Abstract
γ1-Herpesviruses such as Epstein-Barr virus (EBV) have a unique ability to amplify virus loads in vivo through latent growth-transforming infection. Whether they, like α- and β-herpesviruses, have been driven to actively evade immune detection of replicative (lytic) infection remains a moot point. We were prompted to readdress this question by recent work (Pudney, V.A., A.M. Leese, A.B. Rickinson, and A.D. Hislop. 2005. J. Exp. Med. 201:349–360; Ressing, M.E., S.E. Keating, D. van Leeuwen, D. Koppers-Lalic, I.Y. Pappworth, E.J.H.J. Wiertz, and M. Rowe. 2005. J. Immunol. 174:6829–6838) showing that, as EBV-infected cells move through the lytic cycle, their susceptibility to EBV-specific CD8+ T cell recognition falls dramatically, concomitant with a reductions in transporter associated with antigen processing (TAP) function and surface human histocompatibility leukocyte antigen (HLA) class I expression. Screening of genes that are unique to EBV and closely related γ1-herpesviruses of Old World primates identified an early EBV lytic cycle gene, BNLF2a, which efficiently blocks antigen-specific CD8+ T cell recognition through HLA-A–, HLA-B–, and HLA-C–restricting alleles when expressed in target cells in vitro. The small (60–amino acid) BNLF2a protein mediated its effects through interacting with the TAP complex and inhibiting both its peptide- and ATP-binding functions. Furthermore, this targeting of the major histocompatibility complex class I pathway appears to be conserved among the BNLF2a homologues of Old World primate γ1-herpesviruses. Thus, even the acquisition of latent cycle genes endowing unique growth-transforming ability has not liberated these agents from evolutionary pressure to evade CD8+ T cell control over virus replicative foci.
Herpesviruses are an ancient virus family whose members have long histories of coevolution with their host species (1). A hallmark of herpesvirus biology is the ability to colonize a naive host through the productive (lytic) infection of permissive cells, usually at a mucosal site of virus transmission, and thereafter to persist within that host as a nonproductive (latent) infection of a different specialized cell type. Persistence within the now-immune host is achieved through the down-regulation of all viral antigen expression in latently infected cells. Subsequently, occasional reactivations from latency can serve to reestablish the foci of virus replication at mucosal sites, providing a source of infectious virions for transmission to other individuals.
In the best studied system, HSV, a member of the α-herpesvirus subfamily establishing latency in neurons, the possibilities of successful reactivation are increased if the virus initially establishes a high latent virus genome load (2, 3); this in turn is reliant on the level of virus replication initially achieved during primary infection of the naive host (3, 4). Because the host CD8+ T cell response is the principle means of controlling this initial replication, any factor restricting the efficiency of that control would be to the advantage of the virus. Indeed, HSV was the first herpesvirus in which a CD8+ T cell evasion mechanism was recognized. This involves an immediate early protein of the HSV lytic cycle, ICP47, which inhibits the peptide transporter associated with antigen processing (TAP) by acting as a pseudosubstrate preventing peptide binding and transport. Consequently, TAP cannot pump potentially antigenic peptides from proteasomal digestion in the cytoplasm into the endoplasmic reticulum for loading onto MHC class I molecules (5, 6). Other herpesviruses heavily dependent on virus replication to establish a latent viral load, such as the β-subfamily human and mouse cytomegaloviruses, have likewise been found to possess several CD8+ T cell evasion mechanisms targeting either TAP or MHC class I assembly (7, 8).
The situation has been less clear for lymphocryptoviruses (LCVs), the most recently evolved (γ1) herpesvirus genus, whose members are found only in Old World and some New World primate species and whose prototype is the EBV of humans. These viruses are orally transmitted, replicate in oropharyngeal epithelial sites and establish latency in B lymphocytes (for review see reference 9). When first colonizing the B cell system, LCVs use their unique growth-transforming ability to directly drive the expansion of latently infected B cells and only later down-regulate latent antigen expression to establish an immunologically silent infection of the memory B cell pool (10). This ability to expand the latently infected cell reservoir independent of lytic virus replication has raised debate as to whether LCVs might be under less intense immunological pressure to evade T cell control over the lytic cycle (11).
We were prompted to return to this question by two recent findings in the EBV system, both suggestive of an active immune evasion strategy in the lytic cycle. First, cells productively infected with EBV in vitro showed HLA class I down-regulation and reduced TAP function (12). Second, in vitro assays on lytically infected target cells using CD8+ T cell clones specific for immediate early–, early-, or late-expressed EBV antigens indicated much poorer presentation as the lytic cycle progressed (13). In this paper, we show that EBV and other Old World LCVs have indeed acquired an evasion strategy that specifically targets the peptide transporter TAP, thus limiting the supply of antigenic fragments to MHC class I mole cules and, ultimately, their presentation to CD8+ T cells.
RESULTS
Screening of γ1-herpesvirus–specific lytic cycle genes for inhibition of CD8+ T cell recognition
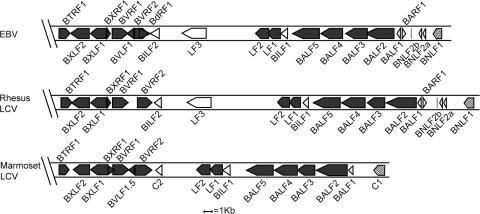
We reasoned that any specific immune evasion function associated with EBV replication would most likely map to a lytic cycle gene restricted to the γ1-herpesvirus (LCV) genus. Genomic sequences of HSV, CMV, and Kaposi's sarcoma–associated herpesvirus, the prototypic α, β, and γ2 human herpesviruses, were aligned alongside those of two Old World γ1 viruses, EBV and rhesus LCV, and with the recently discovered New World γ1 virus of marmosets. This identified 11 LCV-specific genes that were present in the Old World viruses, some of which were also found in the New World virus. 4 out of these 11 genes are centrally positioned in the EBV genome; 2 of the 4, BLLF1 and BZLF2, encode viral en velope glycoproteins known to be involved in B cell entry, whereas the other 2, BLLF2 and BHLF1, were of particular interest because their functions are unknown. The other seven LCV-specific genes all lie within a 35-kb region at the right-hand end of the EBV genome and are identified as open blocks in the genome maps shown in Fig. 1. Two of these, BARF1 and BALF1, have homologies to the cellular CSF-1 receptor and bcl2 genes, respectively, implying functions other than the interception of antigen processing pathways, whereas a third gene, LF3, is deleted in the B95.8 virus strain that displays HLA class I down-regulation in the lytic cycle. The remaining unique genes in this region, BNLF2a, BNLF2b, BILF1, and BILF2, were selected for further study alongside BLLF2 and BHLF1.
Figure 1.
Diagrammatic alignment of the right-hand ends of the sequenced γ1-herpesviruses. ORFs of EBV, the rhesus LCV, and marmoset LCV are shown as boxes; closed symbols represent ORFs with homologues in other herpesviruses; open symbols represent genes found only in γ1-herpesviruses; and hatched symbols represent latent genes.
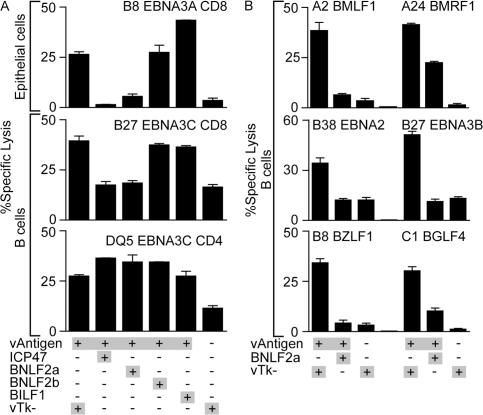
All six genes were separately cloned into recombinant vaccinia virus vectors, as was the immune evasion gene from HSV, ICP47, as a positive control. These recombinants were then tested for their ability to inhibit CD8+ T cell killing when introduced into target cells along with a second vaccinia vector encoding an indicator target antigen. Such experiments were performed in the two cell types that EBV naturally infects in vivo, epithelial cells and B lymphocytes. Fig. 2 A (top) presents the results from a representative assay in the HLA-B*0801–positive epithelial cell line SVK using EBV nuclear antigen (EBNA) 3A as the target antigen and a CD8+ T cell clone specific for an HLA-B*0801–restricted EBNA3A epitope as the effector. Expression of one of the six EBV genes in question, BNLF2a, reduced the level of CD8+ T cell killing almost to background levels, as did the positive control ICP47. None of the other EBV genes tested (including BILF2, BLLF2, and BHLF1; not depicted) ever gave such an effect. Fig. 2 A (bottom) presents data from an essentially similar assay conducted in an EBV-transformed B lymphoblastoid cell line (LCL). In this case, the cells were infected with a vaccinia virus encoding, as an indicator antigen, an invariant chain–tagged form of the EBNA3C protein that is processed by the HLA class I and class II pathways, thereby allowing both CD8+ and CD4+ T cell recognition to be assayed simultaneously on the same target cells. As shown in Fig. 2 A (middle), expres sion of BNLF2a (and of ICP47) reduced target cell killing by an HLA-B*2705–restricted EBNA3C-specific CD8+ T cell clone to the background level seen on unmanipulated LCL targets. This background level represents recognition of preexisting epitopes derived from EBNA3C expressed from the resident EBV genome. In contrast, neither vaccinia BNLF2a nor any of the other proteins tested had any effect on the presentation of invariant chain–tagged EBNA3C to a DQ5-restricted EBNA3C-specific CD4+ T cell clone (Fig. 2 A, bottom).
Figure 2.
Representative cytotoxicity assays testing recognition of target cells coexpressing EBV-unique genes and target proteins. (A) Epithelial cells (top) were infected with vaccinia viruses expressing EBNA3A (vAntigen) and combinations of the indicated viruses expressing genes unique to EBV or a control vaccinia virus lacking an inserted gene (vTk−), before being incubated with HLA-B*0801–restricted CD8+ T cell clones specific for EBNA3A. EBV-transformed B cells (middle and bottom) were coinfected with modified vaccinia Ankara expressing invariant chain–targeted EBNA3C (vAntigen) and the indicated combinations of vaccinia viruses. In parallel assays, the infected B cells were incubated with either HLA-B*2705–restricted CD8+ T cell clones specific for EBNA3C (middle) or CD4+ T cell clones restricted by HLA DQ5 specific for EBNA3C (bottom). (B) EBV-transformed B cells were infected with vaccinia viruses expressing BNLF2a, and the different EBV antigens indicated (vAntigen) encoding epitopes presented through a range of HLA types. Infected cells were incubated with cognate CD8+ T cells specific for an HLA-A*0201 epitope encoded by BMLF1, an HLA-A*2402 epitope encoded by BMRF1, an HLA-B*38 epitope encoded by EBNA2, an HLA-B*2705 epitope encoded by EBNA3B, an HLA-B*0801 epitope encoded by BZLF1, or an HLA-C*0101 epitope encoded by BGLF4. Error bars represent means ± SD.
Fig. 2 B shows the results of subsequent experiments conducted using EBV-transformed B cell lines with different HLA alleles to assess any allele-specific effects of BNLF2a on CD8+ T cell recognition. These assays used several different EBV latent (EBNA2 and EBNA3B) and lytic (BMLF1, BMRF1, BZLF1, and BGLF4) target antigens encoding epitopes recognized in the context of HLA-A (A*0201 and A*2402), HLA-B (B*0801, B*2705, and B*3801), and HLA-C (C*0101) molecules. We observed marked inhibi tion of target cell killing by BNLF2a throughout such experiments (Fig. 2 B).
EBV BNLF2a causes cell-surface HLA class I down-regulation
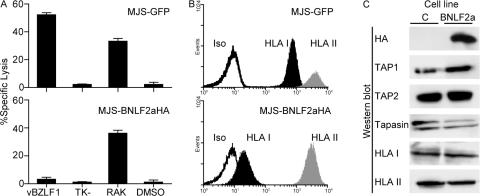
To determine the mechanism of BNLF2a-mediated inhibition of CD8+ T cell recognition, BNLF2a containing a hemagglutinin (HA) epitope tag (BNLF2aHA) was expressed in the melanoma cell line MelJuSo (MJS) through retroviral transduction. Initially, we performed cytotoxicity assays on the transduced cells to confirm that retrovirus-mediated expression of BNLF2aHA in MJS cells recapitulates the BNLF2a-induced phenotype described in the previous section. Fig. 3 A shows the results of a cytotoxicity assay using the HLA-B*0801–positive MJS cells expressing either BNLF2aHA or GFP as targets. Infection of MJS cells with recombinant vaccinia virus expressing the EBV BZLF1 antigen resulted in recognition by BZLF1-specific CD8+ T cells (Fig. 3 A, top), but this was abrogated in BNLF2aHA-expressing MJS cells (Fig. 3 A, bottom). However, when the target cells were sensitized with the relevant synthetic epitope peptide from BZLF1 (RAKFKQLL), the cells were lysed equally well whether or not BNLF2a was present (Fig. 3 A). These data indicate that BNLF2a blocks presentation of antigens that require endogenous processing before being presented to HLA class I–restricted T cells.
Figure 3.
Expression of BNLF2a prevents CD8+ T cell recognition by disrupting antigen presentation. (A) MJS cells were retrovirally transduced to express BNLF2aHA (bottom) or control GFP (top). These cells were either infected with vaccinia viruses expressing BZLF1 or a control TK− virus, or sensitized with RAKFKQLL peptide or DMSO as a control before being incubated with CD8+ T cells specific for the BZLF1-encoded RAKFKQLL epitope in 5-h cytotoxicity assays. Error bars represent means ± SD. (B) Flow cytometry histograms of the same cell lines show surface staining for HLA class I (B9.12.1) and HLA class II (L243), or staining with an isotype control. (C) Lysates of the two cell lines were separated by SDS-PAGE and analyzed by immunoblotting with antibodies specific for the HA tag (12CA5), TAP1 (148.3), TAP2 (435.4), tapasin (7F6), HLA class I heavy chains (HC10), and HLA class II DRα chains (DA6-147).
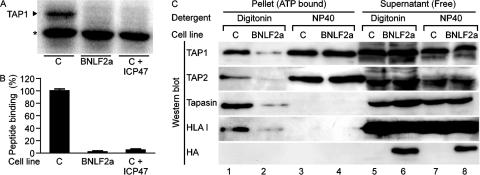
Because HLA class I levels have been found to be markedly reduced during productive EBV infection (12), we next examined whether expression of BNLF2a affected the display of HLA molecules (Fig. 3 B). At the surface of transduced MJS cells, levels of HLA class I were substantially reduced on BNLF2aHA-expressing cells compared with control cells. This was a specific effect, because expression of HLA class II (Fig. 3 B) and transferrin receptor (not depicted) were unaffected.
Down-regulation of surface HLA class I molecules can result from viral interference at specific stages of the class I antigen presentation pathway and may involve the degradation of HLA class I molecules and other components of the HLA class I peptide-loading complex (7, 8). To investigate whether BNLF2a affects levels of the components of the HLA class I peptide-loading complex, lysates of BNLF2aHA or control transduced MJS cells were subjected to Western blot analysis for TAP1, TAP2, tapasin, and HLA class I. No marked differences were observed in the steady-state levels of these proteins (Fig. 3 C). Thus, despite reduced surface HLA class I levels, cells expressing BNLF2aHA contained similar levels of the major components of the HLA class I peptide-loading complex.
EBV BNLF2a blocks peptide transport by TAP
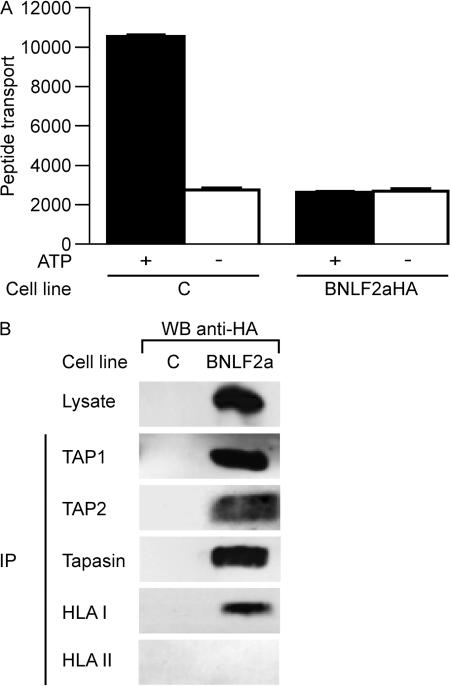
Surface HLA class I down-regulation can also result from the reduced supply of peptides to nascent HLA class I molecules in the endoplasmic reticulum. This is turn relies on the efficient generation of peptides by the proteasome and other peptidases and the transport of these peptides in to the endoplasmic reticulum by the TAP complex. In EBV-infected B cells that have entered the virus lytic cycle, surface HLA class I down-regulation coincides with reduced peptide transport by TAP (12). To evaluate whether the inhibition of HLA class I expression by BNLF2a involves interference with TAP function, we used an in vitro peptide translocation assay to examine TAP activity in BNLF2aHA-expressing and control MJS cells. In control cells, fluorescent model peptides were efficiently transported across the endoplasmic reticulum membrane in an ATP-dependent manner. In contrast, peptide translocation was strongly impaired in BNLF2aHA-expressing cells (Fig. 4 A).
Figure 4.
EBV BNLF2a blocks peptide transport by TAP. (A) TAP-dependent peptide transport in BNLF2aHA-expressing and control MJS cells was assessed by permeabilizing the cells with streptolysin O and incubating them with a fluoresceinated peptide in the presence or absence of ATP. Translocated peptides that had become glycosylated in the endoplasmic reticulum were recovered by adsorption to concanavalin A–sepharose beads. After elution, the recovered peptide was quantitated by fluorometry in arbitrary units. Error bars represent the SEM of triplicates in a representative experiment. (B) Digitonin lysates of BNLF2aHA- expressing and control MJS-GFP cells were subjected to immunoprecipitation (IP) with antibodies specific for TAP1 (148.3), TAP2 (435.4), tapasin (R.gp46C), HLA class I heavy chains (HC10), and HLA class II DRα chains (DA6-147). Cell lysates and immune complexes were separated by SDS-PAGE, followed by Western blot analysis and staining with anti-HA antibody (12CA5) to detect HA-tagged BNLF2a.
We next investigated whether BNLF2a was associated with the peptide-loading complex by performing coimmuno precipitation experiments. BNLF2aHA-expressing or control cells were lysed using the mild detergent digitonin to preserve protein–protein interactions. TAP1, TAP2, tapasin, and HLA class I molecules were immunoprecipitated from the lysates, and all precipitates were probed with an HA-specific mAb to detect BNLF2aHA. Fig. 4 B shows that BNLF2a coprecipitates with TAP1, TAP2, tapasin, and HLA class I molecules, but not with HLA class II and the transferrin receptor (used as controls; TfR data not depicted).
EBV BNLF2a blocks TAP function by preventing the binding of peptides and ATP
TAP-mediated translocation of peptides into the endoplasmic reticulum is initiated by the interaction of peptides with the cytosolic peptide binding domains of TAP, followed by the binding and hydrolysis of ATP, which facilitates the opening of the transmembrane pore and peptide translocation (14, 15). We therefore evaluated the effect of BNLF2aHA on these steps in peptide transfer. The ability of peptides to bind to TAP in the presence of BNLF2a was examined by incubating radiolabeled peptides containing a photoactivatable cross-linker with microsomes derived from BNLF2aHA-expressing or control MJS cells. Peptides were covalently linked to their binding partners by UV irradiation and then separated by PAGE. In control MJS cells, radiolabeled peptides covalently linked to TAP were observed (Fig. 5 A; expressed as 100% in the quantitation depicted in Fig. 5 B), whereas such peptides failed to bind TAP in microsomes from BNLF2aHA-expressing cells (Fig. 5, A and B). As a specificity control, microsomes were incubated with both the radiolabeled reporter peptide and an unlabeled peptide derived from the HSV-1 ICP47 protein that is known to prevent peptide binding to TAP (16, 17). No binding of the radiolabeled peptide to TAP was observed in the presence of the ICP47 peptide (Fig. 5, A and B).
Figure 5.
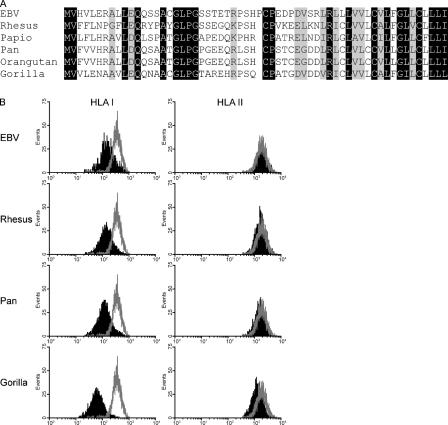
EBV BNLF2a inhibits peptide and ATP binding to TAP. (A) Microsomes prepared from BNLF2aHA-expressing and control MJS cells were incubated with a radiolabeled model peptide; where indicated, an excess of ICP47 competitor peptide was included in this incubation. After UV cross-linking, microsomes were lysed, and the proteins were separated by SDS-PAGE and exposed to a phosphoimaging screen. The position where TAP1 migrated in parallel immunoprecipitation experiments is shown by the arrowhead. The asterisk denotes a nonspecific background band. (B) Quantification of triplicate bands representing TAP-bound peptide. Results are shown as the percentage of peptide binding relative to peptide binding in control MJS cells (set as 100%). Error bars represent means ± SD. (C) Digitonin and NP-40 lysates of MJS-BNLF2aHA and control MJS-GFP cells were incubated with ATP-agarose beads. ATP-agarose–bound (pellet) and unbound (supernatant) protein fractions were separated by SDS-PAGE and immunoblotted. Membranes were probed with antibodies specific for TAP1 (148.3), TAP2 (435.4), tapasin (7F6), HLA class I heavy chain (HC10), and the HA tag (12CA5).
We next asked whether BNLF2a might also affect the binding of ATP to TAP. Lysates of BNLF2aHA-transduced or control cells were made using either the detergents digitonin, which preserves protein interactions within the peptide-loading complex, or 1% NP-40, which releases HLA class I, tapasin, and BNLF2a from TAP (see later in this section). Proteins capable of binding ATP were purified from the lysates by incubation with ATP-agarose beads and pelleting; these fractions, as well as the nonbinding supernatant (free) fractions, were subjected to Western blot analysis for the presence of TAP1, TAP2, tapasin, HLA class I, or BNLF2aHA. In control cells, ATP binds to the TAP subunits solubilized either in digitonin or NP-40 (Fig. 5 C, lanes 1 and 3), whereas tapasin and HLA class I molecules were coisolated only with ATP-bound TAP recovered from digitonin lysates (Fig. 5 C, compare lane 1 with lane 3). In contrast, the ATP-bound fraction from digitonin lysates of BNLF2aHA-expressing cells contained little of the four components of the peptide-loading complex (Fig. 5 C, lane 2), indicating that the EBV protein inhibits the interaction of ATP with the TAP proteins. Accordingly, BNLF2aHA was not detected in the ATP-agarose pellet fraction (lane 2) but was present in the supernatant fraction of BNLF2a-transduced MJS cells (lane 6). Interestingly, BNLF2a was released from TAP in NP-40 lysates, which restored the ATP-binding capacity of the TAP subunits, implying a low-affinity interaction of BNLF2a with TAP that prevents binding to ATP-agarose beads (lane 4).
BNLF2a homologues are encoded by other Old World γ1 primate herpesviruses and down-regulate HLA class I expression
Because earlier genomic alignments had shown that a BNLF2a homologous gene was present in the Rhesus LCV genome, we went on to sequence the equivalent region from the LCV genomes of baboons (papio herpesvirus), chimpanzees (pan herpesvirus), orangutans (orangutan herpesvirus), and gorillas (gorilla herpesvirus). Each virus genome contained an equivalent BNLF2a open reading frame (ORF). Fig. 6 A shows the predicted amino acid sequence of these homologous proteins alongside those of EBV and the rhesus macaque LCV. These proteins show between 53 and 63% overall identity and share similar features; they are small 59–60–amino acid proteins, and each has a hydrophobic C terminus and several sites in the nonhydrophobic region where the sequence is conserved. Homology or alignment searches using programs such as the basic local alignment search tool reveal no obvious matches with any other protein in the database (unpublished data).
Figure 6.
Old World primate γ1-herpesvirus BNLF2a sequence and function. (A) The BNLF2 regions from the indicated herpesviruses were sequenced, and the predicted amino acid sequence of the BNLF2a genes are shown. Black regions represent regions of homology between the different BNLF2a species, whereas gray-shaded regions represent conservative amino acid changes. (B) BNLF2a genes were subcloned into plasmid expression vectors that coexpressed GFP, and these were transiently transfected into MJS cells. At 48 h, surface levels of HLA class I and class II were assessed on GFP-positive cells by staining with the relevant antibody and analyzing the cells by flow cytometry. Black histograms represent surface marker intensity of cells transfected with the relevant BNLF2a, whereas open histograms represent the intensity of cells transfected with the empty vector plasmid.
To test whether the BNLF2a homologues within the nonhuman LCVs had retained BNLF2a-like function, the relevant coding sequences were introduced into plasmid expression vectors under the control of the CMV immediate early promoter and transiently transfected into the human MJS cell line. Plasmid constructs were engineered to coexpress GFP using an internal ribosome entry site (IRES) element allowing identification of transfected cells. Fig. 6 B shows the flow cyto metric analysis of MJS cells that had been transfected with the BNLF2a genes derived from EBV, the rhesus LCV, pan herpesvirus (note that the pan and orangutan BNLF2a genes had identical sequences), or gorilla herpesvirus. Cells were stained for surface HLA class I or surface HLA class II as a control. In each case, expression of BNLF2a derived from the different viruses reproducibly caused down-regulation of surface HLA class I to different degrees depending on the species from which the gene was derived, whereas HLA class II levels were unaffected.
We subsequently extended these studies by transfecting other available Old World primate cell lines with the different BNLF2a genes and measuring the surface MHC class I levels of the transfected cells as before. Fig. S1 (available at http://www.jem.org/cgi/content/full/jem.20070256/DC1) shows histograms of MHC class I staining of transfected LLC-MK2 cells (Rhesus macaque), orangutan B cells transformed with the endogenous orangutan herpesvirus (orangutan LCL), and Cos cells (African green monkey). All BNLF2a expression constructs showed down-regulation of surface MHC class I in LLC-MK2 cells and Cos cells, with a more subtle down-regulation observed in transformed orangutan LCLs. The inhibitory effects of the various LCV BNLF2a proteins therefore appear to be conserved and operate in various cells of Old World primate origin.
DISCUSSION
This study was prompted by recent work (13) looking into target cell recognition by CD8+ T cell clones specific for EBV–lytic cycle antigens. This had shown that recognition was most efficient using clones against immediate early antigens, less so for clones against early antigens, and much less so for late antigen–specific effectors, despite the fact that the latter had the highest avidity in epitope peptide titration assays (13). Therefore, antigen processing function appeared to be progressively impaired as cells moved through the lytic cycle. Two observations argued against this being a secondary effect of general cytopathology associated with virus replication. Thus, in B LCLs in vitro where a small percentage of cells enter the lytic cycle, cells in the late phase of virus replication appear to remain viable for several days. More importantly, surface HLA class I levels on lytically infected cells begin to fall in the early phase of the cycle before other surface proteins, including HLA class II, are affected (12). This implied that EBV was actively impeding lytic cycle antigen presentation to CD8+ T cells.
Noting that immune evasion strategies within the herpesvirus family tend to be virus subfamily or genus specific, we focused on the six lytic cycle genes that were unique to the γ1 genus and of unknown function. This identified one such gene, BNLF2a, as encoding a CD8 immune evasion protein. When expressed from a vaccinia vector in target cells, the BNLF2a protein reproducibly inhibited recognition of various indicator antigens (EBV latent and lytic cycle proteins) by CD8+ T cell clones restricted through several HLA-A, -B, and -C alleles. In contrast, class II antigen presentation in BNLF2a-expressing cells remained intact, and only presentation by the HLA class I pathway was impeded. The BNLF2a protein was found to function by blocking both peptide and ATP binding to the TAP complex, thereby impairing peptide loading of HLA class I molecules and their expression at the cell surface.
It is notable that BNLF2a transcripts initially appear in the early phase of the lytic cycle, peaking 8–12 h after lytic cycle induction in a B cell line (18). The majority of EBV lytic cycle transcripts are expressed in the late phase after BNLF2a expression, at a time when the protein's effects are likely to be well established. This would explain why the late proteins are poorly processed and presented by lytically infected cells. In contrast, the two immediate early proteins, BZLF1 and BRLF1, expressed in the first wave of viral gene expression before BNLF2a are efficiently recognized by their cognate T cells (13). The less efficient presentation of the four early antigens tested in our previous study (13) likely reflects the fact that they are expressed before BNLF2a's inhibitory effects are optimal. These antigens are known to be BZLF1/BRLF1 induced and, therefore, appear very soon after the immediate early-to-early transition, probably just before BNLF2a expression (19–23).
It is also interesting to note how the actions of BNLF2a might explain the highly skewed composition of the lytic antigen-induced CD8+ T cell response to EBV infection, whereby antigens expressed before or at the latest coincident with BNLF2a during the course of the lytic cycle constitute by far the most dominant targets. Thus when we derived CD8+ T cell clones from the expanded CD8+ population of IM patients in an unbiased manner, we often identified strong CD8 reactivities specific for the two immediate early proteins and for a small subset of early proteins, but only rarely to other early proteins or to any of the late proteins analyzed (11, 13). Similarly, when T cells were cloned from the joints of rheumatoid arthritis patients and tested against an EBV-cDNA expression library, only T cells specific for immediate early or a small number of early proteins were detected (24, 25). Thus, very few responses to late proteins have been described and, when they occur, these responses are weak (26). Yet, in principle, all lytic cycle proteins should be available as exogenous antigens for cross-presentation by dendritic cells during the priming of the CD8+ T cell response. We infer that, after priming, the sub sequent numerical dominance of immediate early and early antigen-specific responses reflects the importance of direct contact between primed CD8+ T cells and lytically infected target cells in selectively amplifying responses to epitopes that are well presented on those target cells.
BNLF2a is one of a set of genes, including two others with immunomodulatory function, BCRF1 (viral IL-10 homologue) and BARF1 (viral GCSF-receptor), that have been acquired by the γ1 viruses of Old World primates but are absent from the New World γ1 virus analyzed thus far (27, 28). This implies acquisition, in the last 50 million years or so, after the Old World/New World fork in primate evolution. The Old World primate viruses have also acquired a more complex set of latent growth-transforming genes over this same period. Thus, it may be the combination of these more sophisticated immune-evasion and growth-transforming strategies that underlies the apparently greater prevalence that the Old World viruses achieve in their host population compared with their New World counterpart (29).
BNLF2a is the first TAP-specific inhibitor identified in γ1-herpesviruses, and like the other three herpesvirus TAP inhibitors identified so far, BNLF2a has its own unique structure and mechanism of action. Simplexviruses and varicelloviruses, both members of the α subfamily, use different strategies to inactivate TAP. The simplexviruses HSV1 and 2 encode a small cytosolic protein, ICP47, which acts as a high affinity competitor for peptide binding to TAP yet does not affect ATP binding to the transporter complex (5, 6, 17, 30, 31). In contrast, the varicellovirus bovine herpesvirus 1 and several of its relatives encode the UL49.5 gene product, which renders TAP translocation incompetent without affecting its ability to bind peptide or ATP (32, 33). Ultimately, bovine herpesvirus 1 UL49.5 targets TAP1 and TAP2 for degradation by proteasomes (32, 33). In the case of β-herpesviruses, the prototypic virus of this subfamily, human CMV, inhibits TAP function via the endoplasmic reticulum luminal domain of the transmembrane protein US6. This protein reduces association of ATP to TAP1 and promotes binding to TAP2 but does not interfere with peptide binding, nor does it influence the stability of the transporter complex (34–38). In the case of the γ2-herpesvirus, mouse γ−herpesvirus 68, TAP is targeted secondarily for slow degradation by the virus-encoded ubiquitin ligase mK3, whose primary function is to rapidly target MHC class I heavy chains for proteasomal degradation (39–46).
In contrast to the described TAP inhibitors in the other herpesviruses, BNLF2a disables TAP function by blocking both the association of peptides with the peptide binding site and the interaction of ATP with the nucleotide binding domains of the transporter. How this small 60–amino acid protein, which consists of a hydrophilic N-terminal domain of 42 residues and a hydrophobic domain of 18 residues, achieves both of these functions is currently unclear. Despite the absence of an obvi ous N-terminal signal sequence, preliminary data suggest that BNLF2a is membrane-associated (unpublished data). This may be a consequence of binding to TAP or may result from posttranslational insertion of the hydrophobic C terminus into the endoplasmic reticulum membrane, as has been reported for other proteins (47). The small size of BNLF2a and the relative distance between the peptide and ATP binding sites of TAP may preclude BNLF2a from acting directly on both of these sites. The expected exposure of BNLF2a's N terminus in the cytosol would, however, suggest that this part interacts with cyto solic domains of the TAP complex. BNLF2a may then interfere directly with peptide or ATP binding while simultaneously disrupting subsequent steps of the translocation cycle through an indirect mechanism, e.g., by inhibiting conformational transitions required for proper function of the TAP complex.
The ability of an infected cell to escape CD8+ T cell recognition is likely to be most efficient when multiple points of the antigen processing pathway are targeted by viral immune evasion proteins. In the case of HSV2-infected cells, two viral proteins, the TAP inhibitor ICP47 and the host shutoff protein encoded by UL-41, have been shown to act synergistically to prevent CD8+ T cell lysis (48). In such an infection model, the long-lived TAP complexes are unlikely to be affected in the short term by the virus host shutoff function. Thus, cells infected with mutant HSV viruses lacking either of these two genes show partial inhibition of CD8+ T cell function, whereas cells infected with viruses expressing both genes show a dramatic reduction in their ability to be lysed by CD8+ T cells. Interesting in this context is the recent identification of the EBV-encoded gene BGLF5, which is at least one gene responsible for host protein synthesis shutoff (49). Expression of BGLF5 is sufficient to inhibit synthesis of host proteins, including HLA class I molecules, causing a decrease in surface class I levels. Coexpression of BNLF2a and BGLF5 by EBV could then represent an effective strategy for inhibiting the HLA class I antigen processing pathway (12).
In summary, the prototype γ1-herpesvirus, EBV, evades CD8+ T cell detection in the lytic cycle through a strategy that is unique in its molecular detail yet targets antigen presentation at the same point, TAP-mediated peptide transport, that is targeted by several other herpesviruses. The fact that the TAP inhibitors encoded by the different herpesvirus genera share no obvious homology presents a striking example of convergent evolution. Their existence testifies to the strength of the evolutionary pressure exerted on many herpesviruses by CD8+ T cell responses against virus replicative infections. The present work indicates that, despite their ability to amplify viral loads in vivo through nonreplicative latent infection, EBV and Old World γ1-herpesvirus relatives have indeed been subjected to the same immune pressure.
MATERIALS AND METHODS
CD8+ T cell clones, vaccinia viruses, and cytotoxicity assays.
EBV-specific CD8+ T cell clones were generated from PBMCs of infectious mononucleosis patients by limiting dilution cloning, as previously described (11). CD8+ T cell clones used in this study were specific for the following epitopes derived from the respective EBV gene products: RAKFKQLL from BZLF1 and QAKWRLQTL from EBNA3A (both presented by HLA-B*0801 [50, 51]), GLCTLVAML from BMLF1 (presented by HLA-A*0201 [11]), CYDHAQTHL from BMRF1 (presented by HLA A*2402 [13]), YHLIVDTDSL from EBNA2 (presented by HLA-B*38 [52]), HRCQAIRKK from EBNA3B (presented by HLA-B*2705 [53]), and an undefined epitope derived from BGLF4 (presented by HLA-C*0101 [13]). CD4+ T cell clones specific for the HLA DQ5–presented epitope SDDELPYIDPNMEP from EBNA 3C were produced by the stimulation of PBMCs from healthy donors, with their autologous EBV-transformed B cell line and limiting dilution cloning these cells, as previously described (54). All experiments were approved by the South Birmingham Health Authority Local Research Ethics Committee. Recombinant vaccinia viruses were used to express individual EBV B95.8 strain genes, as previously described, and a vaccinia virus lacking an insert (vTk−) used for control infections (13, 54). Target cells infected with the vaccinia viruses for cytotoxicity assays were either SV40-transformed keratinocytes, HLA-matched EBV-transformed B LCLs, or the melanoma cell line MJS (HLA-A*01, -B*08, and -Cw*07) (55). Target cells were infected with vaccinia viruses at a multiplicity of infection of 10, incubated for 16 h, and used as targets in standard 5-h chromium release assays. Where indicated in the figures, target cells were sensitized with synthetic CTL epitope peptide (Alta Biosciences) at a concentration of 5 μM for 90 min before use in cytotoxicity assays.
Antibodies.
The antibodies used in this study have been described elsewhere (32, 55). Mouse mAbs used to detect human cellular proteins were as follows: W6/32 or B9.12.1, which recognizes β2-microglobulin– associated HLA class I complexes (Immunotech); HC10 (provided by H. Ploegh, Whitehead Institute for Biomedical Research, Cambridge, MA), specific for HLA class I heavy chains; anti–HLA-DR mAb L243 (American Type Culture Collection); DA6-147, specific for HLA-DRα chains (provided by P. Cresswell, Howard Hughes Medical Institute, Yale University School of Medicine, New Haven, CT); and 148.3 directed to TAP1 and 435.4 directed to TAP2 (provided by R. Tampe, Johann Wolfgang Goethe-University, Frankfurt, Germany and by P. van Endert, Hopital Necker, Paris, France, respectively). Antitapasin antibodies used were a rabbit serum, R.gp46C, against the C-terminal region (provided by P. Cresswell) and a rat mAb, 7F6 (provided by R. Tampe). To detect HA-tagged BNLF2a, the influenza-specific mAb 12CA5 was used (Roche Diagnostics).
Retroviral transduction of cell lines.
Retroviral constructs were engineered by cloning the BNLF2a gene into the pLZRS retroviral vector. Immediately downstream from this gene was an IRES, which allowed co expression of a marker gene, truncated nerve growth factor. The BNLF2a sequence was modified at the C terminus to encode four methionines and the influenza HA epitope to allow detection (denoted as BNLF2aHA). Virus was produced using the Phoenix packaging cell line and used to transduce MJS cells, as previously described (55). As a control, MJS cells were transduced with an IRES-GFP–expressing retrovirus.
Flow cytometry analysis of surface HLA class I and II.
Surface levels of HLA class I and II molecules were measured by flow cytometric analysis after staining with the antibodies W6/32 or B9.12.1 for class I molecules or L243 for class II molecules. In all cases, aliquots of cells were stained with the appropriate isotype controls, and bound antibodies were detected using a goat anti–mouse phycoerythrin antibody. Stained cells were analyzed on a flow cytometer (Epics; Beckman Coulter), and data was processed using Win MDI software (Scripps Research Institute).
Peptide transport assay.
TAP-mediated peptide transport assays were conducted by permeablizing aliquots of 3 × 106 MJS cells with 2.5 IU/ml of streptolysin O (Murex Diagnostics) and incubating these cells with 200 nmol of the fluoresceinated peptide CVNKTERAY (provided by W. Benckhuijsen and J.W. Drijfhout, Leiden University Medical Center, Leiden, Netherlands) in the presence or absence of 10 mM ATP at 37°C for 10 min. Translocation was terminated by lysing cells with ice-cold lysis buffer (1% Triton X-100, 500 mM NaCl, 2 mM MgCl2, 50 mM Tris HCl, pH 8), and the nuclei were removed by centrifugation. Peptides that had been glycosylated in the endoplasmic reticulum were isolated from lysates by incubation with concanavalin A–sepharose beads (GE Healthcare). These peptides were eluted from the beads with 500 mM mannopyranoside, 10 mM EDTA, 50 mM Tris HCl, pH 8, and fluorescence was detected using a fluorescence plate reader (Cyto fluor; PerSeptive Biosystems).
Western blot analysis and immunoprecipitations.
SDS-PAGE and immunoblotting were performed as previously reported (32, 56). MJS cells transfected with retroviruses expressing either BNLF2aHA or GFP were lysed using either 1% NP-40 or 1% digitonin made in 50 mm Tris HCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl, 1 mM leupeptin, and 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride. Lysates of solubilized proteins, equivalent to 2 × 105 cells, were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (GE Healthcare). Proteins of interest were detected by incubating the membranes with specific antibodies followed by horseradish peroxidase (HRP)–conjugated secondary antispecies antibodies (Jackson ImmunoResearch Laboratories). Bound HRP-labeled antibodies were visualized using ECL Plus (GE Healthcare).
For immunoprecipitation experiments, cell lysates prepared with 1% digitonin solution were incubated with the different specific antibodies for at least 2 h at 4°C. Protein G–sepharose beads (GE Healthcare) were used to isolate immune complexes, which were washed with 0.1% digitonin and subjected to Western blot analysis. Antibodies that bound the blotted proteins were detected using ExactaCruz (Santa Cruz Biotechnology, Inc.), according to the manufacturer's instructions.
Peptide binding assay.
TAP-containing microsomal membranes were prepared from MJS cells transduced with BNLF2aHA or GFP by homogenization of the cells with a cell cracker (EMBL) and removal of the nuclei by centrifugation. The ability of peptides to bind to TAP complexes in the microsomes was assessed by incubation with an 125I-labeled peptide 5PS2 (ERYDKSE-[BPA]-L [57]) containing a photoactivatable cross-linker in the absence or presence of 2.5 μM of unlabeled HSV-1 ICP47 competitor peptide (16). After incubation for 15 min on ice, microsomes were washed with PBS, and peptide–protein interactions were covalently linked by UV exposure for 10 min on ice at a distance of 2 cm using a UVP lamp (B-100A; Blak-Ray) with a mercury lamp (Par 38; Sylvania). Samples were analyzed by SDS-PAGE, and bound peptide was quantitated by phosphoimaging. Migration of peptide-bound TAP1 was confirmed by immunoprecipitation.
ATP-agarose binding assay.
MJS cells transduced with BNLF2aHA or GFP were lysed with 1% digitonin or 1% NP-40, as described in Western blot analysis and immunoprecipitations. Nuclei were removed, and lysates were incubated with hydrated C-8 ATP-agarose beads (13-μM; final concentration; Sigma-Aldrich) at 4°C for 2 h. The beads were centrifuged to provide a supernatant fraction and a pellet fraction. Proteins bound to the ATP-agarose pellet fraction were eluted with 500 mM EDTA, and these as well as the supernatant fraction were analyzed by Western blot.
Cloning and transient expression of BNLF2a genes.
BNLF2a homologues were cloned from the following LCVs: papio herpesvirus, rhesus herpesvirus, pan herpesvirus, orangutan herpesvirus, and gorilla herpesvirus. PCR primers were designed to hybridize to a sequence flanking the BNLF2 region, which was conserved between EBV and the rhesus herpesvirus. PCR products were amplified from either a genomic DNA fragment of rhesus herpesvirus (provided by F. Wang, Harvard Medical School, Boston, MA) or from DNA extracted from B cell lines spontaneously transformed with the endogenous LCV of each host species. This amplified BNLF2 region of each virus was cloned into the plasmid pCR2.1TOPO (Invitrogen), and inserts were sequenced using standard techniques. Sequences are available from GenBank/EMBL/DDBJ under accession nos. EF207711 (papio herpesvirus), EF207712 (pan herpesvirus), EF207713 (orangutan herpesvirus), and EF207714 (gorilla herpesvirus).
For functional studies, the BNLF2a genes were subcloned into a modified pCDNA3 vector (provided by E. Reits) and expressed under the control of the CMV immediate early promoter. Immediately downstream from BNLF2a was an IRES element followed by the GFP gene, allowing co expression of this marker. Plasmids were transfected into either MJS cells, the Rhesus macaque kidney cell line LLC-MK2, or the African green monkey kidney fibroblast like cell line Cos using Lipofectamine 2000 (Invitrogen), or transfected by electroporation into orangutan B cells transformed with their endogenous LCV. Transfected cells were cultured for 48 h before staining for surface marker expression and flow cytometric analysis, as described in Flow cytometry analysis of surface HLA class I and II.
Online supplemental material.
Fig. S1 shows histograms of the fluorescence intensity of MHC class I staining on Old World primate cells transfected with plasmids expressing BNLF2a homologues. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070256/DC1.
Supplemental Material
Acknowledgments
We thank Paul Lehner for helpful advice and the provision of reagents.
This work was supported by a program grant from the Medical Research Council UK, grant UL 2005-3259 from the Dutch Cancer Society (to D. Horst), the M.W. Beijerinck Virology Fund of the Royal Academy of Arts and Sciences (M.E. Ressing), grant Vidi 917.76.330 from the Netherlands Organisation for Scientific Research (to M.E. Ressing), the Dutch Diabetes Research Foundation (D. Koppers-Lalic), and National Institutes of Health grant AI26296.
The authors have no conflicting financial interests.
Abbreviations used: EBNA, EBV nuclear antigen; HA, hemagglutinin; IRES, internal ribosome entry site; LCL, lymphoblastoid cell line; LCV, lymphocryptovirus; MJS, MelJuSo; ORF, open reading frame; TAP, transporter associated with antigen processing.
A.D. Hislop and M.E. Ressing contributed equally to this work.
References
- 1.McGeoch, D.J., F.J. Rixon, and A.J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90–104. [DOI] [PubMed] [Google Scholar]
- 2.Sawtell, N.M. 1998. The probability of in vivo reactivation of herpes simplex virus type 1 increases with the number of latently infected neurons in the ganglia. J. Virol. 72:6888–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawtell, N.M., R.L. Thompson, L.R. Stanberry, and D.I. Bernstein. 2001. Early intervention with high-dose acyclovir treatment during primary herpes simplex virus infection reduces latency and subsequent reactivation in the nervous system in vivo. J. Infect. Dis. 184:964–971. [DOI] [PubMed] [Google Scholar]
- 4.Sawtell, N. 1997. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J. Virol. 71:5423–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill, A., P. Jugovic, I. York, G. Russ, J. Bennink, J. Yewdell, H. Ploegh, and D. Johnson. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 375:411–415. [DOI] [PubMed] [Google Scholar]
- 6.Fruh, K., K. Ahn, H. Djaballah, P. Sempe, P.M. van Endert, R. Tampe, P.A. Peterson, and Y. Yang. 1995. A viral inhibitor of peptide transporters for antigen presentation. Nature. 375:415–418. [DOI] [PubMed] [Google Scholar]
- 7.Vossen, M.T., E.M. Westerhout, C. Soderberg-Naucler, and E.J. Wiertz. 2002. Viral immune evasion: a masterpiece of evolution. Immunogenetics. 54:527–542. [DOI] [PubMed] [Google Scholar]
- 8.Lilley, B.N., and H.L. Ploegh. 2005. Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond. Immunol. Rev. 207:126–144. [DOI] [PubMed] [Google Scholar]
- 9.Rickinson, A.B., and E. Kieff. 2006. Epstein-Barr Virus. In Fields Virology. Fifth edition. D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, and S.E. Strauss, editors. Lippincott Williams & Wilkins, Philadelphia. 2655–2700.
- 10.Kuppers, R. 2003. B cells under influence: transformation of B cells by Epstein-Barr virus. Nat. Rev. Immunol. 3:801–812. [DOI] [PubMed] [Google Scholar]
- 11.Steven, N.M., N. Annels, A. Kumar, A. Leese, M.G. Kurilla, and A.B. Rickinson. 1997. Immediate early and early lytic cycle proteins are frequent targets of the Epstein-Barr virus–induced cytotoxic T cell response. J. Exp. Med. 185:1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ressing, M.E., S.E. Keating, D. van Leeuwen, D. Koppers-Lalic, I.Y. Pappworth, E.J.H.J. Wiertz, and M. Rowe. 2005. Impaired transporter associated with antigen processing-dependent peptide transport during productive EBV infection. J. Immunol. 174:6829–6838. [DOI] [PubMed] [Google Scholar]
- 13.Pudney, V.A., A.M. Leese, A.B. Rickinson, and A.D. Hislop. 2005. CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells. J. Exp. Med. 201:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reits, E.A., A.C. Griekspoor, and J. Neefjes. 2000. How does TAP pump peptides? Insights from DNA repair and traffic ATPases. Immunol. Today. 21:598–600. [DOI] [PubMed] [Google Scholar]
- 15.van Endert, P.M., L. Saveanu, E.W. Hewitt, and P.J. Lehner. 2002. Powering the peptide pump: TAP crosstalk with energetic nucleotides. Trends Biochem. Sci. 27:454–461. [DOI] [PubMed] [Google Scholar]
- 16.Galocha, B., A. Hill, B.C. Barnett, A. Dolan, A. Raimondi, R.F. Cook, J. Brunner, D.J. McGeoch, and H.L. Ploegh. 1997. The active site of ICP47, a herpes simplex virus–encoded inhibitor of the major histocompatibility complex (MHC)–encoded peptide transporter associated with antigen processing (TAP), maps to the NH2-terminal 35 residues. J. Exp. Med. 185:1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann, L., W. Kraas, S. Uebel, G. Jung, and R. Tampe. 1997. The active domain of the herpes simplex virus protein ICP47: a potent inhibitor of the transporter associated with antigen processing (TAP). J. Mol. Biol. 272:484–492. [DOI] [PubMed] [Google Scholar]
- 18.Yuan, J., E. Cahir-McFarland, B. Zhao, and E. Kieff. 2006. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 80:2548–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rooney, C.M., D.T. Rowe, T. Ragot, and P.J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenney, S., E. Holley-Guthrie, E.-C. Mar, and M. Smith. 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63:3878–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holley-Guthrie, E.A., E.B. Quinlivan, E.-C. Mar, and S. Kenney. 1990. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J. Virol. 64:3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragoczy, T., and G. Miller. 1999. Role of the Epstein-Barr virus Rta protein in activation of distinct classes of viral lytic cycle genes. J. Virol. 73:9858–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C., N. Sista, and J. Pagano. 1996. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J. Virol. 70:2545–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scotet, E., J. David-Ameline, M.-A. Peyrat, A. Moreau-Aubry, D. Pinczon, A. Lim, J. Even, G. Semana, J.M. Berthelot, R. Breathnach, et al. 1996. T cell response to Epstein-Barr virus transactivators in chronic rheumatoid arthritis. J. Exp. Med. 184:1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scotet, E., M.A. Peyrat, X. Saulquin, C. Retiere, C. Couedel, F. Davodeau, N. Dulphy, A. Toubert, J.D. Bignon, A. Lim, et al. 1999. Frequent enrichment for CD8 T cells reactive against common herpes viruses in chronic inflammatory lesions: towards a reassessment of the physiopathological significance of T cell clonal expansions found in auto immune inflammatory processes. Eur. J. Immunol. 29:973–985. [DOI] [PubMed] [Google Scholar]
- 26.Bharadwaj, M., S.R. Burrows, J.M. Burrows, D.J. Moss, M. Catalina, and R. Khanna. 2001. Longitudinal dynamics of antigen-specific CD8+ cytotoxic T lymphocytes following primary Epstein-Barr virus infection. Blood. 98:2588–2589. [DOI] [PubMed] [Google Scholar]
- 27.Cho, Y., J. Ramer, P. Rivailler, C. Quink, R.L. Garber, D. Beier, and F. Wang. 2001. An Epstein-Barr-related herpesvirus from marmoset lymphomas. Proc. Natl. Acad. Sci. USA. 98:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivailler, P., Y.-G. Cho, and F. Wang. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a New World Primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J. Virol. 76:12055–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fogg, M.H., A. Carville, J. Cameron, C. Quink, and F. Wang. 2005. Reduced prevalence of Epstein-Barr virus-related lymphocryptovirus infection in sera from a New World primate. J. Virol. 79:10069–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn, K., T.H. Meyer, S. Uebel, P. Sempe, H. Djaballah, Y. Yang, P.A. Peterson, K. Fruh, and R. Tampe. 1996. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J. 15:3247–3255. [PMC free article] [PubMed] [Google Scholar]
- 31.Tomazin, R., A.B. Hill, P. Jugovic, I. York, P. van Endert, H.L. Ploegh, D.W. Andrews, and D.C. Johnson. 1996. Stable binding of the herpes simplex virus ICP47 protein to the peptide binding site of TAP. EMBO J. 15:3256–3266. [PMC free article] [PubMed] [Google Scholar]
- 32.Koppers-Lalic, D., E.A.J. Reits, M.E. Ressing, A.D. Lipinska, R. Abele, J. Koch, M.M. Rezende, P. Admiraal, D. van Leeuwen, K. Bienkowska-Szewczyk, et al. 2005. Varicelloviruses avoid T cell recognition by UL49.5-mediated inactivation of the transporter associated with antigen processing. Proc. Natl. Acad. Sci. USA. 102:5144–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipinska, A.D., D. Koppers-Lalic, M. Rychlowski, P. Admiraal, F.A.M. Rijsewijk, K. Bienkowska-Szewczyk, and E.J.H.J. Wiertz. 2006. Bovine herpesvirus 1 UL49.5 protein inhibits the transporter associated with antigen processing despite complex formation with glycoprotein M. J. Virol. 80:5822–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn, K., A. Gruhler, B. Galocha, T.R. Jones, E.J.H.J. Wiertz, H.L. Ploegh, P.A. Peterson, Y. Yang, and K. Fruh. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity. 6:613–621. [DOI] [PubMed] [Google Scholar]
- 35.Hengel, H., J.-O. Koopmann, T. Flohr, W. Muranyi, E. Goulmy, G.J. Hammerling, U.H. Koszinowski, and F. Momburg. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity. 6:623–632. [DOI] [PubMed] [Google Scholar]
- 36.Lehner, P.J., J.T. Karttunen, G.W.G. Wilkinson, and P. Cresswell. 1997. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc. Natl. Acad. Sci. USA. 94:6904–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyritsis, C., S. Gorbulev, S. Hutschenreiter, K. Pawlitschko, R. Abele, and R. Tampe. 2001. Molecular mechanism and structural aspects of transporter associated with antigen processing inhibition by the cytomegalovirus protein US6. J. Biol. Chem. 276:48031–48039. [DOI] [PubMed] [Google Scholar]
- 38.Hewitt, E.W., S.S. Gupta, and P.J. Lehner. 2001. The human cytomegalovirus gene product US6 inhibits ATP binding by TAP. EMBO J. 20:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boname, J.M., J.S. May, and P.G. Stevenson. 2005. The murine gamma-herpesvirus-68 MK3 protein causes TAP degradation independent of MHC class I heavy chain degradation. Eur. J. Immunol. 35:171–179. [DOI] [PubMed] [Google Scholar]
- 40.Boname, J.M., and P.G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity. 15:627–636. [DOI] [PubMed] [Google Scholar]
- 41.Boname, J.M., B.D. de Lima, P.J. Lehner, and P.G. Stevenson. 2004. Viral degradation of the MHC class I peptide loading complex. Immunity. 20:305–317. [DOI] [PubMed] [Google Scholar]
- 42.Lybarger, L., X. Wang, M.R. Harris, I. Virgin, W. Herbert, and T.H. Hansen. 2003. Virus subversion of the MHC class I peptide-loading complex. Immunity. 18:121–130. [DOI] [PubMed] [Google Scholar]
- 43.Wang, X., L. Lybarger, R. Connors, M.R. Harris, and T.H. Hansen. 2004. Model for the interaction of gammaherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex. J. Virol. 78:8673–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, Y.Y.L., M.R. Harris, L. Lybarger, L.A. Kimpler, N.B. Myers, H.W. Virgin IV, and T.H. Hansen. 2002. Physical association of the K3 protein of gamma-2 herpesvirus 68 with major histocompatibility complex class I molecules with impaired peptide and beta(2)-microglobulin assembly. J. Virol. 76:2796–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, X., Y. Ye, W. Lencer, and T.H. Hansen. 2006. The viral E3 ubiquitin ligase mK3 uses the Derlin/p97 endoplasmic reticulum-associated degradation pathway to mediate down-regulation of major histocompatibility complex class I proteins. J. Biol. Chem. 281:8636–8644. [DOI] [PubMed] [Google Scholar]
- 46.Wang, X., R. Connors, M.R. Harris, T.H. Hansen, and L. Lybarger. 2005. Requirements for the selective degradation of endoplasmic reticulum-resident major histocompatibility complex class I proteins by the viral immune evasion molecule mK3. J. Virol. 79:4099–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borgese, N., S. Brambillasca, P. Soffientini, M. Yabal, and M. Makarow. 2003. Biogenesis of tail-anchored proteins. Biochem. Soc. Trans. 31:1238–1242. [DOI] [PubMed] [Google Scholar]
- 48.Tigges, M.A., S. Leng, D.C. Johnson, and R.L. Burke. 1996. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J. Immunol. 156:3901–3910. [PubMed] [Google Scholar]
- 49.Rowe, M., B. Glaunsinger, D. van Leeuwen, J. Zuo, D. Sweetman, D. Ganem, J. Middeldorp, E.J. Wiertz, and M.E. Ressing. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. USA. 104:3366–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogedain, C., H. Wolf, S. Modrow, G. Stuber, and W. Jilg. 1995. Specific cytotoxic T-lymphocytes recognize the immediate-early transactivator ZTA of Epstein-Barr virus. J. Virol. 69:4872–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burrows, S.R., J. Gardner, R. Khanna, T. Steward, D.J. Moss, S. Rodda, and A. Suhrbier. 1994. Five new cytotoxic T-cell epitopes identified within Epstein-Barr virus nuclear antigen 3. J. Gen. Virol. 75:2489–2493. [DOI] [PubMed] [Google Scholar]
- 52.Chapman, A.L.N., A.B. Rickinson, W.A. Thomas, R.F. Jarrett, J. Crocker, and S.P. Lee. 2001. Epstein-Barr virus-specific cytotoxic T lymphocyte responses in the blood and tumour site of Hodgkin's disease patients: implications for a T cell-based therapy. Cancer Res. 61:6219–6226. [PubMed] [Google Scholar]
- 53.Rickinson, A.B., and D.J. Moss. 1997. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 15:405–431. [DOI] [PubMed] [Google Scholar]
- 54.Taylor, G.S., H.M. Long, T.A. Haigh, M. Larsen, J. Brooks, and A.B. Rickinson. 2006. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J. Immunol. 177:3746–3756. [DOI] [PubMed] [Google Scholar]
- 55.Ressing, M.E., D. van Leeuwen, F.A.W. Verreck, R. Gomez, B. Heemskerk, M. Toebes, M.M. Mullen, T.S. Jardetzky, R. Longnecker, M.W. Schilham, et al. 2003. Interference with T cell receptor-HLA-DR interactions by Epstein-Barr virus gp42 results in reduced T helper cell recognition. Proc. Natl. Acad. Sci. USA. 100:11583–11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ressing, M.E., D. van Leeuwen, F.A.W. Verreck, S. Keating, R. Gomez, K.L.M.C. Franken, T.H.M. Ottenhoff, M. Spriggs, T.N. Schumacher, L.M. Hutt-Fletcher, et al. 2005. Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion. J. Virol. 79:841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spee, P., J. Subjeck, and J. Neefjes. 1999. Identification of novel peptide binding proteins in the endoplasmic reticulum: ERp72, calnexin, and grp170. Biochemistry. 38:10559–10566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.