Abstract
Immunoglobulin class switch recombination (CSR) deficiencies are rare primary immunodeficiencies, characterized by a lack of switched isotype (IgG, IgA, or IgE) production, variably associated with abnormal somatic hypermutation (SHM). Deficiencies in CD40 ligand, CD40, activation-induced cytidine deaminase, and uracil-N-glycosylase may account for this syndrome. We previously described another Ig CSR deficiency condition, characterized by a defect in CSR downstream of the generation of double-stranded DNA breaks in switch (S) μ regions. Further analysis performed with the cells of five affected patients showed that the Ig CSR deficiency was associated with an abnormal formation of the S junctions characterized by microhomology and with increased cell radiosensitivity. In addition, SHM was skewed toward transitions at G/C residues. Overall, these findings suggest that a unique Ig CSR deficiency phenotype could be related to an as-yet-uncharacterized defect in a DNA repair pathway involved in both CSR and SHM events.
Ig class switch recombination (CSR) deficiencies are rare primary immunodeficiencies, usually called hyper-IgM syndromes, whose frequency is 1 in 100,000 births. They are characterized by a defective Ig CSR, as shown by serum IgM levels that are normal or increased, contrasting with a marked decrease, or absence, of IgG, IgA, and IgE (1). As a consequence of the molecular defect, the defective CSR may be associated with defective generation of somatic hypermutations (SHMs) in the Ig variable (V) region. The definition of several Ig CSR deficiencies made possible a better description of the mechanisms underlying CSR and SHM, both required for the maturation of antibody responses (2).
The maturation of the antibody repertoire produces several antibody isotypes with high affinity for antigen, a necessary feature for an efficient humoral response. Antibody maturation occurs mostly in the germinal centers of the secondary lymphoid organs after antigen and T cell–driven activation: CSR results in the production of antibodies of different isotype (IgG, IgA, and IgE) with the same V(D)J specificity and, therefore, the same antigen affinity (3, 4). SHM commonly introduces stochastic mutations (1/103 bp/cell cycle), mainly in the V region of the Ig, a genetic modification that is followed by the positive selection of B cells harboring a B cell receptor (BCR) with high antigen affinity (5, 6). CSR and SHM occur together in germinal centers under BCR/CD40 activation, but neither is a prerequisite for the other because IgM may be mutated, whereas IgG or IgA may not (7–9).
Mutations in the gene encoding the CD40 ligand molecule (CD40L and CD154; references 10–13), a molecule highly expressed on activated follicular helper T cells (14), results in an Ig CSR deficiency generally associated with reduced SHM generation. This observation, corroborated by the description of a similar phenotype caused by mutations in CD40 (15), has demonstrated an essential role for the CD40 signaling pathway in B cells for both CSR and SHM.
Other Ig CSR deficiencies are a consequence of an intrinsic defect of the CSR machinery (16, 17). The autosomal recessive Ig CSR deficiency, caused by mutations in the AICDA gene encoding the activation-induced cytidine deaminase (AID), is characterized by an impairment of both CSR and SHM (18). This finding, together with the description of the phenotypic characteristics of AID−/− mice (19), has demonstrated a key role for AID in antibody maturation. AID selectively modifies cytosine residues into uracils in Switch (S) and V regions (20–23). The uracil-N-glycosylase (UNG), also mutated in another CSR deficiency (24), removes the uracil residues (introduced by AID) from DNA by base excision repair (25).
However, half the Ig CSR deficiencies caused by an intrinsic B cell defect are not secondary to either AID or UNG deficiency. One subset, named HIGM4 (hyper-IgM 4), has been characterized as a CSR block downstream from the DNA double-stranded breaks (DSBs) in S regions, suggesting a defect during DNA repair (26). SHM, only assessed at the time in six patients with sufficient CD27+ B cells, was normal in both frequency and pattern (26). However, in a further investigation in five more patients with low CD27+ B cell numbers, we observed a skewed pattern of SHM, strongly reminiscent of that found in UNG deficiency, although that condition was excluded. These findings suggest a more global defect in DNA repair affecting both the Ig S and V regions. The functional characteristics of CSR deficiency are now described in greater depth.
RESULTS
Skewed pattern of SHM in patients' CD19+/CD27+ B cells
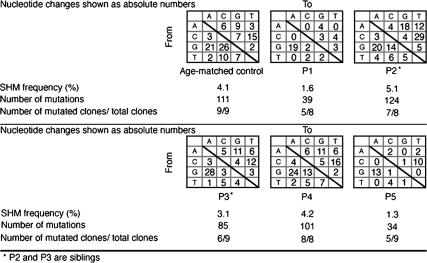
All five patients presented with a defective in vivo and in vitro CSR defect (26). The pattern of SHM was assessed in the IgVH 3–23 region of IgM on purified CD19+CD27+ B lymphocytes (Fig. 1). SHM frequency was either slightly decreased (patients P1 and P5) or normal (P2, P3, and P4), and the ratio of mutated clones to total analyzed clones was generally lower than in controls. G/C nucleotides were preferentially targeted in the two patients with a low SHM frequency, with, respectively, 80 and 74% of mutations on G/C residues (control, 65 ± 4%; range, 55–71%), whereas G/C targeting was normal in P2, P3, and P4. However, in all five patients, although there were variations from one patient to another, including in siblings P2 and P3, SHM showed a skewed pattern of nucleotide substitution on G/C residues: 63–92% mutations were transitions (G > A, C > T), compared with 56 ± 5% (range, 49–62%) in the controls (P < 0.005; nonparametric Mann-Whitney U test). Except for P1, transitions at A/T residues were within normal ranges (Fig. 1).
Figure 1.
SHMs in V regions of patients' CD27+ B cells. SHMs of VH3-23 IgM region were assessed in CD19+CD27+ sorted B cells by RT-PCR using Pfu Taq. Cloned products were sequenced. Nucleotide substitutions are shown as absolute numbers, and SHM frequency is shown as a percentage of mutations occurring among all analyzed nucleotides (at least 2,400). Numbers of mutated clones among all different studied clones are also noted.
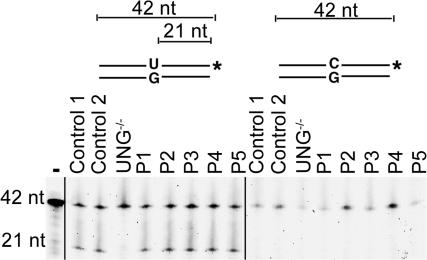
The CSR defect associated with a biased SHM pattern found in these patients was reminiscent of that seen in UNG deficiency (24). The in vitro uracil-DNA glycosylase activity of extracts from patients' cell lines was therefore studied on a double-stranded probe containing an U:G mismatch. Cell-free extracts from all patients were similarly able to undergo base excision, leading to the cleavage of the probe (Fig. 2). As expected, an UNG-deficient B cell line had no detectable activity, and no cleavage was observed using control or patients' cell extracts on a double-stranded DNA probe that did not contain a U/G mismatch. These results, associated with the observation of normal UNG sequences and CSR-induced DSB in Sμ regions (26), excluded a UNG deficiency as the basis of the CSR deficiency found in these patients.
Figure 2.
Normal Uracil incision activity in patients' cell lines. Protein extracts from control fibroblasts (control 1); control EBV B cell line (control 2); P2, P3, and P4 fibroblasts; and P1 and P5 EBV B cell lines were mixed with double-stranded fluorescein-labeled oligonucleotide substrate with or without a single dU/dG mismatch. Cleavage of the probe containing a dU/dG mismatch revealed an efficient base excision activity in patients and controls. No detectable base excision activity was observed with UNG−/− cells or in the absence of protein extracts (−).
Another pathway used for bypassing the UNG-induced abasic site consists of translesion synthesis, involving the REV polymerases (27). Its role in antibody maturation has been recently reported, based on the observation of normal frequency but a skewed pattern of SHM in Rev1-deficient mice (28). Rev1-deficient lines have an increased sensitivity to γ-irradiation and other DNA damaging agents, including methyl methane sulfate (29). Although unlikely, as Rev1−/− mice are not as affected by a CSR defect, REV1, REV3, and REV7 genes were sequenced and found to be normal (unpublished data). The skewed SHM pattern is also reminiscent of mismatch repair (MMR) defect (30). Such a defect, although unlikely because of the absence of cancer during early life (31, 32), was excluded because MSH2, MSH6, MSH5, EXO1, MLH1, and PMS2 RNA transcripts were normally expressed and gene sequence was normal (unpublished data).
Abnormal switch junctions in patients' B cells
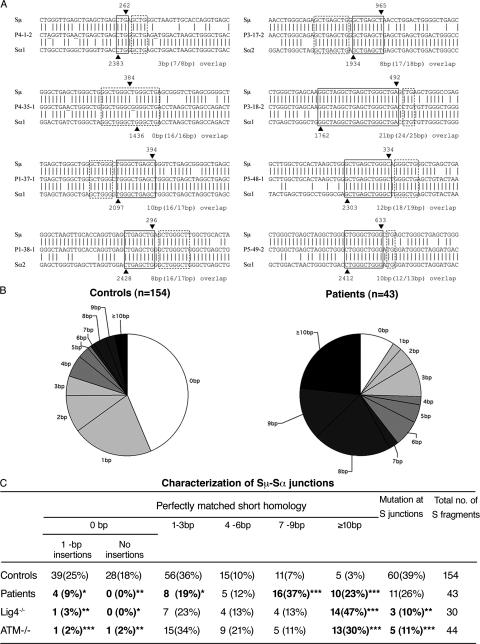
Switch junctions are generated after the processing of DNA ends produced in S regions and several DNA repair defects lead to abnormal structure of these junctions. We therefore characterized switch junctions from patients to detect potential abnormalities. We cloned and sequenced 44 switch fragments (43 Sμ-Sα and 1 Sμ-Sγ-Sα) from B cells of patients P1, P3, P4, and P5. All the switch fragment sequences were unique and therefore represent independent CSR events. Two sequences from each patient are shown in Fig. 3 A. The Sμ-Sα junctions from controls (n = 154), used for comparison, have been previously published (33, 34). There was a significant increase in the extent of donor-acceptor homology at the Sμ-Sα junctions from patients B cells (the mean length of overlap was 7.2 ± 4.7 bp in patients vs. 1.8 ± 3.2 bp in controls; Student's t test, P = 1.2 × 10−9). The majority of junctions (39 out of 43; 91%) from patients displayed a perfectly matched homology (microhomology) of ≥1 bp (i.e., at least one nucleotide is shared by both the Sμ and Sα regions), whereas the remaining four junctions showed a 1-bp insertion and no junction showed precisely joined blunt ends (Fig. 3, B and C). Moreover, 60% of the junctions exhibited a long microhomology of ≥7 bp. When one mismatch was allowed at either side of the switch junction, most of the switch junctions (38 out of 43; 88%) from the patients were flanked by ≥7–8 bp of imperfect repeats (unpublished data). The dramatic shift in using long microhomologies or imperfect repeats in the Sμ-Sα junctions from these patients have previously only been observed in patients with ataxia telangiectasia (A-T) or DNA ligase IV deficiency (Lig4D; references 33, 35). Interestingly, in our patients, the shift was caused by homologies encompassing 7–9 bp or longer, whereas in A-T and Lig4D patients, it was mainly due to an increased usage of microhomologies of ≥10 bp (Fig. 3 C). Of note, a significantly reduced rate of insertions, but not mutations, was observed at or close to the switch junctions of B cells from patients as compared with controls (Fig. 3 C).
Figure 3.
Abnormal pattern of Sμ-Sα junctions in patients' B cells. (A) Sequences of Sμ-Sα junctions. Two sequences from each patient are shown. The Sμ and Sα1 or Sα2 sequences are aligned above and below the recombination switch junctional sequences. Microhomology was determined by identifying the longest region at the switch junction of perfect uninterrupted donor/acceptor identity (boxed with solid lines). Imperfect repeat was determined by identifying the longest overlap region at the switch junction by allowing one mismatch on either side of the breakpoint (the extra nucleotide identified beyond the perfect-matched sequence identity is boxed by dotted lines). The Sμ and Sα breakpoints for each switch fragment are indicated by ▾ and ▴, respectively, and their positions in the germ line sequences are indicated on top of or below the arrowheads. The number of base pairs involved in microhomology and imperfect repeat for each junction is shown at the bottom right of each switch junction. (B) Pie charts demonstrate the perfectly matched short homology usage at Sμ-Sα junctions in controls and patients. The proportion of switch junctions with a given size of perfectly matched short homology is indicated by the size of the slices. (C) Comparison of Sμ-Sα junctions in controls, patients described herein, and A-T and DNA ligase IV–deficient patients. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (χ2 test).
Increased radiosensitivity of patients' cell lines
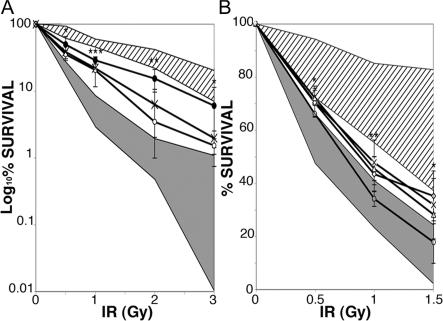
To search for a possible DNA repair defect, the radiosensitivity of patients' fibroblasts was first tested. Fibroblast lines from three patients (P2, P3, and P4) were submitted to increasing doses of γ-irradiation, and their survival was assessed by clonogenic assay. A reproducibly increased radiosensitivity was observed in these three cell lines, including those from the two siblings (P2 and P3). Although the increased radiosensitivity was less marked than that of fibroblasts from patients suffering a DNA repair defect, such as A-T or Artemis deficiency used as controls, it was significantly different from that of healthy fibroblasts (Fig. 4 A; 0.5 and 3 Gy, P < 0.05; 2 Gy, P < 0.005; 1 Gy, P < 0.001 [unpaired two-tailed Student's t test]). These results were confirmed by using Epstein-Barr virus (EBV) B cell lines from P1, P2, P4, and P5 that were more radiosensitive than the control cell lines (Fig. 4 B; 0.5 and 1 Gy, P < 0.05; 2 Gy, P < 0.005 [unpaired two-tailed Student's t test]). In contrast, increased radiosensitivity of AID or UNG-deficient cell lines or cell lines from patients with other forms of Ig CSR deficiency was not observed (Fig. 4). These results suggest a defect in double-stranded DNA break repair.
Figure 4.
Increased radiosensitivity of patients' cell lines. (A) Fibroblasts from patients (P2 [X], P3 [•], and P4 [○]) were irradiated at 0.5–3 Gy. Survival was assessed after 14 d of culture as the number of colony-forming cells compared with nonirradiated cells. Fibroblasts from two age-matched controls and fibroblasts from two Ig CSR–deficient patients with normal SHM (diagonal lines), one ARTEMIS−/−, and one A-T cell line (gray) were positive and negative controls. Results are expressed in log scale. *, P < 0.05; **, P < 0.005; ***, P < 0.001 (unpaired two-tailed Student's t test). (B) EBV B cell lines from patients (P1 [▵], P2 [X], P4 [○], and P5 [⋄]) were irradiated at 0.5–1.5 Gy. After 10 d of culture, survival was assessed as the number of positive wells (defined as viable cell colonies containing >32 cells) for each plate containing irradiated cells compared with number of positive wells for each plate containing unirradiated cells. EBV B cell lines from four controls, three AID-deficient patients, two UNG-deficient patients, and six patients with Ig CSR deficiency with normal SHM (diagonal lines), and two A-T–EBV B cell lines (gray) were used as positive and negative controls. *, P < 0.05; **, P < 0.005 (unpaired two-tailed Student's t test). Radiosensitivity of cell lines was assessed two to four times each. Results are expressed as mean ± SD.
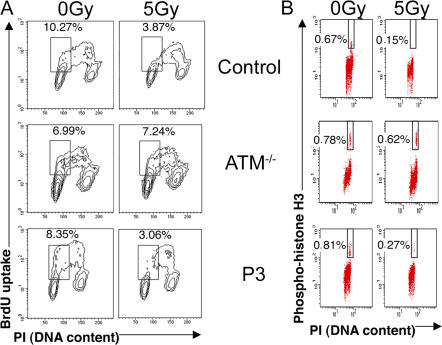
Normal irradiation-induced cell cycle progression arrest in patients' fibroblasts
Another event occurring rapidly after DNA damage sensing in dividing cells is cell cycle progression arrest. We therefore studied the irradiation-induced inhibition of cycle progression in fibroblasts from patients P2, P3, and P4. Arrest of entry into S phase (G1/S checkpoint) was studied 10 h after a 5-Gy irradiation, whereas entry into mitosis (G2/M checkpoint) was assessed after 1 h. Arrested cell cycle progression was observed in the patients' cells in contrast to A-T fibroblasts, which exhibited, as expected, a drastic defect in both checkpoints (Fig. 5). These results show that the increased sensitivity of cells to γ-irradiation does not result from a defect in the cell cycle checkpoints induced by DNA damage.
Figure 5.
Normal irradiation-induced cell cycle progression arrest in patients' fibroblasts. (A) The G1/S cell cycle checkpoint was assessed by BrdU incorporation and DNA content quantification of fibroblasts from a control patient, an A-T patient, and P3 after 5 Gy of irradiation or no irradiation. (B) The G2/M cell cycle checkpoint was assessed by FACS analysis of phosphorylation of histone H3 and DNA content in either untreated or 5 Gy–irradiated fibroblasts from a control patient, an A-T patient, and P3. The same results were obtained in P2 and P4 fibroblasts. Percentages of G1/S and G2/M cells are indicated.
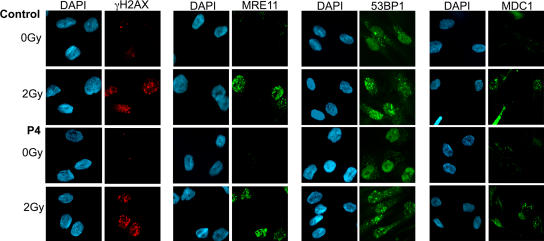
Normal irradiation-induced foci formation in patients' cell lines
Excessive radiosensitivity could be due to a DNA repair defect caused by an impaired recruitment of proteins to double-stranded DNA break sites. We, therefore, studied one of the earliest responses to DNA damage, namely, the induction of histone H2AX phosphorylation (γH2AX; references 36, 37). γH2AX is essential for keeping DNA ends together and for stabilizing the association of DNA repair factors, such as the MRE11–RAD50–NBS1 complex, 53BP1 (tumor.protein.p53-binding.protein.1), and mediator of DNA damage checkpoint 1 (MDC1), at the site of the damage (38). DNA repair foci, including γH2AX, MRE11, 53BP1, and MDC1, were equally recruited 2 h after a 2-Gy irradiation in control and patients' cells, both in fibroblasts (P2, P3, and P4) and in EBV B cell lines (P1, P2, P4, and P5; Fig. 6 and not depicted). FACS analysis did not demonstrate the persistence of γH2AX in nuclei of fibroblasts (P2 and P4) or of EBV B cell lines (P1, P2, P4, and P5) as analyzed at different time points after irradiation (unpublished data). These results indicate that the increased sensitivity to irradiation does not result from a defect in the initial DNA damage sensing, or in a major DNA repair, pathway. Alternatively, the molecular defect may lead to the unrepair of only a fraction of irradiation-induced DNA damage, not detectable in these experiments, as observed in Artemis deficiency (39).
Figure 6.
Normal irradiation-induced foci formation in patients' fibroblasts. Primary fibroblasts from P4 either untreated or irradiated with 2 Gy were labeled with anti-MRE11, anti-53BP1, or anti-γH2AX mouse monoclonal antibodies and anti-MDC1 rabbit polyclonal antibody followed by anti-mouse Alexa Fluor 488, anti-mouse Alexa Fluor 546,or anti-rabbit Alexa Fluor 488. Nuclei were stained with DAPI. The same results were obtained with fibroblasts from patients P2, P3, and P4 and EBV B cell lines from patients P1, P2, and P5. Similar foci formation was observed at 5 Gy of irradiation.
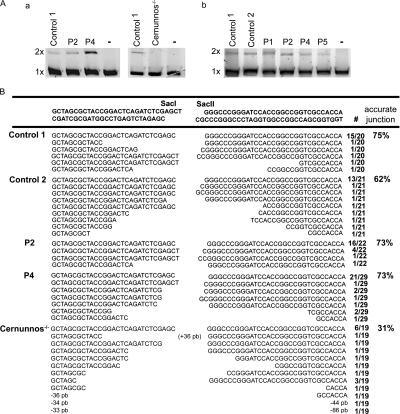
Normal nonhomologous end joining (NHEJ) in patients' cell lines
The major DSB DNA repair pathway used in mammals is the NHEJ pathway. Some NHEJ factors have been shown to be necessary during CSR (40, 41). To study the ability of patients' cells to join double-stranded DNA ends by this pathway, we analyzed the in vitro end joining of linearized plasmid DNA by using patients' fibroblast and/or EBV B cell line extracts by the methods of Baumann and West (42) and Buck et al. (43). The DNA-end ligation assay resulted in the formation of DNA concatemers when extracts from both patient and controls were used (Fig. 7 A). Expectedly, Cernunnos-deficient fibroblast extracts did not lead to concatemer formation. We next examined the ability of patients' cells to join incompatible DNA DSB formed in vivo and, thus, requiring DNA break processing before ligation. P2 and P4 fibroblasts were transfected with restriction enzyme– digested, linearized plasmids containing incompatible 3′-3′ overhang ends. Recircularized plasmids were recovered 72 h after transfection, and their junctions were studied by DNA sequencing (43). Most junctions in plasmids recovered from both patient and control fibroblasts showed similarly accurate repair, in contrast to what is observed in Cernunnos- deficient fibroblasts (Fig. 7 B). Because a subtle defect may be undetectable in these assays, we excluded hypomorphic defects in DNA-dependent protein kinase, Ku complex, Cernunnos, Artemis, XRCC4 (X-ray repair complementing defective in chinese hamster), and ligase IV either by gene sequencing in patients or by study of segregation with polymorphic markers in both informative families (P2, P3, and P5; unpublished data).
Figure 7.
Normal in vitro and in vivo NHEJ assays in patients. (A) In vitro NHEJ activity was assessed by testing ligation of a linearized plasmid incubated with protein extracts from fibroblasts from two age-matched controls, from patients P2 and P4, and one Cernunnos-deficient patient as a negative control (a), and from EBV B cell lines from two controls and patients P1, P2, P4, and P5 (b). The reaction products were run on agarose gel and stained with SYBR green. Dimerized DNA molecules (2×) were formed with controls and P1–P5 extracts, but not, as expected, with the Cernunnos-deficient fibroblast extracts or without WCEs (–). (B) To test the in vivo function of the NHEJ pathway, fibroblasts from two age-matched controls, patients P2 and P4, and one Cernunnos-deficient patient were transfected with a 3′-3′ linearized plasmid. After 72 h of culture, plasmids were extracted, and the ligation junctions were PCR amplified, cloned, and sequenced. NHEJ was found to be normal in patients' cells, as shown by a similar number of accurate junctions in control and patients' fibroblasts. The frequency of each sequence among analyzed clones is indicated.
DISCUSSION
We have described here a new primary immunodeficiency, characterized by a defect in Ig class switching associated with a reduced memory B cell population and a skewed nucleotide substitution in SHM. The observation of a female born to consanguineous parents and of two affected siblings suggests an autosomal recessive inheritance pattern. Although all patients exhibited the same CSR defect with increased radiosensitivity, there were some differences between patients in regard to SHM pattern and frequency. Because the underlying molecular basis of this condition is still unknown, we do not know so far whether all patients are affected by distinct molecular defects or by distinct mutations of the same gene. Other genetic and/or environmental factors could also be involved, according to P2 and P3 phenotypic differences. However, the overall phenotype was unique in these five patients. Several hypotheses, including one of defective survival of switched B cells, or of a defect in the CSR/SHM machinery involving DNA repair, could account for the condition.
Abnormalities in survival signaling of switched B cells could underlie this condition. Molecular interactions are known to be essential for B cell survival, including that of B cell activating factor (BAFF) with its receptor on B cells, BAFF-R (44–46). The observation of fewer switched CD27+ B cells could fit this model. However, the observed in vitro defective CD40-dependent CSR cannot be accounted for by a BAFF-R or BAFF abnormality. A response to DNA damage leading to inappropriate cell death should also be considered. In cells other than germinal center B cells, DSBs activate p53 and p21, resulting in cell cycle arrest and apoptosis. In contrast, in germinal centers, the p53 response to DNA damage is directly inhibited by the highly expressed transcriptional regulator B cell lymphoma 6 (BCL6), whereas p21-induced cell cycle arrest is suppressed through interaction of its transcriptional activator Miz1 (protein inhibitor of activated STAT2) with BCL6. Both these events enable intense proliferation of B cells undergoing CSR (47). Fitting in with this observation, BCL6-deficient mice are depleted of germinal centers because of a strong B cell apoptosis (48). Such a defect in transcriptional repression of proteins involved in cell cycle arrest induced by DNA damage could also underlie this Ig CSR deficiency. Another interesting hypothesis is related to the recently described role of phosphoinositide-3 kinase (PI3K) acting as a negative regulator of CSR (49). PI3K-induced CSR inhibition is dependent on the inhibition of AICDA transcription via B lymphocyte–induced maturation protein 1 overexpression and inactivation of the Forkhead Box family (FOXO) of transcription factors by the serine threonine kinase AKT. Activated B cells from the patients in this study expressed the AICDA gene transcripts and AID protein normally. However, PI3K also exerts a downstream function in AID activity regulation, as shown by the observation that overexpressed AID does not fully compensate for the inhibitory effect of PI3K on CSR. Thus, any abnormal signal leading to increased PI3K activity in patients' B cells could induce a CSR deficiency.
Alternatively, a defect in the complex machinery underlying the SHM and the CSR processes, and especially the DNA repair, appears more likely because of abnormal S junctions and increased radiosensitivity of fibroblasts and EBV B cell lines. AID and UNG-induced DNA lesions are repaired differently in S and V regions. The CSR-induced DSB repair requires phosphorylation of the H2AX histone (γH2AX), as well as the MRE11–RAD50–NBS1 complex and the 53BP1 and MDC1 proteins, as shown by the phenotype of mice or B cells depleted of each of these molecules (50–54). The observed normal accumulation of γH2AX, MRE11–RAD50–NBS1 complex, and 53BP1 and MDC1 proteins on DNA repair foci of patients' cells excluded a defect in the first step after DNA damage sensing, leading to DNA repair. In addition, the H2AX, 53BP1, and MDC1 genes were sequenced and found to be normal. MRE11 and NBS1 mutations in man lead to well-known syndromes, A-T–like disease (55) and Nijmegen syndrome (56), respectively. ATM (A-T mutated) is involved in DNA repair of S regions (33, 57). Nevertheless, as A-T patients exhibit an Ig CSR deficiency (reference 58; unpublished data), a potential role for ATM could be envisaged. The observation of a normal irradiation-induced cell cycle progression arrest excludes an abnormal ATM-mediated cell cycle checkpoint pathway. Moreover, the switched junctions, although based on microhomologies, were slightly different from those observed in A-T. The NHEJ enzymes have been shown to be required for CSR (35, 40, 41, 59). However, such deficiency of these factors is unlikely because of normal TCR and BCR expression and function in patients' cells, as well as the absence of γH2AX persistence after irradiation and the normal results obtained for both NHEJ assays. However, we cannot definitively rule out a defect in a NHEJ factor redundant with other factors in mediating the V(D)J recombination process that remains undetectable in the experimental assays used. It should be stressed that the nature of the switch junctions and the bias to transitions at G/C residues do not fit well with defective NHEJ. The SHM abnormality could be the consequence of a defective repair of V region breaks. The nature of the DNA breaks in V regions and their repair have not been completely elucidated (60). The recent observation that the MRE11–RAD50–NBS1 complex localizes on V regions is compatible with DSB generation in V regions during SHM introduction (61). The MMR enzymes play a role in CSR and SHM in mice, as shown by the slightly defective CSR and skewed pattern of SHM observed in MMR-deficient mice (62, 63). In addition, abnormality of S junctions observed in the B cells from our patients is reminiscent of that observed in Mlh1- or Pms2-deficient mice (30), both defects, however, that are excluded by gene sequencing. Thus, an as-yet- uncharacterized defect in a DNA repair pathway can be postulated to account for a unique phenotype characterized by defective CSR and SHM, associated with an abnormality of the switch junction repair and increased cell radiosensitivity. This factor could be required for efficient NHEJ in S regions and DNA repair of V regions. It could also be NHEJ independent. Of note, AID-dependent illegitimate recombination events occurring between the IgH locus and c-myc in B cell lymphomagenesis have been shown to be mediated by an as-yet-unknown NHEJ-independent process (64). It is, thus, attractive to consider that this as-yet-uncharacterized DNA repair pathway might be physiologically involved in the CSR and SHM processes.
MATERIALS AND METHODS
Patients.
We studied five patients (4 males and 1 female), 4–8 yr of age, from four unrelated families. P2 and P3 were siblings, and P5 was born to a consanguineous family. All patients suffered from recurrent bacterial infections. P4 also presented with enlarged lymph nodes, and P4 and P5 had severe autoimmune hemolytic anemia. P3 died from severe hepatitis, and P5 suddenly died at 6 yr of age. A diagnosis of defective Ig CSR was made on the basis of low-serum IgG, IgA, and IgE concentrations and high-serum IgM concentrations (Table I). Cases P1, P2, P3, and P4 were previously reported (26). B cell and T cell counts were similar to those of age-matched controls, and T cell functions were found to be normal (unpublished data). However, the percentage of memory B cells was strongly decreased, and switched B cells were virtually absent (Fig. 1), whereas the in vitro IgE production of B cells upon soluble CD40L+ IL4 activation was negative, though B cells proliferated normally in that setting. The in vitro CSR defect was shown to occur downstream of DNA DSBs (26). AID and UNG deficiencies were excluded by normal gene sequence. AID protein was detectable by Western blot in activated B cells whenever tested (EBV B cell lines from P1, P2, P4, and P5 and soluble CD40L+IL4-activated B cells from P1 and P4 (unpublished data). Immunological and genetic studies were performed after the informed consent of parents. The study was approved by the ethics committee.
Table I.
Phenotype of patients
| Characteristic | P1 | P2a | P3a | P4 | P5 | Age-matched controls |
|---|---|---|---|---|---|---|
| Age at diagnosis (yr) | 6 | 6 | 8 | 4 | 4 | — |
| Serum Ig levels (g/l) | — | |||||
| IgM | 16 | 4.1 | 2.6 | 2.8 | 0.9 | 0.5–1.2 |
| IgG | 0.6 | 3.6 | 5.7 | 0.7 | 1.4 | 6.8–12.5 |
| IgA | 0.3 | <0.03 | <0.03 | <0.03 | <0.03 | 0.6–1.6 |
| T lymphocyte counts/μl | 2,400 | 1,420 | 1,238 | 1,612 | 1,300 | 1,200–2,600 |
| B lymphocyte counts/μl | 452 | 120 | 170 | 462 | 440 | 110–570 |
| IgD+/CD19+ (%)b | 99.9 | 99.2 | ND | 99.8 | ND | 83–93 |
| CD27+/CD19+ (%)b | 1.5 | 3.3 | 3.1 | 4.7 | 3.1 | 13–58 |
P2 and P3 are siblings.
Gated on CD19+ B cells.
Analysis of SHM in variable gene of Ig.
SHM in VH3-23 IgM gene in CD19+CD27+ sorted B cells was assessed as described previously (18). RT-PCR was performed with 0.5 U Pfu polymerase (Stratagene) and the primers V3-23 leader exon and CμB (35 cycles at 94°C for 45 s, 60°C for 1.5 min, and 72°C for 2 min). Products were cloned and sequenced.
Uracil incision assay.
Whole-cell extracts (WCEs) were prepared by cells lysis in buffer 1 (10 mM TRIS, pH 8, 1 mM EDTA, 5 mM dithiothreitol [DTT], and protease inhibitors) followed by the addition of 0.5 volume of buffer 2 (50 mM Tris, pH 8.0, 1 M Kcl, 2 mM EDTA, and 2 mM DTT) and submitted to three freeze–thaw cycles. After centrifugation, supernatants were dialyzed against dialysis buffer (20 mM Tris, pH 8.0, 20% glycerol, 0.1 M K [OAc], 0.5 mM EDTA, and 1 mM DTT).
For the uracil incision assay, adapted from Di Noia (65), 10 μg WCEs were mixed in reaction buffer with 1 pmol of double-stranded, FITC- labeled oligonucleotide containing or not containing a single dU/dG mismatch in 20 μl. After 2 h at 37°C, the reaction was stopped with 10 μl of formamide loading dye (GE Healthcare) and products run on a denaturing gel were visualized via FluorImager (FLA3000; Fujifilm).
Study of switch junctions.
The amplification of Sμ-Sα fragments from in vivo switched B cells was performed as described previously (33, 66). The PCR-amplified switch fragments were gel purified, cloned, and sequenced. The switch breakpoints were determined by aligning the switch fragment sequences with the Sμ (X54713), Sα1 (L191219), or Sα2 (AF030305) sequences. Analysis of microhomology usage at the junctions and mutations ± 15 bp around the junction and upstream Sμ region were performed as described previously (33, 58). Data from controls, A-T, and DNA ligase IV–deficient patients were described previously (33–35).
Ionizing radiation sensitivity assay.
Primary fibroblasts were irradiated (137Cs source) with different doses (0, 0.5, 1, 2, or 3 Gy), and serial dilutions were cultured for 14 d in a 10-cm culture dish. The number of colonies for each dose was assessed, and ratios referring to the dilutions of the nonirradiated cells were determined (67).
Irradiated (0, 0.5, 1, or 1.5 Gy) EBV B cell lines were serially diluted and cultured in a 96-well plate for 10 d (68). Each well containing viable cell colonies, dark blue stained after a 4-h incubation in 1 mg/ml 3-(4,5dimethylthiazol-2-4)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich), with >32 cells was scored as positive.
G1/S and G2/M checkpoint cell cycle analysis.
For G1/S checkpoint, primary fibroblasts were subjected to 5-Gy γ-rays, followed by a 1-h pulse of BrdU incorporation 10 h after irradiation, and incubated with FITC-conjugated anti-BrdU antibody (Becton Dickinson; reference 69). G2/M checkpoint was performed in immortalized SV40-transformed fibroblasts harvested 1 h after 5 Gy of γ-irradiation labeled with a rabbit polyclonal anti-phosphohistone H3 (Upstate Biotechnology) detected with FITC- conjugated goat anti–rabbit IgG antibody (Becton Dickinson; reference 70). Determination of the cellular proportions in G1/S and G2/M phases and DNA content measured by propidium iodide were analyzed by FACS (Becton Dickinson).
DNA repair foci detection.
2 h after 2–5 Gy of γ-irradiation, primary fibroblasts or EBV B cell lines were labeled with the mouse monoclonal anti-γH2AX (Ser139; clone JBW103; Upstate Biotechnology), mouse monoclonal anti-MRE11 (clone 12D7; Abcam), mouse monoclonal anti-53BP1 (provided by I. Ward, Mayo Clinic College of Medicine, Rochester, MN), or rabbit polyclonal anti-MDC1 (provided by S. Grant, Baylor College of Medicine, Houston, TX) antibodies. Cells were incubated with the secondary antibodies goat anti-mouse Alexa Fluor 488, goat anti-mouse Alexa Fluor 546, and goat anti-rabbit Alexa Fluor 488 (Invitrogen). Slides were counterstained with DAPI and analyzed by epifluorescence microscopy (Axioplan; Carl Zeiss MicroImaging, Inc.).
In vitro and in vivo NHEJ assay.
Functional activity of the NHEJ system repair was assessed in vitro (43) by incubation of 5 μg WCEs with 25 ng EcoRI-digested pEGFPN2 in ligation buffer for 1 h at 37°C. Reactions were treated with 1 mg/ml RNase followed by deproteination. Samples were run on agarose gels and stained by SYBR gold (Invitrogen), and fluorescence was detected via a FluorImager.
In vivo NHEJ assay was performed as described previously (43), and 5 μg SacI–SacII linearized 3′-3′ overhang-ends EGFP-N2 plasmid (CLONTECH Laboratories, Inc.) were introduced into primary fibroblasts by electroporation. After 72 h, recircularized plasmids were extracted. The junctions were PCR amplified, cloned, and sequenced.
Acknowledgments
We are indebted to Dr. I. Ward and to Dr. G. Stewart for the kind gift of mouse monoclonal anti-53BP1 and for rabbit polyclonal anti-MDC1 antibodies and Drs. J. Duguet (Le Mans, France), I. Tezscan, and F. Erzoy (Ankara, Turkey) for care of the patients. We thank Mrs. Monique Forveille, Mrs. Françoise Selz, and M. Luc Nonnenmacher for their excellent technical assistance and Malika Sifouane for skillful secretarial assistance. We are grateful to Amgen for kindly providing the soluble CD40L.
This work was supported in part by grants from l'Institut National de la Santé et de la Recherche Médicale, l'Association de la Recherche Contre le Cancer, la Ligue Contre le Cancer, the European Community (no. PL 006411 [EUROPOLICY-PID]; Sixth Programme Cadre de Recherche et Développement), the French Rare Disease Program, the Association Nationale pour la Recherche, the Institut National du Cancer, and the Hungarian Research Fund (OTKA T49017). P. Revy is a scientist from Centre National de la Recherche Scientifique.
The authors have no conflicting financial interests.
Abbreviations used: AID, activation-induced cytidine deaminase; A-T, ataxia telangiectasia; ATM, A-T mutated; BCR, B cell receptor; CSR, class switch recombination; DSB, double-stranded break; EBV, Epstein-Barr virus; MDC1, mediator of DNA damage checkpoint 1; MMR, mismatch repair; NHEJ, nonhomologous end joining; SHM, somatic hypermutation; UNG, uracil-N-glycosylase; WCE, whole-cell extract; 53BP1, tumor.protein.p53-binding.protein.1.
References
- 1.Notarangelo, L.D., M. Duse, and A.G. Ugazio. 1992. Immunodeficiency with hyper-IgM (HIM). Immunodefic. Rev. 3:101–121. [PubMed] [Google Scholar]
- 2.Durandy, A., S. Peron, and A. Fischer. 2006. Hyper-IgM syndromes. Curr. Opin. Rheumatol. 18:369–376. [DOI] [PubMed] [Google Scholar]
- 3.Iwasato, T., A. Shimizu, T. Honjo, and H. Yamagishi. 1990. Circular DNA is excised by immunoglobulin class switch recombination. Cell. 62:143–149. [DOI] [PubMed] [Google Scholar]
- 4.Kinoshita, K., and T. Honjo. 2000. Unique and unprecedented recombination mechanisms in class switching. Curr. Opin. Immunol. 12:195–198. [DOI] [PubMed] [Google Scholar]
- 5.Storb, U., A. Peters, E. Klotz, N. Kim, H.M. Shen, J. Hackett, B. Rogerson, and T.E. Martin. 1998. Cis-acting sequences that affect somatic hypermutation of Ig genes. Immunol. Rev. 162:153–160. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs, H., K. Rajewsky, Y. Fukita, and L. Bross. 2001. Indirect and direct evidence for DNA double-strand breaks in hypermutating immunoglobulin genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaartinen, M., G.M. Griffiths, A.F. Markham, and C. Milstein. 1983. mRNA sequences define an unusually restricted IgG response to 2-phenyloxazolone and its early diversification. Nature. 304:320–324. [DOI] [PubMed] [Google Scholar]
- 8.Jacob, J., and G. Kelsoe. 1992. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath-associated foci and germinal centers. J. Exp. Med. 176:679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, Y.J., F. Malisan, O. de Bouteiller, C. Guret, S. Lebecque, J. Banchereau, F.C. Mills, E.E. Max, and H. Martinez-Valdez. 1996. Within germinal centers, isotype switching of immunoglobulin genes occurs after the onset of somatic mutation. Immunity. 4:241–250. [DOI] [PubMed] [Google Scholar]
- 10.Korthauer, U., D. Graf, H.W. Mages, F. Briere, M. Padayachee, S. Malcolm, A.G. Ugazio, L.D. Notarangelo, R.J. Levinsky, and R.A. Kroczek. 1993. Defective expression of T-cell CD40 ligand causes X-linked immunodeficiency with hyper-IgM. Nature. 361:539–541. [DOI] [PubMed] [Google Scholar]
- 11.DiSanto, J.P., J.Y. Bonnefoy, J.F. Gauchat, A. Fischer, and G. de Saint Basile. 1993. CD40 ligand mutations in X-linked immunodeficiency with hyper-IgM. Nature. 361:541–543. [DOI] [PubMed] [Google Scholar]
- 12.Aruffo, A., M. Farrington, D. Hollenbaugh, X. Li, A. Milatovich, S. Nonoyama, J. Bajorath, L.S. Grosmaire, R. Stenkamp, M. Neubauer, et al. 1993. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 72:291–300. [DOI] [PubMed] [Google Scholar]
- 13.Allen, R.C., R.J. Armitage, M.E. Conley, H. Rosenblatt, N.A. Jenkins, N.G. Copeland, M.A. Bedell, S. Edelhoff, C.M. Disteche, D.K. Simoneaux, et al. 1993. CD40 ligand gene defects responsible for X-linked hyper-IgM syndrome. Science. 259:990–993. [DOI] [PubMed] [Google Scholar]
- 14.Breitfeld, D., L. Ohl, E. Kremmer, J. Ellwart, F. Sallusto, M. Lipp, and R. Forster. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 192:1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrari, S., S. Giliani, A. Insalaco, A. Al-Ghonaium, A.R. Soresina, M. Loubser, M.A. Avanzini, M. Marconi, R. Badolato, A.G. Ugazio, et al. 2001. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc. Natl. Acad. Sci. USA. 98:12614–12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callard, R.E., R.J. Armitage, W.C. Fanslow, and M.K. Spriggs. 1993. CD40 ligand and its role in X-linked hyper-IgM syndrome. Immunol. Today. 14:559–564. [DOI] [PubMed] [Google Scholar]
- 17.Durandy, A., C. Hivroz, F. Mazerolles, C. Schiff, F. Bernard, E. Jouanguy, P. Revy, J.P. DiSanto, J.F. Gauchat, J.Y. Bonnefoy, et al. 1997. Abnormal CD40-mediated activation pathway in B lymphocytes from patients with hyper-IgM syndrome and normal CD40 ligand expression. J. Immunol. 158:2576–2584. [PubMed] [Google Scholar]
- 18.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. [DOI] [PubMed] [Google Scholar]
- 19.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhury, J., M. Tian, C. Khuong, E. Pinaud, and F.W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. [DOI] [PubMed] [Google Scholar]
- 21.Bransteitter, R., P. Pham, M.D. Scharff, and M.F. Goodman. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA. 100:4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. [DOI] [PubMed] [Google Scholar]
- 23.Dickerson, S.K., E. Market, E. Besmer, and F.N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai, K., G. Slupphaug, W.I. Lee, P. Revy, S. Nonoyama, N. Catalan, L. Yel, M. Forveille, B. Kavli, H.E. Krokan, et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 4:1023–1028. [DOI] [PubMed] [Google Scholar]
- 25.Rada, C., G.T. Williams, H. Nilsen, D.E. Barnes, T. Lindahl, and M.S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 26.Imai, K., N. Catalan, A. Plebani, L. Marodi, O. Sanal, S. Kumaki, V. Nagendran, P. Wood, C. Glastre, F. Sarrot-Reynauld, et al. 2003. Hyper-IgM syndrome type 4 with a B lymphocyte-intrinsic selective deficiency in Ig class-switch recombination. J. Clin. Invest. 112:136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, Y., X. Wu, O. Rechkoblit, N.E. Geacintov, J.S. Taylor, and Z. Wang. 2002. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 30:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansen, J.G., P. Langerak, A. Tsaalbi-Shtylik, P. van den Berk, H. Jacobs, and N. de Wind. 2006. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 203:319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada, T., E. Sonoda, M. Yoshimura, Y. Kawano, H. Saya, M. Kohzaki, and S. Takeda. 2005. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol. Cell. Biol. 25:6103–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrader, C.E., J. Vardo, and J. Stavnezer. 2002. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J. Exp. Med. 195:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Q., C. Lasset, F. Desseigne, D. Frappaz, C. Bergeron, C. Navarro, E. Ruano, and A. Puisieux. 1999. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res. 59:294–297. [PubMed] [Google Scholar]
- 32.Whiteside, D., R. McLeod, G. Graham, J.L. Steckley, K. Booth, M.J. Somerville, and S.E. Andrew. 2002. A homozygous germ-line mutation in the human MSH2 gene predisposes to hematological malignancy and multiple cafe-au-lait spots. Cancer Res. 62:359–362. [PubMed] [Google Scholar]
- 33.Pan, Q., C. Petit-Frere, A. Lahdesmaki, H. Gregorek, K.H. Chrzanowska, and L. Hammarstrom. 2002. Alternative end joining during switch recombination in patients with ataxia-telangiectasia. Eur. J. Immunol. 32:1300–1308. [DOI] [PubMed] [Google Scholar]
- 34.Lahdesmaki, A., A.M. Taylor, K.H. Chrzanowska, and Q. Pan-Hammarstrom. 2004. Delineation of the role of the Mre11 complex in class switch recombination. J. Biol. Chem. 279:16479–16487. [DOI] [PubMed] [Google Scholar]
- 35.Pan-Hammarstrom, Q., A.M. Jones, A. Lahdesmaki, W. Zhou, R.A. Gatti, L. Hammarstrom, A.R. Gennery, and M.R. Ehrenstein. 2005. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J. Exp. Med. 201:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paull, T.T., E.P. Rogakou, V. Yamazaki, C.U. Kirchgessner, M. Gellert, and W.M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886–895. [DOI] [PubMed] [Google Scholar]
- 37.Rogakou, E.P., D.R. Pilch, A.H. Orr, V.S. Ivanova, and W.M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 38.Bassing, C.H., and F.W. Alt. 2004. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 3:149–153. [DOI] [PubMed] [Google Scholar]
- 39.Riballo, E., M. Kuhne, N. Rief, A. Doherty, G.C. Smith, M.J. Recio, C. Reis, K. Dahm, A. Fricke, A. Krempler, et al. 2004. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 16:715–724. [DOI] [PubMed] [Google Scholar]
- 40.Casellas, R., A. Nussenzweig, R. Wuerffel, R. Pelanda, A. Reichlin, H. Suh, X.F. Qin, E. Besmer, A. Kenter, K. Rajewsky, and M.C. Nussenzweig. 1998. Ku80 is required for immunoglobulin isotype switching. EMBO J. 17:2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manis, J.P., N. van der Stoep, M. Tian, R. Ferrini, L. Davidson, A. Bottaro, and F.W. Alt. 1998. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 188:1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumann, P., and S.C. West. 1998. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. USA. 95:14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buck, D., L. Malivert, R. de Chasseval, A. Barraud, M.C. Fondaneche, O. Sanal, A. Plebani, J.L. Stephan, M. Hufnagel, F. le Deist, et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 124:287–299. [DOI] [PubMed] [Google Scholar]
- 44.Schiemann, B., J.L. Gommerman, K. Vora, T.G. Cachero, S. Shulga-Morskaya, M. Dobles, E. Frew, and M.L. Scott. 2001. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 293:2111–2114. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J.S., S.A. Bixler, F. Qian, K. Vora, M.L. Scott, T.G. Cachero, C. Hession, P. Schneider, I.D. Sizing, C. Mullen, et al. 2001. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science. 293:2108–2111. [DOI] [PubMed] [Google Scholar]
- 46.Kayagaki, N., M. Yan, D. Seshasayee, H. Wang, W. Lee, D.M. French, I.S. Grewal, A.G. Cochran, N.C. Gordon, J. Yin, et al. 2002. BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2. Immunity. 17:515–524. [DOI] [PubMed] [Google Scholar]
- 47.Phan, R.T., M. Saito, K. Basso, H. Niu, and R. Dalla-Favera. 2005. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat. Immunol. 6:1054–1060. [DOI] [PubMed] [Google Scholar]
- 48.Ye, B.H., G. Cattoretti, Q. Shen, J. Zhang, N. Hawe, R. de Waard, C. Leung, M. Nouri-Shirazi, A. Orazi, R.S. Chaganti, et al. 1997. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 16:161–170. [DOI] [PubMed] [Google Scholar]
- 49.Omori, S.A., M.H. Cato, A. Anzelon-Mills, K.D. Puri, M. Shapiro-Shelef, K. Calame, and R.C. Rickert. 2006. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 25:545–557. [DOI] [PubMed] [Google Scholar]
- 50.Celeste, A., S. Petersen, P.J. Romanienko, O. Fernandez-Capetillo, H.T. Chen, O.A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M.J. Difilippantonio, et al. 2002. Genomic instability in mice lacking histone H2AX. Science. 296:922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theunissen, J.W., M.I. Kaplan, P.A. Hunt, B.R. Williams, D.O. Ferguson, F.W. Alt, and J.H. Petrini. 2003. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell. 12:1511–1523. [DOI] [PubMed] [Google Scholar]
- 52.Ward, I.M., K. Minn, J. van Deursen, and J. Chen. 2003. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reina-San-Martin, B., M.C. Nussenzweig, A. Nussenzweig, and S. Difilippantonio. 2005. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc. Natl. Acad. Sci. USA. 102:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lou, Z., K. Minter-Dykhouse, S. Franco, M. Gostissa, M.A. Rivera, A. Celeste, J.P. Manis, J. van Deursen, A. Nussenzweig, T.T. Paull, et al. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 21:187–200. [DOI] [PubMed] [Google Scholar]
- 55.Stewart, G.S., R.S. Maser, T. Stankovic, D.A. Bressan, M.I. Kaplan, N.G. Jaspers, A. Raams, P.J. Byrd, J.H. Petrini, and A.M. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 99:577–587. [DOI] [PubMed] [Google Scholar]
- 56.Varon, R., C. Vissinga, M. Platzer, K.M. Cerosaletti, K.H. Chrzanowska, K. Saar, G. Beckmann, E. Seemanova, P.R. Cooper, N.J. Nowak, et al. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 93:467–476. [DOI] [PubMed] [Google Scholar]
- 57.Reina-San-Martin, B., H.T. Chen, A. Nussenzweig, and M.C. Nussenzweig. 2004. ATM is required for efficient recombination between immunoglobulin switch regions. J. Exp. Med. 200:1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan-Hammarstrom, Q., S. Dai, Y. Zhao, I.F. van Dijk-Hard, R.A. Gatti, A.L. Borresen-Dale, and L. Hammarstrom. 2003. ATM is not required in somatic hypermutation of VH, but is involved in the introduction of mutations in the switch mu region. J. Immunol. 170:3707–3716. [DOI] [PubMed] [Google Scholar]
- 59.Rolink, A., F. Melchers, and J. Andersson. 1996. The SCID but not the RAG-2 gene product is required for S mu-S epsilon heavy chain class switching. Immunity. 5:319–330. [DOI] [PubMed] [Google Scholar]
- 60.Odegard, V.H., and D.G. Schatz. 2006. Targeting of somatic hypermutation. Nat. Rev. Immunol. 6:573–583. [DOI] [PubMed] [Google Scholar]
- 61.Yabuki, M., M.M. Fujii, and N. Maizels. 2005. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat. Immunol. 6:730–736. [DOI] [PubMed] [Google Scholar]
- 62.Ehrenstein, M.R., and M.S. Neuberger. 1999. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 18:3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schrader, C.E., W. Edelmann, R. Kucherlapati, and J. Stavnezer. 1999. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 190:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramiro, A.R., M. Jankovic, E. Callen, S. Difilippantonio, H.T. Chen, K.M. McBride, T.R. Eisenreich, J. Chen, R.A. Dickins, S.W. Lowe, et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Noia, J., and M.S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48. [DOI] [PubMed] [Google Scholar]
- 66.Pan, Q., C. Petit-Frere, J. Stavnezer, and L. Hammarstrom. 2000. Regulation of the promoter for human immunoglobulin gamma3 germ-line transcription and its interaction with the 3′alpha enhancer. Eur. J. Immunol. 30:1019–1029. [DOI] [PubMed] [Google Scholar]
- 67.Nicolas, N., D. Moshous, M. Cavazzana-Calvo, D. Papadopoulo, R. de Chasseval, F. Le Deist, A. Fischer, and J.P. de Villartay. 1998. A human severe combined immunodeficiency (SCID) condition with increased sensitivity to ionizing radiations and impaired V(D)J rearrangements defines a new DNA recombination/repair deficiency. J. Exp. Med. 188:627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huo, Y.K., Z. Wang, J.H. Hong, L. Chessa, W.H. McBride, S.L. Perlman, and R.A. Gatti. 1994. Radiosensitivity of ataxia-telangiectasia, X-linked agammaglobulinemia, and related syndromes using a modified colony survival assay. Cancer Res. 54:2544–2547. [PubMed] [Google Scholar]
- 69.Moshous, D., C. Pannetier, R. de Chasseval, F. le Deist, M. Cavazzana-Calvo, S. Romana, E. Macintyre, D. Canioni, N. Brousse, A. Fischer, et al. 2003. Partial T and B lymphocyte immunodeficiency and predisposition to lymphoma in patients with hypomorphic mutations in Artemis. J. Clin. Invest. 111:381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu, B., S.-t. Kim, and M.B. Kastan. 2001. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]