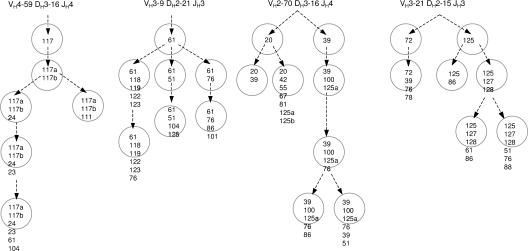
Consistent with an active SHM mechanism, we detected footprints of ongoing subclonal diversification of VH gene segments in 11 of 46 cases of Ph+ ALL (Table S2). Reflecting their clonal evolution, several early mutations are common among many subclones, which differ from each other by subsequently introduced diversifying mutations. As an example, genealogic trees for four VH gene rearrangements amplified from Ph+ ALL cell lines are shown in Fig. 4. To test whether AID expression and aberrant SHM are indeed causally linked in Ph+ ALL, we studied DNA-SSB within rearranged Ig VH genes and the tumor suppressor gene CDKN2B in the presence and absence of AID.
Figure 4.
Tracing the clonal evolution of Ph+ ALL cells by VH region gene mutations. Genealogic trees of ongoing SHM of VH gene segments amplified from the Ph+ ALL cell lines NALM1 and BV173 are shown. Numbers indicate the mutated codons within the rearranged V region. Each circle represents one VH sequence amplified from a leukemia subclone, and a and b denote distinct mutations within the same codon.
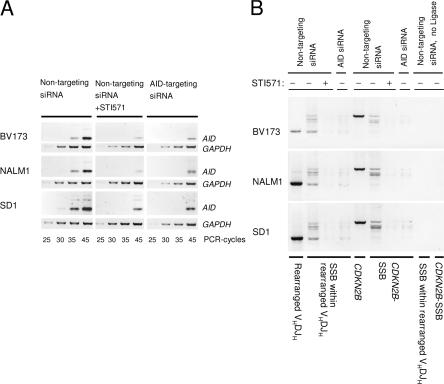
CDKN2B encodes the p15/INK4B tumor suppressor, which is frequently deleted in Ph+ ALL and lymphoid blast crisis (LBC) of Ph+ chronic myeloid leukemia (18, 19). Therefore, we silenced AID expression in three Ph+ ALL cell lines by RNA interference (Fig. 5 A) and studied the effect of AID knockdown on DNA-SSB in rearranged VH genes and the CDKN2B locus. AID targeting and nontargeting small interfering RNA (siRNA) duplexes were fluorochrome labeled and repeatedly transfected into Ph+ ALL cells by nucleofection. At a transfection efficiency between 3 and 10%, fluorochrome-labeled cells were sorted by flow cytometry and subjected to further analysis. RNA interference substantially reduced AID mRNA levels in all three Ph+ ALL cell lines (Fig. 5 A). Comparing Ph+ ALL cells carrying nontargeting siRNAs with Ph+ ALL cells carrying AID-specific siRNA duplexes, DNA-SSB were detected within rearranged VH gene segments as well as within the CDKN2B gene in the former but not in the latter case (Fig. 5 B). Likewise, inhibition of BCR-ABL1 kinase activity by STI571, leading to down-regulation of AID (Fig. 1), largely reduced the frequency of DNA-SSB within VH and CDKN2B genes (Fig. 5 B). We conclude that DNA-SSB introduced into both rearranged VH region genes and the tumor suppressor gene CDKN2B require AID expression. To search for actual somatic mutations within the CDKN2B and the immediately adjacent CDKN2A genes, we attempted to amplify genomic fragments of these genes from multiple Ph+ and Ph− leukemia cell lines. Consistent with previous findings, however (18, 19), we found that one or even both genes were already deleted in Ph+ (BV173, K562, Nalm1, SUP-B15) and Ph− (REH, RS4;11) cell lines, which precluded a comprehensive sequence analysis (not depicted). CDKN2A (INK4A, P16) and CDKN2B (INK4B, P15) belong to a group of genes that encode the INK4 family of tumor suppressors. Interestingly, a recent study demonstrated that deletion of another member of these gene families, CDKN2D (INK4D, P19/ARF), was sufficient to render otherwise normal B cells highly susceptible to aberrant chromosomal rearrangements during AID-mediated CSR (7). Therefore, the loss of one or more of these INK4 tumor suppressors in the context of aberrant AID expression in Ph+ ALL may further enhance genetic instability in these leukemia cells.
Figure 5.
AID induces DNA-SSB in Ig and non-Ig genes. To establish a causative link between AID function and the occurrence of DNA-SSB, AID mRNA expression was silenced in three Ph+ ALL cell lines by RNA interference using fluorochrome-labeled siRNAs against AID or nontargeting siRNA duplexes as a control. Fluorochrome-labeled cells were sorted and analyzed for silencing efficiency and specificity by RT-PCR (A), and for DNA-SSB within rearranged VH gene segments and the CDKN2B gene by LM-PCR (B). For BV173, Nalm1, and SD1 cells, DNA-SSB intermediates in rearranged VH3-21, VH3-9, and VH3-30 gene segments were amplified, respectively. As a loading control for genomic DNA, VH gene rearrangements and a genomic fragment of the CDKN2B gene were amplified (B). The CDKN2A gene at chromosome 9p21 immediately adjacent to CDKN2B was partly or entirely deleted in BV173 and Nalm1 cells, precluding LM-PCR analysis of this locus in these cell lines (not depicted).
DISCUSSION
In summary, these findings identify AID as a mutator within but also outside the Ig gene loci in Ph+ ALL cells. Aberrant expression of AID leading to SHM of Ig and non-Ig genes is driven by oncogenic BCR-ABL1 kinase activity and likely contributes to the particularly unfavorable prognosis of Ph+ leukemia (9). Ph+ leukemias typically carry secondary genetic aberrations (Table S2) and develop resistance to chemotherapy and inhibition of BCR-ABL1 kinase activity within a short period of time (9). Further studies that address a specific role of AID in the acquisition of mutations leading to drug resistance in Ph+ ALL are currently under way. Of note, a small fraction of cases of Ph− pre–B ALL carry mutated VH region genes (and presumably express AID) in the absence of BCR-ABL1. This indicates that other aberrations besides the Ph may likewise induce aberrant SHM in ALL cells. Given that BCR-ABL1–induced up-regulation of AID involves repression of ID2 and indirect activation of E2A, one would envision that other factors that influence the balance between ID2 and E2A may have a similar effect in Ph− leukemia cells.
MATERIALS AND METHODS
Patient samples and cell lines.
Normal pro–B cells (CD19+ CD34+ μ chain−) and pre–B cells (CD19+ VpreB+ μ chain+) were sorted from human bone marrow (from four healthy donors; Cambrex) by flow cytometry using antibodies from BD Biosciences and a FACSVantage SE cell sorter (BD Biosciences). B1 cells (CD19+ CD5+), naive B cells (CD19+ CD27− IgD+), and memory B cells (CD19+ CD27+ IgD−) as well as plasma cells (CD19+ CD20− CD138+) were sorted from the peripheral blood of 12 healthy donors (buffy coats were obtained from the Institute for Blood Transfusion, Heinrich-Heine-Universität Düsseldorf) by flow cytometry using the same FACS sorter. Human germinal center B cells were isolated from tonsillar resectates provided by T. Hoffmann (Heinrich-Heine-Universität Düsseldorf). To this end, tonsillar B cells were preenriched by MACS using immunomagnetic beads against CD19 (Miltenyi Biotec). Thereafter, CD20+ CD38+ germinal center B cells were isolated by flow cytometry as described above using antibodies from BD Biosciences.
In total, 108 cases of ALL were analyzed for AID expression. 28 cases of Ph+ leukemia, including seven cell lines (BV173, CMLT1, K562, NALM1, SD1, SUP-B15, and TOM1; DSMZ,) and 80 cases of Ph− ALL, including eight cell lines (BEL1, HBP-NULL, KASUMI2, MHH-CALL3, NALM6, REH, RS4;11, and 697; DSMZ) were analyzed by RT-PCR (see below). Ph+ leukemia also included five cases of LBC CML (cases 21–25 in Table S2). In these cases, CD19+ CD34+ B lymphoid leukemia cells were sorted by flow cytometry from leukemic bone marrow samples using antibodies from BD Biosciences.
VH gene rearrangements were amplified and sequenced from 106 cases of ALL, including 46 Ph+ and 60 Ph− cases. For 28 cases (18 Ph+, 10 Ph−), information on both AID expression and VH region gene sequence was available. Cytogenetic data on the patient samples and cell lines studied are given in Tables S1 and S2. Patient samples were provided from the Department of Hematology and Oncology, Universität Frankfurt (W.-K. Hofmann) and the Department of Medical Biosciences, Pathology, Umea University, Umea, Sweden (A. Li) in compliance with Institutional Review Board regulations. Murine Abelson-MuLV–transformed pre–B cells (300–19) were provided by M. Reth (Max-Planck-Institute for Immunobiology, Freiburg, Germany). Human Ph+ ALL cells and v-abl–transformed mouse pre–B cells were cultured in the presence or absence of 10 μmol/l STI571 (human ALL) or 1 μmol/l STI571 (murine pre–B cells), respectively. STI571 was provided by Novartis. Germinal center–derived B cell lines (MHH-PREB1, MN60, Karpas-422, MC116, JEKO-1, and SJO) were obtained from DSMZ.
Induced expression of BCR-ABL1 and ID2.
A murine IL-3–dependent pro–B cell line, TONB210, which carries an inducible BCR-ABL1 transgene under the control of a doxycycline-dependent promoter (provided by G.Q. Daley, Harvard Medical School, Boston, MA), and Ph− ALL cells transiently transfected with pMIG-GFP or pMIG-GFP/BCR-ABL1 vectors were studied in cell culture experiments as described previously (20). pMIG-GFP or pMIG-GFP/BCR-ABL1 vectors encode either GFP only or GFP and BCRABL1 and were transfected by electroporation (250 V and 950 μF). For both transfections, GFP+ and GFP− cells were sorted after 24 h and subjected to further analysis. BV173 and SUP-B15 cells were transduced with the lentiviral vector pCL1 (provided by H. Hanenberg, Heinrich-Heine-Universität Düsseldorf, Germany) encoding GFP or GFP and ID2 as described previously (21). The coding sequence of the ID2 cDNA (provided by E. Hara, Science University of Tokyo, Noda, Japan) was excised with BamHI and XhoI and subcloned into pIRESEGFP via BglII and XhoI. The expression cassette containing ID2-IRES-EGFP was digested with NheI and BsrGI and cloned into the lentiviral vector pCL1. 10 d after lentiviral transduction, GFP+ cells were sorted by flow cytometry and further analyzed or kept under cell culture conditions.
Sequence analysis of VH and CH region genes and semiquantitative RT-PCR.
To characterize the configuration of VH and CH region genes, two primer sets were used to amplify the V region alone (using VH- and JH-specific primers) or the V region together with the constant region (using VH- and CH-specific primers) of Ig heavy chain transcripts as described previously (22) in two rounds of PCR using the primers listed in Table S4, which is available at http://www.jem.org/cgi/content/full/jem.20062662/DC1. PCR products were then cloned and sequenced.
Mutation analysis of BCL6 and MYC genes.
For mutation analysis of BCL6 and MYC genes, genomic fragments were amplified and sequenced as described previously (3) using PFU DNA polymerase. For each PCR product, both DNA strands were sequenced and mutations were only counted if they were found both in the forward and reverse sequence. PCR primers used for amplification of BCL6 and MYC fragments are listed in Table S4.
Mutation analysis of TCRB and TCRG V region genes.
TCRB and TCRG gene rearrangements were amplified from multiple leukemia samples, including Ph+ ALL cell lines, primary leukemia cells from Ph+ LBC CML (CD19+ CD34+ B lymphoid cells were sorted from leukemic bone marrow samples), and Ph− ALL cell lines. As controls, TCRB and TCRG gene rearrangements were amplified from normal CD3+ T cells (purified by CD3+ MACS from peripheral blood) using the primers listed in Table S4.
Affymetrix GeneChip analysis and semiquantitative RT-PCR.
Total RNA from cells used for microarray or RT-PCR analysis was isolated by RNeasy (QIAGEN) purification. For microarray analysis, two human Ph+ ALL cell lines (BV173, SUP-B15) were cultured for 16 h in the presence or absence of 10 μmol/l STI571 (Novartis). Double-strand cDNA was generated from 5 μg of total RNA using a poly(dT) oligonucleotide that contains a T7 RNA polymerase initiation site and the SuperScript III Reverse Transcriptase (Invitrogen). Biotinylated cRNA was generated and fragmented according to the Affymetrix protocol and hybridized to U133A 2.0 microarrays (Affymetrix). After scanning (scanner from Affymetrix), the expression values for the genes were determined using Affymetrix GeneChip software. For semiquantitative RT-PCR analysis of human AID, ID2, GAPDH, and BCR-ABL1 and for RT-PCR analysis of murine Aid, Oct2, Obf1, and Hprt transcripts, PCR primers are listed in Table S4.
Western blotting and flow cytometry.
For the detection of AID by Western blot, an antibody against human AID (L7E7; Cell Signaling Technology) was used together with the WesternBreeze immunodetection system (Invitrogen). Detection of EIF4e was used as a loading control (Santa Cruz Biotechnology, Inc.). For analysis of AID and ID2 expression by flow cytometry, antibodies against ID2 (rabbit anti–human ID2 IgG; C-20; Santa Cruz Biotechnology, Inc.) and AID (mouse anti–human AID IgG1; L7E7; Cell Signaling Technology) were used together with secondary antibodies (goat anti–rabbit IgG Cy2 and goat anti–mouse IgG Cy3; Jackson ImmunoResearch Laboratories). Before staining, cells were fixed with 0.4% paraformaldehyde and incubated for 10 min in 90% methanol on ice.
Silencing of AID mRNA expression by RNA interference.
For silencing of AID mRNA expression, one previously validated siRNA (23) and a nontargeting siRNA duplex were used. All siRNA duplexes (for sequences see Table S4) were labeled with fluorescein using an siRNA labeling kit (Ambion) according to the manufacturer's protocol. Fluorochrome-labeled siRNA duplexes were transfected into Ph+ ALL cell lines (BV173, Nalm1, SD1) by nucleofection according to the manufacturer's protocol (Amaxa). Transfection was repeated after 48 h and transfected fluorescein+ cells were sorted by FACS after 72 h as described previously (24). RNA interference–mediated knockdown of AID mRNA expression was verified by RT-PCR.
Ligation-mediated PCR (LM-PCR) for detection of DNA-SSB.
Genomic DNA from 2.5 × 106 cells containing a nick on the lower strand was denatured for 10 min at 95°C. Thereafter, a gene-specific primer (Table S4) was hybridized and extended to the position of the nick as described previously (first strand extension; reference 25), leaving a blunt end using Vent DNA polymerase (New England Biolabs, Inc.). Next, a double-stranded linker was ligated to the newly created blunt end using T4 DNA ligase (Invitrogen) at 14°C overnight. The linker was constructed by annealing of the oligonucleotides 5′- TTTCTGCTCGAATTCAAGCTTCTAACGATGTACGGGGACATG 3′ and 3′ amino (C7)- GACGAGCTTAAGTTCGAAGATTGCTACATGCCCCT-5′, and protruding 3′ overhangs were removed by 3′→5′ exonuclease activity of the Klenow fragment of Escherichia coli DNA polymerase I (Invitrogen). LM-PCR (26) was performed with modifications as described previously (27). In two semi-nested rounds of amplification at an annealing temperature of 59°C, linker-ligated intermediates of DNA-SSB within various genes were amplified using gene-specific primers together with two linker-specific primers (Table S4).
Online supplemental material.
Fig. S1 shows the morphology measurement of mRNA levels of Aid in normal mouse splenocytes that were stimulated with IL-4 and LPS and treated with or without STI571. Fig. S2 shows mRNA levels in human B cell lymphoma cells that constitutively express AID after treatment with or without STI571. Tables S1–S4 and Figs. S1 and S2 are available at http://www.jem.org/cgi/content/full/jem.20062662/DC1.
Supplemental Material
Acknowledgments
We would like to thank Nora Heisterkamp, John Groffen (Los Angeles, CA), Janet D. Rowley (Chicago, IL), and Michael Reth (Freiburg) for critical discussions; Nora Heisterkamp and John Groffen for provision of BCR-ABL1–transgenic mice; Shahab Asgharzadeh (Los Angeles, CA) for his support with quantitative RT-PCR; Kornelia Linnenbrinck and Gabi Tillmann (Düsseldorf, Germany) for help with sequencing; Christoph Göttlinger (Köln, Germany) for cell sorting; and Dr. Gernot Röder (Düsseldorf, Germany) for performing Affymetrix GeneChip hybridizations and analysis.
N. Feldhahn is supported by a fellowship award from the Deutsche José-Carreras-Leukemia Foundation. This work is supported by grants from the Stem Cell Network North-Rhine-Westphalia (to M. Müschen), the Deutsche Forschungsgemeinschaft (through Emmy-Noether-Program; to M. Müschen), the German José-Carreras-Leukemia Foundation (grant to M. Müschen), the Deutsche Krebshilfe (program project grant; to M. Müschen), and the T.J. Martell Foundation.
The authors have no conflicting financial interests.
Abbreviations used: Abelson-MuLV, Abelson murine leukemia virus; AID, activation-induced cytidine deaminase; ALL, acute lymphoblastic leukemia; CSR, class-switch recombination; DNA-SSB, DNA single-strand breaks; LBC, lymphoid blast crisis; LM-PCR, ligation-mediated PCR; Ph, Philadelphia chromosome; SHM, somatic hypermutation; siRNA, small interfering RNA; V, variable.
References
- 1.MacLennan, I.C., and D. Gray. 1986. Antigen-driven selection of virgin and memory B cells. Immunol. Rev. 91:61–85. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 3.Pasqualucci, L., P. Neumeister, T. Goossens, G. Nanjangud, R.S. Chaganti, R. Küppers, and R. Dalla-Favera. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346. [DOI] [PubMed] [Google Scholar]
- 4.Shen, H.M., N. Michael, N. Kim, and U. Storb. 2000. The TATA binding protein, c-Myc and survivin genes are not somatically hypermutated, while Ig and BCL6 genes are hypermutated in human memory B cells. Int. Immunol. 12:1085–1093. [DOI] [PubMed] [Google Scholar]
- 5.Wang, C.L., R.A. Harper, and M. Wabl. 2004. Genome-wide somatic hypermutation. Proc. Natl. Acad. Sci. USA. 101:7352–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramiro, A.R., M. Jankovic, T. Eisenreich, S. Difilippantonio, S. Chen-Kiang, M. Muramatsu, T. Honjo, A. Nussenzweig, and M.C. Nussenzweig. 2004. AID is required for c-Myc/Igh chromosome translocations in vivo. Cell. 118:431–438. [DOI] [PubMed] [Google Scholar]
- 7.Ramiro, A.R., M. Jankovic, E. Callen, S. Difilippantonio, H.T. Chen, K.M. McBride, T.R. Eisenreich, J. Chen, R.A. Dickins, S.W. Lowe, et al. 2006. Role of genomic instability and p53 in AID-induced c-Myc-Igh translocations. Nature. 440:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bird, J., N. Galili, M. Link, D. Stites, and J. Sklar. 1988. Continuing rearrangement but absence of somatic hypermutation in immunoglobulin genes of human B cell precursor leukemia. J. Exp. Med. 168:229–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arico, M., M.G. Valsecchi, B. Camitta, M. Schrappe, J. Chessells, A. Baruchel, P. Gaynon, L. Silverman, G. Janka-Schaub, W. Kamps, et al. 2000. Outcome of treatment in children with Philadelphia chromosome-positive acute lymphoblastic leukemia. N. Engl. J. Med. 342:998–1006. [DOI] [PubMed] [Google Scholar]
- 10.Beishuizen, A., M.A. Verhoeven, E.R. van Wering, K. Hahlen, H. Hooijkaas, and J.J. van Dongen. 1994. Analysis of Ig and T-cell receptor genes in 40 childhood acute lymphoblastic leukemias at diagnosis and subsequent relapse: implications for the detection of minimal residual disease by polymerase chain reaction analysis. Blood. 83:2238–2247. [PubMed] [Google Scholar]
- 11.Brumpt, C., E. Delabesse, K. Beldjord, F. Davi, J.M. Cayuela, C. Millien, P. Villarese, P. Quartier, A. Buzyn, F. Valensi, and E. Macintyre. 2000. The incidence of clonal T-cell receptor rearrangements in B-cell precursor acute lymphoblastic leukemia varies with age and genotype. Blood. 96:2254–2261. [PubMed] [Google Scholar]
- 12.Okazaki, I.M., H. Hiai, N. Kakazu, S. Yamada, M. Muramatsu, K. Kinoshita, and T. Honjo. 2003. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197:1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride, K.M., A. Gazumyan, E.M. Woo, V.M. Barreto, D.F. Robbiani, B.T. Chait, and M.C. Nussenzweig. 2006. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl. Acad. Sci. USA. 103:8798–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gourzi, P., T. Leonova, and F.N. Papavasiliou. 2006. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 24:779–786. [DOI] [PubMed] [Google Scholar]
- 15.Gonda, H., M. Sugai, Y. Nambu, T. Katakai, Y. Agata, K.J. Mori, Y. Yokota, and A. Shimizu. 2003. The balance between Pax5 and Id2 activities is the key to AID gene expression. J. Exp. Med. 198:1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayegh, C.E., M.W. Quong, Y. Agata, and C. Murre. 2003. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 4:586–593. [DOI] [PubMed] [Google Scholar]
- 17.Muljo, S.A., and M.S. Schlissel. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol. 4:31–37. [DOI] [PubMed] [Google Scholar]
- 18.Sill, H., J.M. Goldman, and N.C. Cross. 1995. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of CML. Blood. 85:2013–2016. [PubMed] [Google Scholar]
- 19.Haidar, M.A., X.B. Cao, T. Manshouri, L.L. Chan, A. Glassman, H.M. Kantarjian, M.J. Keating, M.S. Beran, and M. Albitar. 1995. p16INK4A and p15INK4B gene deletions in primary leukemias. Blood. 86:311–315. [PubMed] [Google Scholar]
- 20.Klein, F., N. Feldhahn, S. Herzog, M. Sprangers, J.L. Mooster, H. Jumaa, and M. Müschen. 2006. BCR-ABL1 induces aberrant splicing of IKAROS and lineage infidelity in pre-B lymphoblastic leukemia cells. Oncogene. 25:1118–1124. [DOI] [PubMed] [Google Scholar]
- 21.Feldhahn, N., P. Rio, B.N. Soh, S. Liedtke, M. Sprangers, F. Klein, P. Wernet, H. Jumaa, W.K. Hofmann, H. Hanenberg, et al. 2005. Deficiency of Bruton's tyrosine kinase in B cell precursor leukemia cells. Proc. Natl. Acad. Sci. USA. 102:13266–13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein, F., N. Feldhahn, L. Harder, H. Wang, M. Wartenberg, W.K. Hofmann, P. Wernet, R. Siebert, and M. Müschen. 2004. The BCR-ABL1 kinase bypasses selection for the expression of a pre–B cell receptor in pre–B acute lymphoblastic leukemia cells. J. Exp. Med. 199:673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machida, K., K.T. Cheng, V.M. Sung, S. Shimodaira, K.L. Lindsay, A.M. Levine, M.Y. Lai, and M.M. Lai. 2004. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl. Acad. Sci. USA. 101:4262–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldhahn, N., F. Klein, J.L. Mooster, P. Hadweh, M. Sprangers, M. Wartenberg, M.M. Bekhite, W.K. Hofmann, S. Herzog, H. Jumaa, et al. 2005. Mimicry of a constitutively active pre–B cell receptor in acute lymphoblastic leukemia cells. J. Exp. Med. 201:1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arudchandran, A., R.M. Bernstein, and E.E. Max. 2004. Single-stranded DNA breaks at cytosines occur during Ig gene class switch recombination. J. Immunol. 173:3223–3229. [DOI] [PubMed] [Google Scholar]
- 26.Schlissel, M., A. Constantinescu, T. Morrow, M. Baxter, and A. Peng. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7:2520–2532. [DOI] [PubMed] [Google Scholar]
- 27.Klein, F., N. Feldhahn, S. Lee, H. Wang, F. Ciuffi, M. von Elstermann, M.L. Toribio, H. Sauer, M. Wartenberg, V.S. Barath, et al. 2003. T lymphoid differentiation in human bone marrow. Proc. Natl. Acad. Sci. USA. 100:6747–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.