Abstract
Vascular disrupting agents (VDAs) represent a novel approach to the treatment of cancer, resulting in the collapse of tumor vasculature and tumor death. 5,6-dimethylxanthenone-4-acetic acid (DMXAA) is a VDA currently in advanced phase II clinical trials, yet its precise mechanism of action is unknown despite extensive preclinical and clinical investigations. Our data demonstrate that DMXAA is a novel and specific activator of the TANK-binding kinase 1 (TBK1)–interferon (IFN) regulatory factor 3 (IRF-3) signaling pathway. DMXAA treatment of primary mouse macrophages resulted in robust IRF-3 activation and ∼750-fold increase in IFN-β mRNA, and in contrast to the potent Toll-like receptor 4 (TLR4) agonist lipopolysaccharide (LPS), signaling was independent of mitogen-activated protein kinase (MAPK) activation and elicited minimal nuclear factor κB–dependent gene expression. DMXAA-induced signaling was critically dependent on the IRF-3 kinase, TBK1, and IRF-3 but was myeloid differentiation factor 88–, Toll–interleukin 1 receptor domain–containing adaptor inducing IFN-β–, IFN promoter-stimulator 1–, and inhibitor of κB kinase–independent, thus excluding all known TLRs and cytosolic helicase receptors. DMXAA pretreatment of mouse macrophages induced a state of tolerance to LPS and vice versa. In contrast to LPS stimulation, DMXAA-induced IRF-3 dimerization and IFN-β expression were inhibited by salicylic acid. These findings detail a novel pathway for TBK1-mediated IRF-3 activation and provide new insights into the mechanism of this new class of chemotherapeutic drugs.
For years, a primary goal of tumor immunologists has been to trigger an anticancer response by the patient's own immune system, directed largely at engaging the adaptive immune system to mount a tumor-specific response (1–3). However, a considerable body of evidence suggests that nonlymphocytic immune cells also play an important role in eradicating tumors (4, 5). A new class of low molecular mass chemotherapeutic agents, vascular disrupting agents (VDAs), stimulate a variety of cell types, including cells of the monocyte/macrophage lineage, to undergo morphological and functional changes that lead to cytokine release, increased vascular permeability, and rapid and sustained tumor vascular collapse (6–9).
One class of VDAs includes flavone acetic acid and its derivatives, e.g., 5,6-dimethylxanthenone-4-acetic acid (DMXAA). Although flavone acetic acid was found to exert extraordinary antitumor effects in mice, failed clinical trials revealed the species-specific nature of this compound (10, 11). In contrast, DMXAA is currently in advanced phase II clinical trials and has shown great promise in the treatment of a variety of malignancies (12, 13). The molecular mechanisms of action of flavonoid VDAs are largely unknown; however, induction of cytokines has been implicated as a proximal event by which these agents induce tumor necrosis (14, 15).
Early studies revealed differences in gene induction patterns elicited in mouse macrophages stimulated by DMXAA versus the highly potent Toll-like receptor 4 (TLR4) agonist, Escherichia coli LPS (7, 16). Perera et al. reported that DMXAA potently induced a subset of LPS-inducible genes that included both IFN-inducible protein 10 (IP-10) and IFN-β but poorly induced expression of proinflammatory genes such as TNF-α (7). Although TNF-α was initially suspected to induce tumor necrosis after DMXAA, TNF-α receptor–deficient mice displayed only a partially diminished capacity to reject tumor explants when treated with DMXAA, and serum from human subjects treated with DMXAA contained no detectable TNF-α (17, 18). Jassar et al. later showed that macrophages are among the first cells to infiltrate the tumor after DMXAA treatment and are responsible for secreting large amounts of cytokines (19). Moreover, they express high levels of chemokines that may recruit cells into the tumor. Although the mechanism of action of DMXAA remains unknown, it is apparent from these studies that the macrophage response to DMXAA is important and requires further clarification.
Major advances have led to a detailed understanding of many of the signaling molecules involved in activation of the cells of the innate immune system (20). Among these, TLRs compose a major receptor family that enables pathogens to be sensed by the host. TLRs are expressed either on the surface or on an endosomal membrane of immune cells, where they detect conserved pathogen-associated molecular patterns (PAMPs). PAMP-induced oligomerization of TLRs recruits intracellular adaptor molecules to the C-terminal domain. Differential engagement of PAMPs through the N terminus, coupled with differential recruitment and utilization of individual adaptor molecules by the different TLRs, provides the basis for the specificity with which cells respond to different PAMPs with different patterns of gene expression (21).
To date, four adaptors (myeloid differentiation factor 88 [MyD88], Toll–IL-1 resistance [TIR] domain–containing adaptor protein, TRIF-related adaptor molecule [TRAM], and TIR domain–containing adaptor inducing IFN-β [TRIF] [22–25]) have been associated with TLR signaling. MyD88 is absolutely required for the response to PAMPs detected by all known TLRs, with the exception of TLRs 3 and 4 (22, 26–29). In the case of TLR4, all four adaptors are used, and the intracellular signaling cascade bifurcates into MyD88-dependent (i.e., MyD88 and TIR domain–containing adaptor protein mediated) and MyD88-independent (i.e., TRAM and TRIF mediated) arms (26). MyD88-dependent signaling leads to rapid recruitment of the family of IL-1R–associated kinases, phosphorylation of inhibitor of κB (IκB) α, nuclear translocation of NF-κB, and expression of proinflammatory genes such as TNF-α and IL-1β (20). In the case of TLR4, the MyD88-independent pathway utilizes TRAM to recruit TRIF that, in turn, recruits two noncannonical IκB kinases (IKKs), TANK-binding kinase 1 (TBK1) and IKKɛ (25, 30). Both phosphorylate the transcription factor IFN regulatory factor 3 (IRF-3) and result in a later wave of NF-κB translocation (31). Once phosphorylated, IRF-3 and NF-κB translocate to the nucleus, where they activate genes such as IFN-β.
In 2004, Yoneyama et al. described a TLR-independent pathway leading to IFN-β expression (32). Rather than a TLR, a cytosolic RNA helicase, retinoic acid–inducible gene I (RIG-I), detects double-stranded viral RNA via its helicase domain. RIG-I binds to an adaptor molecule, IFN-β promoter-stimulator 1 (IPS-1; also known as MAVS, Cardif, and VISA), that leads to TBK1/IKKɛ activation, IRF-3 phosphorylation, and transcription of IFN-β (33). Another RIG-I–like molecule, melanoma differentiation–associated gene 5, has also been previously described (34). RIG-I and melanoma differentiation–associated gene 5 distinguish between different RNA viruses, but both use IPS-1 (35, 36). Stetson et al. recently described yet another pathway leading to IRF-3 activation (37). Although the molecular sensor was not identified, cytosolic DNA was found to activate IRF-3 and induce IFN-β in the absence of detectable NF-κB or mitogen-activated protein kinase (MAPK) activation.
In this study, we detail a novel IFN-β–inducing pathway that is activated by DMXAA. DMXAA dramatically up-regulates IRF-3–dependent gene expression in a TLR- and IPS-1–independent manner. The response was completely dependent on both TBK1 and IRF-3 but elicited no detectable MAPK activation and minimal, delayed NF-κB DNA binding activity. Additionally, we show that although DMXAA does not lead to measurable IκBα degradation, it results in phosphorylation of p65 in a TBK1-dependent, but IKKβ-independent, manner. We also find that pretreatment of macrophages with either DMXAA or LPS induces a state of “cross-tolerance” for subsequent stimulation by DMXAA or LPS, suggesting shared utilization of signaling molecules. Interestingly, we also show that salicylic acid (SA) inhibits DMXAA- but not LPS-induced IRF-3 signaling in macrophages. Collectively, these data establish DMXAA as a novel, potent, and specific activator of the TBK1–IRF-3 signaling cascade.
RESULTS
DMXAA is a specific activator of IRF-3–mediated gene expression
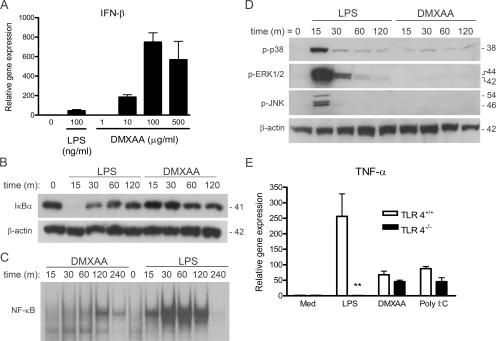
It has been previously reported that DMXAA is a much more potent inducer of IFN-β protein and IP-10 mRNA in mouse macrophages than LPS, whereas LPS stimulation results in much higher levels of proinflammatory cytokines, e.g., TNF-α and IL-1β (7). Fig. 1 confirms and extends these findings. Using real-time PCR to quantify mRNA expression of these genes in peritoneal exudate macrophages, DMXAA induced ∼10-fold more IFN-β steady-state mRNA than LPS (Fig. 1 A). Although LPS stimulation led to the rapid disappearance of IκBα (Fig. 1 B) and NF-κB translocation (Fig. 1 C) in primary macrophages and the RAW 264.7 macrophage-like cell line, respectively, treatment with DMXAA had a minimal effect on the level of IκBα protein and on NF-κB binding activity in EMSAs. Peak NF-κB activation in DMXAA-stimulated cells was observed at 120 min and was both delayed and less abundant than that seen in LPS-stimulated cells. Furthermore, under conditions in which LPS strongly activated p38, extracellular signal-regulated kinase, and c-Jun N-terminal kinase MAPK signaling cascades within 15 min of treatment, treatment with DMXAA had no measurable effect on these signaling intermediates over a 2-h time course (Fig. 1 D).
Figure 1.
DMXAA preferentially induces IRF-3–mediated gene expression. (A) Peritoneal macrophages from C57BL/6 mice were stimulated for 2 h with 100 ng/ml LPS or increasing concentrations of DMXAA, as shown. Total RNA was extracted and subjected to reverse transcription followed by quantitative real-time PCR, as described in Materials and methods. (B) Primary mouse macrophages were stimulated with medium alone, 100 ng/ml LPS, or 100 μg/ml DMXAA for the indicated times. Total protein was collected and subjected to SDS-PAGE, followed by Western blotting with anti-IκBα antibody. Protein molecular masses (in kD) appear at the right. (C) RAW 264.7 macrophages were stimulated with medium alone, 10 ng/ml LPS, or 1 mg/ml DMXAA for the indicated times. Nuclear extracts were prepared and subjected to EMSA with an NF-κB–specific labeled oligonucleotide. (D) Peritoneal macrophages from C57BL/6 mice were stimulated with medium alone, 100 ng/ml LPS, or 100 μg/ml DMXAA for the indicated times. Western blotting for activated MAPK was performed on whole-cell lysates. β-Actin was used as a loading control. Protein masses (in kD) appear at the right. (E) Peritoneal macrophages from TLR4+/+ and TLR4−/− were stimulated with medium alone, 100 ng/ml LPS, or 100 μg/ml DMXAA for 2 h. Total RNA was collected and analyzed by quantitative real-time PCR, as in A. Results represent the mean ± SE for at least three independent experiments. **, P < 0.01.
The partial overlap of gene expression profiles for LPS- and DMXAA-stimulated macrophages led us to ask whether DMXAA might preferentially activate the MyD88-independent pathway via a unique interaction with TLR4, leading to activation of the transacting factor IRF-3. To address this possibility, macrophages from TLR4+/+ or background-matched TLR4−/− mice were stimulated with LPS or DMXAA, and gene expression was measured. Although LPS failed to induce TNF-α mRNA expression in the absence of TLR4 (P < 0.01), levels of TNF-α mRNA induced by DMXAA or the TLR3 agonist poly I:C were not statistically different (Fig. 1 E). Collectively, these observations led us to hypothesize that DMXAA-induced signaling drives primarily on IRF-3, rather than NF-κB or the MAPK signaling cascades.
To extend these findings at the level of gene expression, macrophages were stimulated with medium only or DMXAA for 3 h, and mRNA was subjected to Affymetrix microarray analysis. Of ∼14,000 genes analyzed, DMXAA resulted in a ≥3-fold change in expression of 136 genes (110 and 26 genes were up-regulated or down-regulated, respectively), compared with the response of medium-treated cells (microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession no. GSE7194; Table S1, available at http://www.jem.org/cgi/content/full/jem.20061845/DC1). Because many of the genes that were modulated greater than or equal to threefold by DMXAA, such as Mx1, are known to be IFN-β–dependent, we also performed the same analysis in IFN-β−/− macrophages (38). A comparison of the results from these two strains revealed that 77 out of the 136 genes modulated by DMXAA in wild-type macrophages were IFN-β dependent (Table S1, WT/KO column), based on a threefold difference. As TRIF is an adaptor required for IRF-3 activation after LPS stimulation (39), genes identified as poorly LPS inducible in TRIF−/− macrophages represent a reliable surrogate for IRF-3–dependent gene induction. Many of the same genes induced by DMXAA in our microarray analysis were identified as being poorly inducible by LPS in macrophages derived from TRIF-null mice (40); e.g., Rantes (Ccl5), Ifit1, Ccl4, and Oasl were shown by Hirotani et al. to be highly TRIF dependent in LPS-treated macrophages. Thus, these data support the hypothesis that DMXAA preferentially induces IRF-3–dependent genes.
DMXAA is a potent and specific activator of TBK1
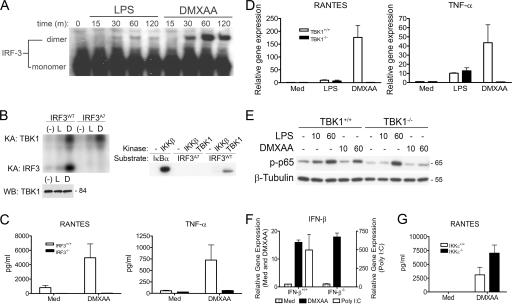
The IRF family of transcription factors has been shown to be integral to the regulation of the type I IFN response (41, 42). Phosphorylation of IRF-3 leads to the formation of IRF-3 dimers, followed by the nuclear translocation and transcription of genes such as IFN-β and regulated on activation, normal T expressed and secreted (RANTES) (43). To study the capacity of DMXAA to activate IRF-3, cell lysates from peritoneal macrophages exposed to either LPS or DMXAA were subjected to native PAGE to preserve fragile IRF-3 dimers. Proteins were transferred to polyvinylidene difluoride and subjected to Western blot analysis with an anti–IRF-3 antibody. Activated IRF-3 dimers were much more abundant and longer lived in DMXAA- versus LPS-stimulated macrophages (Fig. 2 A).
Figure 2.
DMXAA is a potent and specific activator of TBK1. (A) Peritoneal macrophages from C57BL/6 mice were stimulated with medium alone, 100 ng/ml LPS, or 100 μg/ml DMXAA for the indicated times. Total protein was collected and subjected to native PAGE, followed by Western blotting with an anti–IRF-3 antibody. (B) Bone marrow–derived macrophages were stimulated for 90 min with medium alone, 200 ng/ml LPS, or 100 μg/ml DMXAA. Whole-cell lysates were subjected to immunoprecipitation with anti-TBK1 pAb, and immunoprecipitates were subjected to an in vitro kinase assay using the GST–C-terminal IRF-3 wild-type or A7 mutant as the substrate, as described in Materials and methods. Levels of TBK1 in immunoprecipitates were examined by Western blotting using anti-TBK1 mAb. Recombinant TBK1 and IKKβ were also included as controls with GST–IRF-3 or IκBα substrates. Protein molecular mass appears at the right. Results shown are representative of four independent experiments. (C) Peritoneal macrophages from IRF-3+/+ and IRF-3−/− were exposed to medium alone or 100 μg/ml DMXAA for 24 h. Supernatants were collected, and cytokine concentrations were determined by Luminex bead assay. Results are representative of three independent experiments. (D) TBK1+/+ and TBK1−/− MEFs were stimulated with medium alone, 100 ng/ml LPS, or 100 μg/ml DMXAA for 2 h. Total RNA was harvested and subjected to reverse transcription. Relative gene expression was measured by quantitative real-time PCR. Results represent the mean ± SE of at least three separate experiments. (E) TBK1+/+ and TBK1−/− MEFs were stimulated with 10 μg/ml LPS or 100 μg/ml DMXAA for 10 or 60 min. Whole-cell lysates were subjected to SDS-PAGE and probed with the indicated antibodies. Protein molecular masses appear at the right. Results are representative of three independent experiments. (F) IKKβ+/+ and IKKβ−/− MEFs were stimulated for 3 h with medium alone, 100 μg/ml DMXAA, 10 μg/ml or poly I:C in the presence of Fugene 6. IFN-β expression was measured by real-time PCR. Results are representative of three independent experiments. (G) Peritoneal macrophages isolated from IKKɛ+/+ or IKKɛ−/− mice were stimulated with 100 μg/ml DMXAA for 24 h. Supernatants were collected, and RANTES expression was determined by ELISA. Results shown are mean ± SD for three independent experiments.
To demonstrate the ability of DMXAA to activate TBK1 kinase activity in macrophages, TBK1 was immunoprecipitated (using a highly specific pAb raised against the C terminus of TBK1) from macrophages that had been stimulated for 90 min with either LPS or DMXAA. Immunoprecipitated TBK1 complexes were subjected to an in vitro kinase assay using purified glutathione S transferase (GST)–IRF-3 (C-terminal aa 380–427), and kinase activity was measured by autoradiography. To ensure comparability of levels of TBK1 in the immunoprecipitates, TBK1 was detected by Western blotting with an anti-TBK1 mAb. As seen in Fig. 2 B, DMXAA potently activated endogenous TBK1 kinase activity and induced clear phosphorylation of both TBK1 itself (left, top) and the wild-type GST–IRF-3 substrate (left, bottom). Consistent with the results of the IRF-3 dimerization assay, DMXAA-induced TBK1 kinase activity was considerably more potent than that observed after stimulation with LPS. Importantly, a mutant version of IRF-3, in which seven serine/threonine residues were mutated to alanine (S385, S386, S396, S398, S402, T404, and S405→A; GST–IRF-3–A7), was not phosphorylated by endogenous TBK1 under conditions in which TBK1 autophosphorylation was intact. In addition, an in vitro kinase assay revealed that recombinant TBK1 phosphorylated the wild-type GST-IRF-3, but not the A7 mutant, whereas recombinant IKKβ, which potently phosphorylated IκBα, failed to phosphorylate GST–IRF-3 measurably, consistent with previously published data (44). Collectively, these results clearly demonstrate that DMXAA is a potent activator of the TBK1–IRF-3 signaling axis.
To address the possibility that IRF-3 was required for activation of cells by DMXAA, peritoneal macrophages from wild-type and IRF-3−/− mice were cultured in medium only or DMXAA. Supernatants collected at 24 h were analyzed for cytokine production. Consistent with the robust IRF-3 activation observed in DMXAA-treated cells (Fig. 2, A and B), IRF-3−/− macrophages failed to produce RANTES, the product of a known IRF-3–dependent gene (45). Surprisingly, secretion of TNF-α was also reduced to background levels in IRF-3–deficient macrophages (Fig. 2 C). To evaluate further the role of activated IRF-3 in DMXAA-induced signaling, we exposed wild-type or TBK1-deficient mouse embryonic fibroblasts (MEFs) to medium only, LPS, or DMXAA and measured gene expression. Interestingly, we found that, in contrast to experiments with macrophages, DMXAA induced much more robust responses in MEFs than did LPS, an observation that is consistent with the diminished LPS sensitivity that has been observed in MEFs by others (46). In agreement with previous work (46), LPS-stimulated, TBK1−/− MEFs produced wild-type levels of RANTES and TNF-α mRNA (Fig. 2 D). However, TBK1−/− MEFs failed to express either RANTES or TNF-α mRNA in response to DMXAA. These results suggest that, in addition to being a potent activator of TBK1, DMXAA is critically dependent on both TBK1 and its downstream target, IRF-3, for gene expression.
Although TBK1 seems to function primarily as an IRF-3 kinase, it has also been shown that, under certain circumstances, TBK1 may phosphorylate the NF-κB subunit p65 on serine 536 (47). This phosphorylation event is believed to play a role in p65 transactivation, because cells lacking TBK1 show a defect in NF-κB–dependent gene expression despite normal IκBα degradation and NF-κB binding activity (48). Because DMXAA is a relatively poor inducer of both IκBα degradation and NF-κB binding activity when compared with LPS (Fig. 1, B and C) but has previously been shown to induce NF-κB–dependent gene expression (49), we sought to examine the phosphorylation status of p65 in LPS versus DMXAA-stimulated cells. In wild-type MEFs, LPS-induced phosphorylation of p65 on S536 was observed at 10 min and peaked at 60 min, whereas DMXAA-induced p65 phosphorylation was undetectable at 10 min but measurable at 60 min. Surprisingly, in contrast to LPS-induced phospho-p65, DMXAA-induced p65 phosphorylation was ablated in TBK1-null MEFs at 60 min (Fig. 2 E). In further support of the assertion that DMXAA is a specific activator of the TBK1–IRF-3 signaling axis, we tested the ability of DMXAA to induce IFN-β in MEFs deficient in the NF-κB–activating kinase IKKβ. Remarkably, under conditions in which transfected poly I:C, a known inducer of NF-κB (32), failed to activate IFN-β expression in IKKβ-null MEFs, DMXAA-induced IFN-β was found to be independent of IKKβ (Fig. 2 F). Collectively, these findings suggest that DMXAA activates NF-κB in a manner that is both independent of IKKβ but completely dependent on TBK1.
To address a possible role for IKKɛ, the only other IRF-3 kinase identified thus far, in DMXAA-induced signaling, we compared the response of macrophages isolated from wild-type and IKKɛ-deficient mice after treatment with DMXAA. Induction of RANTES protein was not inhibited in IKKɛ-null cells (Fig. 2 G). Collectively, these data support the conclusion that DMXAA activates a pathway that is dependent on both IRF-3 and TBK1 but is independent of both IKKβ and IKKɛ.
DMXAA-induced gene expression is TLR- and IPS-1–independent
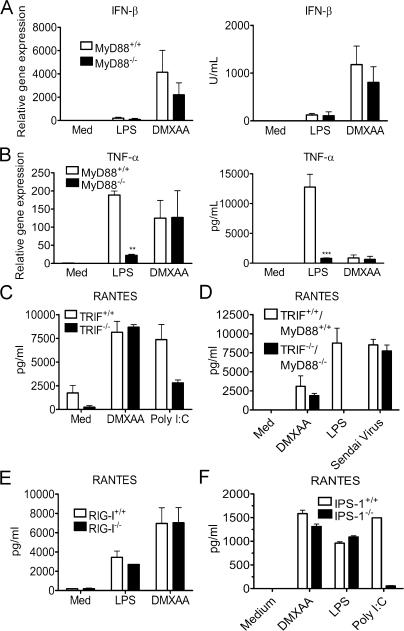
Because all known TLRs, with the exception of TLRs 3 and 4, have an absolute requirement for MyD88 to induce gene expression, we tested the ability of DMXAA to induce signaling in MyD88−/− macrophages. Consistent with previous reports (22, 26), LPS-induced IFN-β mRNA and protein (Fig. 3 A) were not significantly decreased by MyD88 deficiency, whereas levels of TNF-α were dramatically inhibited in the MyD88−/− macrophages (Fig. 3 B). In contrast, DMXAA-induced IFN-β and TNF-α mRNA and protein were not significantly different in wild-type and MyD88−/− cells. Thus, DMXAA-induced gene expression is MyD88 independent.
Figure 3.
DMXAA signaling is TLR- and IPS-1–independent. (A and B) Peritoneal macrophages from MyD88+/+ or MyD88−/− mice were stimulated with 100 ng/ml LPS or 100 μg/ml DMXAA. After a 2-h incubation (left), total RNA was collected, and gene expression was assessed by quantitative real-time PCR. ELISA results (right) were generated from supernatants collected 24 h after exposure to medium, 100 ng/ml LPS, or 100 μg/ml DMXAA. (C) TRIF+/+ and TRIF−/− MEFs were stimulated with 100 μg/ml DMXAA or 100 μg/ml poly I:C for 24 h. Supernatants were collected, and RANTES levels were measured by ELISA. (D) Wild-type or MyD88−/−/TRIF−/− peritoneal macrophages were stimulated with 100 mg/ml DMXAA, 100 ng/ml LPS, or 200 hemagglutination U/ml Sendai virus for 24 h. RANTES expression was measured by ELISA. RIG-I+/+ and RIG-I−/− (E) or IPS-1+/+ and IPS-1−/− (F) MEFs were treated for 24 h with medium alone, 1 μg/ml LPS, 100 μg/ml DMXAA, or 10 μg/ml poly I:C in the presence of Fugene 6. Supernatants were collected, and RANTES levels were assessed by ELISA. Results shown are the mean ± SE of at least three independent experiments. **, P < 0.01; ***, P < 0.001.
TLRs 3 and 4 share the ability to activate IRF-3 and induce IFN-β via another adaptor, TRIF. To directly address the possibility that DMXAA utilizes the MyD88-independent pathway mediated by TRIF, background-matched, wild-type, and TRIF−/− MEFs were stimulated with DMXAA or the TLR3 agonist poly I:C. Fig. 3 C illustrates that compared with poly I:C, a known TRIF-dependent inducer of RANTES (25), DMXAA-induced RANTES was unaffected by the absence of TRIF. In further support of the conclusion that DMXAA does not require any known TLR for activity, macrophages deficient in both MyD88 and TRIF responded to DMXAA by making RANTES protein at a level that was not statistically different from that made by wild-type cells, whereas LPS-induced RANTES was reduced to baseline levels in TRIF−/−/MyD88−/−-deficient macrophages (Fig. 3 D). Because DMXAA is, therefore, neither MyD88- nor TRIF-dependent, these data indicate that none of the known TLRs serve as a receptor for DMXAA, because all require MyD88 and/or TRIF to mediate signaling.
Because our data implied that DMXAA does not require known TLRs to activate IRF-3–inducible genes, we postulated that DMXAA might engage the recently identified cytosolic RNA helicases RIG-I or Mda5. Therefore, we first examined the response of background-matched wild-type and RIG-I−/− MEFs, and in accordance with previous work (50), the latter failed to respond to Newcastle disease virus (unpublished data). However, when stimulated with LPS or DMXAA, RANTES secretion was intact in the RIG-I−/− MEFs (Fig. 3 E). Thus, DMXAA-activated IRF-3 and IRF-3–dependent gene expression is RIG-I independent.
Both RIG-I and another RNA helicase, Mda5, use a downstream adaptor molecule, IPS-1, to induce gene expression. To determine if Mda5 might contribute to DMXAA-induced signaling, we stimulated IPS-1–deficient MEFs with either LPS, DMXAA, or cytosolic (i.e., transfected) poly I:C. As shown in Fig. 3 F, under conditions in which the cytosolic poly I:C–induced RANTES expression was reduced to near-background levels, DMXAA- and LPS-induced RANTES were unaffected. Collectively, the results in Fig. 3 indicate that DMXAA does not require any known TLR or RNA helicase for a cellular response.
LPS and DMXAA induce cross-tolerance
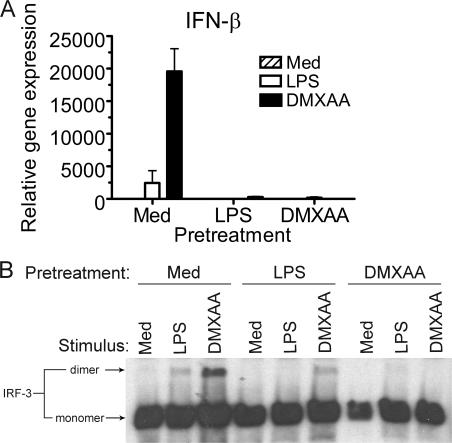
Endotoxin tolerance is a poorly understood phenomenon that has been described as a transient state of LPS hyporesponsiveness induced by prior exposure to a low level of LPS both in vitro in macrophages and in vivo. Moreover, “TLR heterotolerance” can be induced (i.e., prior exposure to one TLR agonist diminishes responsiveness to challenge through a distinct TLR [51]), and LPS and IL-1β “cross-tolerize” (52). The ability to induce heterotolerance or cross-tolerance has been suggested to be caused by the disruption of shared signaling pathway molecules between distinct receptor systems (51). To determine if LPS and DMXAA can cross-tolerize, peritoneal macrophages were pretreated with medium, LPS, or DMXAA. After 24 h, cells were washed and restimulated for 1 h with LPS or DMXAA. Protein was subjected to native PAGE and Western blotting for IRF-3, and IFN-β mRNA was quantified by real-time PCR.
LPS pretreatment of cells resulted in a diminished response to a second LPS exposure, both at the level of IFN-β mRNA (Fig. 4 A) and IRF-3 dimerization (Fig. 4 B), indicating that classical endotoxin tolerance was induced. LPS pretreatment of macrophages also mitigated the subsequent response to DMXAA. Conversely, pretreatment with DMXAA induced a state of refractoriness to restimulation with either LPS or DMXAA. These results suggest that signaling elements rendered hypoactive by pretreatment with LPS are also used by DMXAA and vice versa.
Figure 4.
DMXAA pretreatment of macrophages induces a state of refractoriness upon reexposure to either LPS or DMXAA. Total RNA (A) or protein (B) from C57BL/6 peritoneal macrophages pretreated with medium alone, 100 ng/ml LPS, or 100 μg/ml DMXAA, washed, and restimulated with medium, LPS, or DMXAA was collected and subjected to quantitative real-time PCR (A) or native PAGE, followed by Western blotting for IRF-3 (B). Results shown are the mean ± SE of one representative experiment (n = 3).
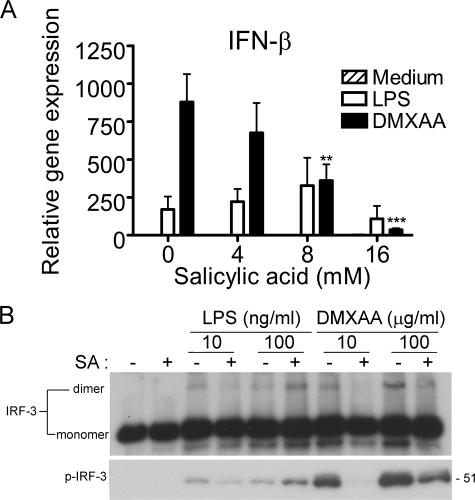
SA selectively inhibits DMXAA-induced IFN-β
SA has been reported to inhibit IKKβ (53, 54) and has been shown to inhibit TNF-α in human mononuclear cells when DMXAA is combined with anti-CD14 antibodies or deacylated LPS (8). Because IRF-3–dependent gene expression had not been shown previously to be SA sensitive, we sought to test the hypothesis that SA might down-regulate DMXAA-induced IFN-β expression. To address this hypothesis, peritoneal macrophages were pretreated with increasing concentrations of SA, followed by stimulation with LPS or DMXAA. Fig. 5 A shows that SA dramatically reduced DMXAA-induced IFN-β expression, whereas LPS-induced IFN-β mRNA expression was essentially unaffected. Furthermore, pretreatment of macrophages with SA also inhibited both IRF-3 dimer formation and phosphorylation of S396 of IRF-3 in response to DMXAA but not LPS (Fig. 5 B). In toto, these data support the hypothesis that DMXAA activates a unique signaling pathway leading to the TBK1-dependent induction of IRF-3– and phospho-p65–mediated gene expression.
Figure 5.
Sensitivity of DMXAA-induced gene expression and IRF-3 activation to SA. (A) Peritoneal macrophages from C57BL/6 mice were pretreated with medium alone or increasing concentrations of SA for 1 h and stimulated with LPS or DMXAA for an additional 2 h. Total RNA was harvested, and gene expression was assessed by quantitative PCR. Results represent the mean ± SE of at least three separate experiments. **, P < 0.01; ***, P < 0.001. (B) Macrophages were pretreated with or without 16 mM SA for 1 h and stimulated with 10 or 100 ng/ml LPS, or 10 or 100 μg/ml DMXAA, for an additional 1 h. Total protein was harvested and subjected to native PAGE (top) or SDS-PAGE (bottom), followed by Western blot analysis for IRF-3 and anti–phospho–IRF-3, respectively. Protein molecular mass appears at the right. Results shown are representative of three independent experiments.
DISCUSSION
DMXAA is currently in advanced phase II clinical trials for efficacy against lung, prostate, and ovarian cancers. It is well tolerated at therapeutic doses (13, 55) and induces measurable changes in tumor blood flow within 30 min of i.v. administration (56). In this paper, we have detailed as of yet unexplored aspects of the flavonoid class of VDAs: the dramatic up-regulation of IFN-β by DMXAA and the demonstration of the TBK1–IRF-3 axis as an absolute requirement for gene induction by DMXAA in macrophages and MEFs. The link between this promising new chemotherapeutic agent and its remarkable capacity to induce IFN-β provides a new rationale for exploring the complex role played by type I IFNs in the host against cancer.
A previous study (7) showed that stimulation of mouse macrophages with DMXAA led to the rapid up-regulation of IFN-β, findings confirmed and extended by our data. Importantly, the robust expression of IFN-β was not accompanied by activation of the MAPKs and led to NF-κB activation at dramatically reduced levels and delayed kinetics compared with LPS (Fig. 1). Early studies performed by Maniatis et al. detailed the assembly of a multiprotein complex termed the “enhanceosome” (57). The components of the enhanceosome (NF-κB, IRF-3, IRF-7, and ATF2–c-Jun) bind to adjacent regulatory elements in the IFN-β promoter termed positive regulatory domain (PRD) II, PRD III-I, and PRD IV, respectively, and there is compelling evidence that suggests LPS-induced IFN-β is driven by the assembly of the enhanceosome (58). Indeed, after treatment with LPS, we observed robust activation of all three members of the enhanceosome, and this activation coincided with IFN-β gene transactivation. In contrast, DMXAA-induced activation of both NF-κB and the MAPK cascades was considerably less pronounced than that observed in LPS-stimulated cells despite a more potent induction of IFN-β. Moreover, IKKβ-deficient MEFs respond normally to DMXAA by producing wild-type levels of IFN-β mRNA (Fig. 2 F), suggesting that DMXAA does not use the classical NF-κB pathway upstream of IFN-β transcription. Interestingly, however, DMXAA induces phosphorylation of p65 at S536 at levels comparable with those achieved with LPS (Fig. 2 E). Phosphorylation of S536 has been suggested by others to enhance the transactivation potential of the p65 subunit (for review see reference 59). Thus, phosphorylation of p65 on S536 may increase the “gain” of NF-κB, providing a plausible explanation for DMXAA's ability to induce robust IFN-β expression despite very little IκBα degradation. In other words, it is possible that the relatively small amount of activated NF-κB available after treatment with DMXAA is sufficient to complete the IFN-β enhanceosome or is compensated for by its increased transactivation potential. Finally, in contrast to LPS treatment, DMXAA-induced p65 phosphorylation is abolished in TBK1-deficient MEFs, providing further support for the conclusion that DMXAA is a novel and specific activator of the TBK1–IRF-3 signaling axis.
This claim is further supported by our results derived from TBK1- and IRF-3–deficient mice. DMXAA-induced expression of RANTES, a heavily IRF-3–dependent gene (45), was observed to be completely dependent on the TBK1–IRF-3 axis (Fig. 2). Surprisingly, this dependence on TBK1 and IRF-3 extended to genes not normally considered to be dependent on IRF-3, such as TNF-α. Under conditions where LPS-induced TNF-α was unaffected, IRF-3–deficient cells failed to induce TNF-α mRNA in response to DMXAA. This suggests that DMXAA-induced TNF-α expression is strictly IRF-3–dependent. Although it is possible that the failure of DMXAA-treated TBK1-null MEFs to phosphorylate p65 contributes to reduced availability of NF-κB for induction of genes such as TNF-α, our DNA microarray data (Table S1) revealed that TNF-α expression in response to DMXAA is diminished in IFN-β–null macrophages. These results support the alternative possibility that TNF-α is part of an IFN-β–dependent “second wave” of gene expression after DMXAA treatment.
Although the role of type I IFN in both tumor immunity and the treatment of cancer has been studied for decades, the direct involvement of IRF-3 is considerably less well understood. However, it was recently shown that IRF-3 drives the up-regulation of TNF-related apoptosis-inducing ligand in virally infected cells, as well as directing cells into p53-dependent cell-cycle arrest and senescence (60, 61). Perhaps even more pertinent to the current work are recent studies by Duguay et al. with human IRF-3–expressing B16 melanoma tumors (62). In their study, tumors expressing IRF-3 grew more slowly than those that had been mock transduced. Furthermore, the IRF-3–positive tumors demonstrated a robust up-regulation of a variety of chemokines in vivo, including RANTES, macrophage inflammatory protein 1β, and IP-10. Accordingly, IRF-3–expressing tumors recruited considerably more neutrophils and lymphocytes and showed signs of retarded tumor growth, including a larger capsule, fewer blood vessels, and areas of necrosis. Importantly, the results reported by Duguay et al. mirror those of Jassar et al. (19) in which implanted tumors showed dramatically increased levels of IP-10 and RANTES, as well as necrosis, after i.v. DMXAA administration. The results presented herein provide a plausible link between the direct antitumor results of IRF-3 overexpression and those after treatment with DMXAA.
The ability of DMXAA to activate IRF-3 and induce IRF-3–mediated gene expression led us to address the involvement of established pattern recognition receptors in DMXAA signaling. DMXAA-induced signaling was found to be intact in both MyD88−/−/TRIF−/− and IPS-1−/− cells, thereby eliminating the possibility of involvement of all known TLRs and RNA helicases. However, a third non–TLR-dependent pathway leading to expression of IFN-β was recently described by Stetson et al. in which the presence of cytosolic, non–CpG-containing DNA stimulated high levels of type I IFN (37). In that study, however, the molecular sensor of the stimulus was not identified. Notably, the authors reported that the type I IFNs induced by cytosolic DNA were not accompanied by either MAPK activation or NF-κB translocation, consistent with our observations. With a molecular mass of 304 daltons, DMXAA is much smaller than the DNA used in that study. However, it remains possible that DMXAA may engage the molecular sensor or a downstream signaling component of this novel pathway to initiate signaling leading to IRF-3 activation. Studies to identify the nature of this sensor are currently in progress.
Previous studies have shown that pretreatment of macrophages or mice with LPS results in a transient desensitization to subsequent stimulation by LPS, other TLR agonists, or IL-1β. The mechanisms that underlie tolerance are clearly multifactorial; however, interference with signal transduction appears to be a common mechanism (51, 63). DMXAA, like LPS, induced a state of tolerance in macrophages to subsequent stimulation with either DMXAA or LPS, as indicated by inhibition of not only IFN-β gene expression but also IRF-3 dimer formation (Fig. 4). This implies that disrupted signaling in LPS- or DMXAA-tolerized cells is a consequence of an event that occurs early in the signaling pathway, because IRF-3 dimers are detected within 15 min after agonist stimulation. Although we have shown that LPS and DMXAA seem to engage distinct signaling pathways, both lead to IRF-3 activation via TBK1. Thus, it seems plausible that one signaling component tolerized by pretreatment with LPS or DMXAA is TBK1 itself. Studies are ongoing to address the role of TBK1 expression levels and enzymatic activity in the induction of cross-tolerance by LPS and DMXAA.
During the course of these studies, we extended previous findings that demonstrated SA as an inhibitor of DMXAA. Although an inhibitory effect of SA on DMXAA-induced TNF-α expression had been previously reported (8), our results identify a possible explanation for the role played by SA in DMXAA inhibition. Pretreatment of macrophages with SA blocked DMXAA-induced phosphorylation of IRF-3 at residue S396, IRF-3 dimerization, and IFN-β expression (Fig. 5). However, all three events were unaffected by SA in LPS-stimulated cells. These results support our conclusion that the pathways leading to IFN-β gene expression by these two stimuli differ.
In conclusion, we present data that firmly establishes the clinically important VDA DMXAA as a potent and specific activator of the TBK1–IRF-3 axis. The link between heightened activity of this signaling pathway and a systemic antitumor response likely involves myriad and divergent events. However, by identifying a key signaling pathway with known antitumor potential as critical to the response to DMXAA, we hope to further our understanding of both the mechanism of action of this promising new chemotherapeutic agent as well as the role of the innate immune response in defending the host against cancer.
MATERIALS AND METHODS
Mice.
5–6-wk-old C57BL/6J females were purchased from the Jackson Laboratory. IRF-3−/− mice were a gift of T. Taniguchi (University of Tokyo, Tokyo, Japan). IFN-β−/− mice (>N8 on a C57BL/6 background) were a gift of E. Fish (University of Toronto, Toronto, Canada). MyD88−/−/TRIF−/− mice were bred from MyD88−/− (64) and TRIF−/− (39) mice. IKKɛ−/− mice were generated at Millennium Pharmaceuticals. TBK1−/− mice were a gift of W.-C. Yeh (University of Toronto, Toronto, Canada) and were bred with TNFR1−/− mice at the University of Massachusetts Medical School. All experiments were conducted with Institutional Animal Care and Use Committee approval.
Reagents and virus.
DMXAA (sodium salt; 304 daltons) was synthesized at the Auckland Cancer Society Research Centre (65). Poly I:C (GE Healthcare) was used exogenously as a TLR3 agonist. For triggering intracellular RNA helicases, poly I:C was transfected as follows: 10 μg/ml poly I:C was mixed with a transfection reagent (Fugene 6; Roche) at a ratio of 1:1 (volume/weight) in OptiMEM (Invitrogen) and incubated for 15 min before stimulation. Sendai virus was used at 200 hemagglutination U/ml. Protein-free E. coli K235 LPS (66) was used as a TLR4 agonist. SA was obtained from Sigma-Aldrich. C-terminal GST fusions of IRF-3 (wild-type aa 380–427 and IRF-3–A7 mutant in which amino acids S385, S386, S396, S398, S402, T404, and S405 were all mutated to alanine) were purified according to standard protocols (GE Healthcare). pAb to TBK1 (raised against the C terminus of human TBK1) was provided by T. Maniatis (Millenium Pharmaceuticals, Cambridge, MA). Anti-TBK1 mAb (IMG-139, raised against aa 563–577 of human TBK1) was obtained from Imgenex.
Cell culture.
Thioglycollate-elicited mouse peritoneal macrophages were obtained and cultured as previously described (52). Bone marrow–derived macrophages were generated from bone marrow cells cultured in L929-conditioned media for 10 d and were examined by FACS and found to be >99% F4/80 and CD11b double positive. Mouse macrophage-like RAW 264.7 cells were purchased from the American Type Culture Collection. Embryonic fibroblasts from TBK1+/+ and TBK1−/− mice were a gift of W.-C. Yeh. RIG-I (50) and IPS-1 (36) knockout MEFs have been described elsewhere. Embryonic fibroblasts from IKKβ+/+ and IKKβ−/− mice were a gift of J. DiDonato (Cleveland Clinic, Cleveland, Ohio). RAW 264.7 macrophages and embryonic fibroblasts were cultured in DMEM (BioWhittaker), supplemented with 10% (vol/vol) FBS (HyClone Laboratories), 10,000 U/ml penicillin, and 10,000 μg/ml streptomycin at 37°C in 5% CO2 in air. The endotoxin content in the medium was <0.01 EU/ml, according to the manufacturer's specifications. Only cells passaged ≤20 times were used.
Quantitative real-time PCR.
Primers for detection of IFN-β, RANTES, TNF-α, and hypoxanthine phosphoribosyltransferase (HPRT) mRNAs were designed using the Primer Express program (version 2.0; Applied Biosystems). 31.25 ng of total cDNA was used as starting material for real-time PCR quantitation with SYBR Green (Applied Biosystems) on a real-time PCR system (7900HT; Applied Biosystems). Ct values were compared using the ΔΔ-Ct method using HPRT as a housekeeping gene (67).
Cytokine analysis.
IFN-β protein in cell culture supernatants was measured using a custom ELISA originally described elsewhere (68), with few modifications. In brief, 96-well polystyrene plates (Maxisorp; Nunc International) were coated overnight with a 1:4,000 dilution of rat anti–mouse IFN-β mAb (Yamasa) in 0.1 M sodium carbonate at 4°C. Plates were blocked with 10% FCS in 1× PBS for 2 h at room temperature. Samples and a mouse IFN-β standard (National Institutes of Health [NIH]) were added to wells and incubated overnight at 4°C. Plates were washed 3 times with 1% FCS/PBS-T, followed by incubation with a 1:2,000 dilution of rabbit anti–mouse IFN-β pAb (PBL Biomedical Laboratories) in 10% FCS-PBS overnight at 4°C. Wells were washed 3 times, followed by incubation with a 1:2,000 dilution of goat anti–rabbit horseradish peroxidase (HRP; Cell Signaling Technologies) in 10% FCS-PBS for 1 h at room temperature. Plates were washed 3 times and developed with TMB substrate (KPL). The reaction was stopped by addition of 1 N H2SO4, and plates were read at 450 nm. For quantification of RANTES and TNF-α, Luminex bead-based colorimetric assays were performed by the Cytokine Core Laboratory (University of Maryland, Baltimore).
EMSA.
Oligonucleotides containing the DNA sequence corresponding to the prototypic NF-κB binding site in the mouse Igκ light chain gene enhancer (5′-AGCTCAGAGGGGACTTTCCGAGAG-3′ and 3′-GTCTCCCCTGAAAGGCTCTCTCGA-5′; NF-κB binding sites are bolded) was annealed in a buffer containing 10 mM Tris HCl (pH 8), 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol. 50 ng of the annealed oligonucleotide was labeled with an oligolabeling kit, according to the manufacturer's instructions (GE Healthcare). After the labeling, unincorporated nucleotides were removed with a Bio-spin column (Bio-Rad Laboratories). For each DNA–protein binding reaction, 5 μg of nuclear extract was used in the presence of 0.2 ng of labeled probe in a 25-μl reaction mixture. The DNA–protein binding buffer contained 1 μg of (poly dI:dC) per ml, 10% glycerol, 10 mM Tris HCl (pH 8), 1 mM EDTA, 40 mM KCl, and 1 mM dithiothreitol. All DNA–protein binding reactions were allowed to proceed at room temperature for 30 min. Samples were loaded onto a nondenaturing 4% polyacrylamide gel. After electrophoresis, the gel was transferred to chromatography paper and dried at 80°C. The dried gel was exposed for signal development to film (XAR-5; Kodak) in the presence of an intensifying screen at −70°C.
Antibodies and native PAGE.
Polyclonal rabbit anti–mouse IκBα was purchased from Santa Cruz Biotechnology, Inc. Rabbit anti–mouse MAPK pAbs and rabbit anti–human phospho–IRF-3 pAbs were purchased from Cell Signaling Technologies. Rabbit anti–mouse IRF-3 pAb was purchased from Zymed Laboratories. Anti-TBK1 was a gift of T. Maniatis. Native PAGE for the detection of IRF-3 dimers was performed as previously described (69). In brief, thioglycollate-elicited peritoneal macrophages were lysed after stimulation with either LPS or DMXAA, as indicated in the figures. Proteins were separated in the absence of SDS in 7.5% Tris-Glycine gels (Bio-Rad Laboratories) and transferred to polyvinylidene difluoride. Membranes were probed with a 1:250 dilution of rabbit anti–mouse IRF-3 for 1 h at room temperature. Goat anti–rabbit IgG-HRP at a 1:2,000 dilution was used as the secondary antibody. Blots were developed with ECL Plus (GE Healthcare).
In vitro kinase assays.
Bone marrow–derived macrophages were differentiated ex vivo, plated, allowed to rest overnight, and stimulated with medium alone, 200 ng/ml LPS, or 100 μg/ml DMXAA for 90 min. Cells were lysed, and 500 μg of whole-cell lysate was subjected to immunoprecipitation with anti-TBK1 pAb together with protein G beads (Sigma-Aldrich). Immunoprecipitates were washed three times and examined for TBK1 protein levels by Western blotting with anti-TBK1 mAb and TBK1 kinase activity by an in vitro kinase assay. For in vitro kinase assays, TBK1 immunoprecipitates were incubated with a wild-type C-terminal (aa 380–427) GST–IRF-3 or GST–IRF-3–A7 mutant (1 μg). Recombinant TBK1 and IKKβ were also examined for their ability to phosphorylate wild-type GST–IRF-3 and the GST–IRF-3–A7 mutant (40 ng). GST-IκBα was used as a positive control for IKKβ kinase activity. Kinase reactions were performed in kinase buffer (20 mM Hepes, 50 mM NaCl, 10 mM MgCl2, 20 mM β-glycerol phosphate, 1 mM sodium orthovanadate, 1 mM dithiothreitol) for 30 min at 30°C in the presence of γ-[32P]ATP using methods previously outlined (44). Proteins were separated by SDS-PAGE and visualized via autoradiography.
Online supplemental material.
Table S1 shows the results of a microarray analysis performed using Affymetrix mouse array 430A_2 exposed to total RNA prepared from C57BL/6J or IFN-β−/− macrophages that had been treated with medium alone or DMXAA for 3 h. Fold induction was calculated using GeneChip operating software (Affymetrix). A ≥3-fold increase or decrease between inducible and basal mRNA levels was set as the criteria for inclusion of a gene as modulated. Complete microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession no. GSE7194. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20061845/DC1.
Supplemental Material
Acknowledgments
This work was supported in part by NIH grants T32 AI007540 (to Z.J. Roberts), AI067497 (to K.A. Fitzgerald), and AI18797 and AI44936 (to S.N. Vogel), the Center for Cancer Research, the National Cancer Institute, the NIH (R. Savan, D. van Echo, and H.A. Young), and the Health Research Council of New Zealand project no. 05-237 (L.-M. Ching).
The authors have no conflicting financial interests.
Abbreviations used: DMXAA, 5,6-dimethylxanthenone-4-acetic acid; GST, glutathione S transferase; IκB, inhibitor of κB; IKK, IκB kinase; IP-10, IFN-inducible protein 10; IPS-1, IFN-β promoter-stimulator 1; IRF-3, IFN regulatory factor 3; MAPK, mitogen-activated protein kinase; MEF, mouse embryonic fibroblast; MyD88, myeloid differentiation factor 88; PAMP, pathogen-associated molecular pattern; PRD, positive regulatory domain; RANTES, regulated on activation, normal T expressed and secreted; RIG-I, retinoic acid–inducible gene I; SA, salicylic acid; TBK1, TANK-binding kinase 1; TIR, Toll–IL-1 resistance; TLR, Toll-like receptor; TRAM, TRIF-related adaptor molecule; TRIF, TIR domain–containing adaptor inducing IFN-β; VDA, vascular disrupting agent.
References
- 1.Kaplan, D.H., V. Shankaran, A.S. Dighe, E. Stockert, M. Aguet, L.J. Old, and R.D. Schreiber. 1998. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. USA. 95:7556–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shankaran, V., H. Ikeda, A.T. Bruce, J.M. White, P.E. Swanson, L.J. Old, and R.D. Schreiber. 2001. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 410:1107–1111. [DOI] [PubMed] [Google Scholar]
- 3.van den Broek, M.E., D. Kagi, F. Ossendorp, R. Toes, S. Vamvakas, W.K. Lutz, C.J. Melief, R.M. Zinkernagel, and H. Hengartner. 1996. Decreased tumor surveillance in perforin-deficient mice. J. Exp. Med. 184:1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pace, J.L., S.W. Russell, P.A. LeBlanc, and D.M. Murasko. 1985. Comparative effects of various classes of mouse interferons on macrophage activation for tumor cell killing. J. Immunol. 134:977–981. [PubMed] [Google Scholar]
- 5.Tsung, K., J.P. Dolan, Y.L. Tsung, and J.A. Norton. 2002. Macrophages as effector cells in interleukin 12-induced T cell-dependent tumor rejection. Cancer Res. 62:5069–5075. [PubMed] [Google Scholar]
- 6.Galbraith, S.M., D.J. Chaplin, F. Lee, M.R. Stratford, R.J. Locke, B. Vojnovic, and G.M. Tozer. 2001. Effects of combretastatin A4 phosphate on endothelial cell morphology in vitro and relationship to tumour vascular targeting activity in vivo. Anticancer Res. 21:93–102. [PubMed] [Google Scholar]
- 7.Perera, P.Y., S.A. Barber, L.M. Ching, and S.N. Vogel. 1994. Activation of LPS-inducible genes by the antitumor agent 5,6-dimethylxanthenone-4-acetic acid in primary murine macrophages. Dissection of signaling pathways leading to gene induction and tyrosine phosphorylation. J. Immunol. 153:4684–4693. [PubMed] [Google Scholar]
- 8.Philpott, M., L.M. Ching, and B.C. Baguley. 2001. The antitumour agent 5,6-dimethylxanthenone-4-acetic acid acts in vitro on human mononuclear cells as a co-stimulator with other inducers of tumour necrosis factor. Eur. J. Cancer. 37:1930–1937. [DOI] [PubMed] [Google Scholar]
- 9.Zhao, L., L.M. Ching, P. Kestell, L.R. Kelland, and B.C. Baguley. 2005. Mechanisms of tumor vascular shutdown induced by 5,6-dimethylxanthenone-4-acetic acid (DMXAA): Increased tumor vascular permeability. Int. J. Cancer. 116:322–326. [DOI] [PubMed] [Google Scholar]
- 10.Havlin, K.A., J.G. Kuhn, J.B. Craig, D.H. Boldt, G.R. Weiss, J. Koeller, G. Harman, R. Schwartz, G.N. Clark, and D.D. Von Hoff. 1991. Phase I clinical and pharmacokinetic trial of flavone acetic acid. J. Natl. Cancer Inst. 83:124–128. [DOI] [PubMed] [Google Scholar]
- 11.Kerr, D.J., T. Maughan, E. Newlands, G. Rustin, N.M. Bleehen, C. Lewis, and S.B. Kaye. 1989. Phase II trials of flavone acetic acid in advanced malignant melanoma and colorectal carcinoma. Br. J. Cancer. 60:104–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tozer, G.M., C. Kanthou, and B.C. Baguley. 2005. Disrupting tumour blood vessels. Nat. Rev. Cancer. 5:423–435. [DOI] [PubMed] [Google Scholar]
- 13.Jameson, M.B., P.I. Thompson, B.C. Baguley, B.D. Evans, V.J. Harvey, D.J. Porter, M.R. McCrystal, M. Small, K. Bellenger, L. Gumbrell, et al. 2003. Clinical aspects of a phase I trial of 5,6-dimethylxanthenone-4-acetic acid (DMXAA), a novel antivascular agent. Br. J. Cancer. 88:1844–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, L.C., C.B. Reddy, B.C. Baguley, P. Kestell, R. Sutherland, and L.M. Ching. 2004. Induction of tumour necrosis factor and interferon-gamma in cultured murine splenocytes by the antivascular agent DMXAA and its metabolites. Biochem. Pharmacol. 67:937–945. [DOI] [PubMed] [Google Scholar]
- 15.Cao, Z., B.C. Baguley, and L.M. Ching. 2001. Interferon-inducible protein 10 induction and inhibition of angiogenesis in vivo by the antitumor agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA). Cancer Res. 61:1517–1521. [PubMed] [Google Scholar]
- 16.Ching, L.M., H.A. Young, K. Eberly, and C.R. Yu. 1999. Induction of STAT and NFkappaB activation by the antitumor agents 5,6-dimethylxanthenone-4-acetic acid and flavone acetic acid in a murine macrophage cell line. Biochem. Pharmacol. 58:1173–1181. [DOI] [PubMed] [Google Scholar]
- 17.Rustin, G.J., C. Bradley, S. Galbraith, M. Stratford, P. Loadman, S. Waller, K. Bellenger, L. Gumbrell, L. Folkes, and G. Halbert. 2003. 5,6-dimethylxanthenone-4-acetic acid (DMXAA), a novel antivascular agent: phase I clinical and pharmacokinetic study. Br. J. Cancer. 88:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao, L., L.M. Ching, P. Kestell, and B.C. Baguley. 2002. The antitumour activity of 5,6-dimethylxanthenone-4-acetic acid (DMXAA) in TNF receptor-1 knockout mice. Br. J. Cancer. 87:465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jassar, A.S., E. Suzuki, V. Kapoor, J. Sun, M.B. Silverberg, L. Cheung, M.D. Burdick, R.M. Strieter, L.M. Ching, L.R. Kaiser, and S.M. Albelda. 2005. Activation of tumor-associated macrophages by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid induces an effective CD8+ T-cell-mediated antitumor immune response in murine models of lung cancer and mesothelioma. Cancer Res. 65:11752–11761. [DOI] [PubMed] [Google Scholar]
- 20.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. [DOI] [PubMed] [Google Scholar]
- 21.Vogel, S.N., K.A. Fitzgerald, and M.J. Fenton. 2003. TLRs: differential adapter utilization by toll-like receptors mediates TLR-specific patterns of gene expression. Mol. Interv. 3:466–477. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, T., O. Adachi, T. Ogawa, K. Takeda, and S. Akira. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 11:115–122. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald, K.A., E.M. Palsson-McDermott, A.G. Bowie, C.A. Jefferies, A.S. Mansell, G. Brady, E. Brint, A. Dunne, P. Gray, M.T. Harte, et al. 2001. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 413:78–83. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald, K.A., D.C. Rowe, B.J. Barnes, D.R. Caffrey, A. Visintin, E. Latz, B. Monks, P.M. Pitha, and D.T. Golenbock. 2003. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the Toll adapters TRAM and TRIF. J. Exp. Med. 198:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668–6672. [DOI] [PubMed] [Google Scholar]
- 26.Toshchakov, V., B.W. Jones, P.Y. Perera, K. Thomas, M.J. Cody, S. Zhang, B.R. Williams, J. Major, T.A. Hamilton, M.J. Fenton, and S.N. Vogel. 2002. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat. Immunol. 3:392–398. [DOI] [PubMed] [Google Scholar]
- 27.Schnare, M., A.C. Holt, K. Takeda, S. Akira, and R. Medzhitov. 2000. Recognition of CpG DNA is mediated by signaling pathways dependent on the adaptor protein MyD88. Curr. Biol. 10:1139–1142. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi, F., K.D. Smith, A. Ozinsky, T.R. Hawn, E.C. Yi, D.R. Goodlett, J.K. Eng, S. Akira, D.M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 410:1099–1103. [DOI] [PubMed] [Google Scholar]
- 29.Hemmi, H., T. Kaisho, O. Takeuchi, S. Sato, H. Sanjo, K. Hoshino, T. Horiuchi, H. Tomizawa, K. Takeda, and S. Akira. 2002. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 3:196–200. [DOI] [PubMed] [Google Scholar]
- 30.Fitzgerald, K.A., S.M. McWhirter, K.L. Faia, D.C. Rowe, E. Latz, D.T. Golenbock, A.J. Coyle, S.M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496. [DOI] [PubMed] [Google Scholar]
- 31.Covert, M.W., T.H. Leung, J.E. Gaston, and D. Baltimore. 2005. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 309:1854–1857. [DOI] [PubMed] [Google Scholar]
- 32.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, C.L., and M. Gale Jr. 2006. CARD games between virus and host get a new player. Trends Immunol. 27:1–4. [DOI] [PubMed] [Google Scholar]
- 34.Kang, D.C., R.V. Gopalkrishnan, Q. Wu, E. Jankowsky, A.M. Pyle, and P.B. Fisher. 2002. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA. 99:637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K.J. Ishii, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105. [DOI] [PubMed] [Google Scholar]
- 36.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K.J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. [DOI] [PubMed] [Google Scholar]
- 37.Stetson, D.B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 24:93–103. [DOI] [PubMed] [Google Scholar]
- 38.Deonarain, R., A. Alcami, M. Alexiou, M.J. Dallman, D.R. Gewert, and A.C. Porter. 2000. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J. Virol. 74:3404–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643. [DOI] [PubMed] [Google Scholar]
- 40.Hirotani, T., M. Yamamoto, Y. Kumagai, S. Uematsu, I. Kawase, O. Takeuchi, and S. Akira. 2005. Regulation of lipopolysaccharide-inducible genes by MyD88 and Toll/IL-1 domain containing adaptor inducing IFN-beta. Biochem. Biophys. Res. Commun. 328:383–392. [DOI] [PubMed] [Google Scholar]
- 41.Schafer, S.L., R. Lin, P.A. Moore, J. Hiscott, and P.M. Pitha. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J. Biol. Chem. 273:2714–2720. [DOI] [PubMed] [Google Scholar]
- 42.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777. [DOI] [PubMed] [Google Scholar]
- 43.Au, W.C., P.A. Moore, W. Lowther, Y.T. Juang, and P.M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA. 92:11657–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McWhirter, S.M., K.A. Fitzgerald, J. Rosains, D.C. Rowe, D.T. Golenbock, and T. Maniatis. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA. 101:233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, R., C. Heylbroeck, P. Genin, P.M. Pitha, and J. Hiscott. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19:959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemmi, H., O. Takeuchi, S. Sato, M. Yamamoto, T. Kaisho, H. Sanjo, T. Kawai, K. Hoshino, K. Takeda, and S. Akira. 2004. The roles of two IκB kinase–related kinases in lipopolysaccharide and double-stranded RNA signaling and viral infection. J. Exp. Med. 199:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buss, H., A. Dorrie, M.L. Schmitz, E. Hoffmann, K. Resch, and M. Kracht. 2004. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-{kappa}B at serine 536 is mediated by multiple protein kinases including I{kappa}B kinase (IKK)-{alpha}, IKK{beta}, IKK{epsilon}, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein- associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 279:55633–55643. [DOI] [PubMed] [Google Scholar]
- 48.Bonnard, M., C. Mirtsos, S. Suzuki, K. Graham, J. Huang, M. Ng, A. Itie, A. Wakeham, A. Shahinian, W.J. Henzel, et al. 2000. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 19:4976–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woon, S.T., S. Zwain, M.A. Schooltink, A.L. Newth, B.C. Baguley, and L.M. Ching. 2003. NF-kappa B activation in vivo in both host and tumour cells by the antivascular agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA). Eur. J. Cancer. 39:1176–1183. [DOI] [PubMed] [Google Scholar]
- 50.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 23:19–28. [DOI] [PubMed] [Google Scholar]
- 51.Dobrovolskaia, M.A., A.E. Medvedev, K.E. Thomas, N. Cuesta, V. Toshchakov, T. Ren, M.J. Cody, S.M. Michalek, N.R. Rice, and S.N. Vogel. 2003. Induction of in vitro reprogramming by Toll-like receptor (TLR)2 and TLR4 agonists in murine macrophages: effects of TLR “homotolerance” versus “heterotolerance” on NF-kappa B signaling pathway components. J. Immunol. 170:508–519. [DOI] [PubMed] [Google Scholar]
- 52.Medvedev, A.E., K.M. Kopydlowski, and S.N. Vogel. 2000. Inhibition of lipopolysaccharide-induced signal transduction in endotoxin-tolerized mouse macrophages: dysregulation of cytokine, chemokine, and toll-like receptor 2 and 4 gene expression. J. Immunol. 164:5564–5574. [DOI] [PubMed] [Google Scholar]
- 53.Kopp, E., and S. Ghosh. 1994. Inhibition of NF-kappa B by sodium salicylate and aspirin. Science. 265:956–959. [DOI] [PubMed] [Google Scholar]
- 54.Yin, M.J., Y. Yamamoto, and R.B. Gaynor. 1998. The anti-inflammatory agents aspirin and salicylate inhibit the activity of I(kappa)B kinase-beta. Nature. 396:77–80. [DOI] [PubMed] [Google Scholar]
- 55.McKeage, M.J., P. Fong, M. Jeffery, B.C. Baguley, P. Kestell, M. Ravic, and M.B. Jameson. 2006. 5,6-dimethylxanthenone-4-acetic acid in the treatment of refractory tumors: a phase I safety study of a vascular disrupting agent. Clin. Cancer Res. 12:1776–1784. [DOI] [PubMed] [Google Scholar]
- 56.Zwi, L.J., B.C. Baguley, J.B. Gavin, and W.R. Wilson. 1994. The morphological effects of the anti-tumor agents flavone acetic acid and 5,6-dimethyl xanthenone acetic acid on the colon 38 mouse tumor. Pathology. 26:161–169. [DOI] [PubMed] [Google Scholar]
- 57.Maniatis, T., J.V. Falvo, T.H. Kim, T.K. Kim, C.H. Lin, B.S. Parekh, and M.G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 63:609–620. [DOI] [PubMed] [Google Scholar]
- 58.Honda, K., H. Yanai, A. Takaoka, and T. Taniguchi. 2005. Regulation of the type I IFN induction: a current view. Int. Immunol. 17:1367–1378. [DOI] [PubMed] [Google Scholar]
- 59.Viatour, P., M.P. Merville, V. Bours, and A. Chariot. 2005. Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 30:43–52. [DOI] [PubMed] [Google Scholar]
- 60.Kirshner, J.R., A.Y. Karpova, M. Kops, and P.M. Howley. 2005. Identification of TRAIL as an interferon regulatory factor 3 transcriptional target. J. Virol. 79:9320–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim, T.K., J.S. Lee, J.E. Jung, S.Y. Oh, S. Kwak, X. Jin, S.Y. Lee, J.B. Lee, Y.G. Chung, Y.K. Choi, et al. 2006. Interferon regulatory factor 3 activates p53-dependent cell growth inhibition. Cancer Lett. 242:215–221. [DOI] [PubMed] [Google Scholar]
- 62.Duguay, D., F. Mercier, J. Stagg, D. Martineau, J. Bramson, M. Servant, R. Lin, J. Galipeau, and J. Hiscott. 2002. In vivo interferon regulatory factor 3 tumor suppressor activity in B16 melanoma tumors. Cancer Res. 62:5148–5152. [PubMed] [Google Scholar]
- 63.Medvedev, A.E., A. Lentschat, L.M. Wahl, D.T. Golenbock, and S.N. Vogel. 2002. Dysregulation of LPS-induced Toll-like receptor 4-MyD88 complex formation and IL-1 receptor-associated kinase 1 activation in endotoxin-tolerant cells. J. Immunol. 169:5209–5216. [DOI] [PubMed] [Google Scholar]
- 64.Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 9:143–150. [DOI] [PubMed] [Google Scholar]
- 65.Rewcastle, G.W., G.J. Atwell, B.C. Baguley, S.B. Calveley, and W.A. Denny. 1989. Potential antitumor agents. 58. Synthesis and structure-activity relationships of substituted xanthenone-4-acetic acids active against the colon 38 tumor in vivo. J. Med. Chem. 32:793–799. [DOI] [PubMed] [Google Scholar]
- 66.McIntire, F.C., M.P. Hargie, J.R. Schenck, R.A. Finley, H.W. Sievert, E.T. Rietschel, and D.L. Rosenstreich. 1976. Biologic properties of nontoxic derivatives of a lipopolysaccharide from Escherichia coli K235. J. Immunol. 117:674–678. [PubMed] [Google Scholar]
- 67.Livak, K.J., and T.D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. [DOI] [PubMed] [Google Scholar]
- 68.Weinstein, S.L., A.J. Finn, S.H. Dave, F. Meng, C.A. Lowell, J.S. Sanghera, and A.L. DeFranco. 2000. Phosphatidylinositol 3-kinase and mTOR mediate lipopolysaccharide-stimulated nitric oxide production in macrophages via interferon-beta. J. Leukoc. Biol. 67:405–414. [DOI] [PubMed] [Google Scholar]
- 69.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells. 6:375–388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.